Abstract
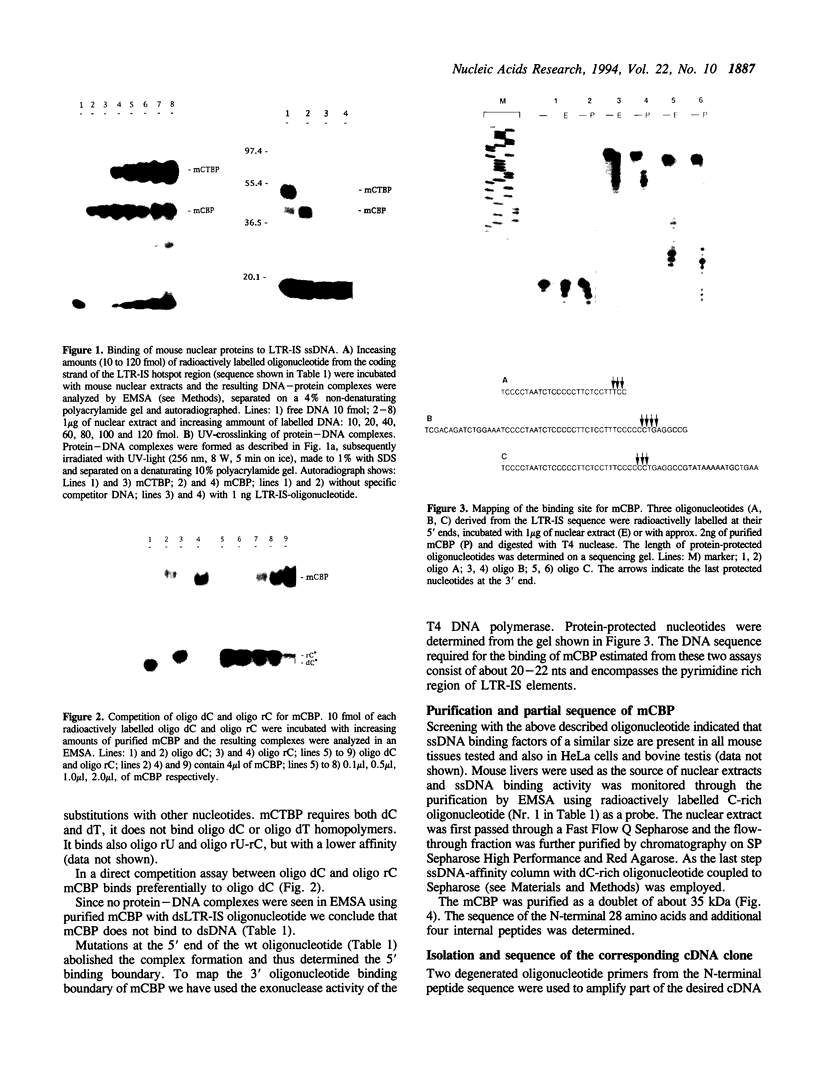
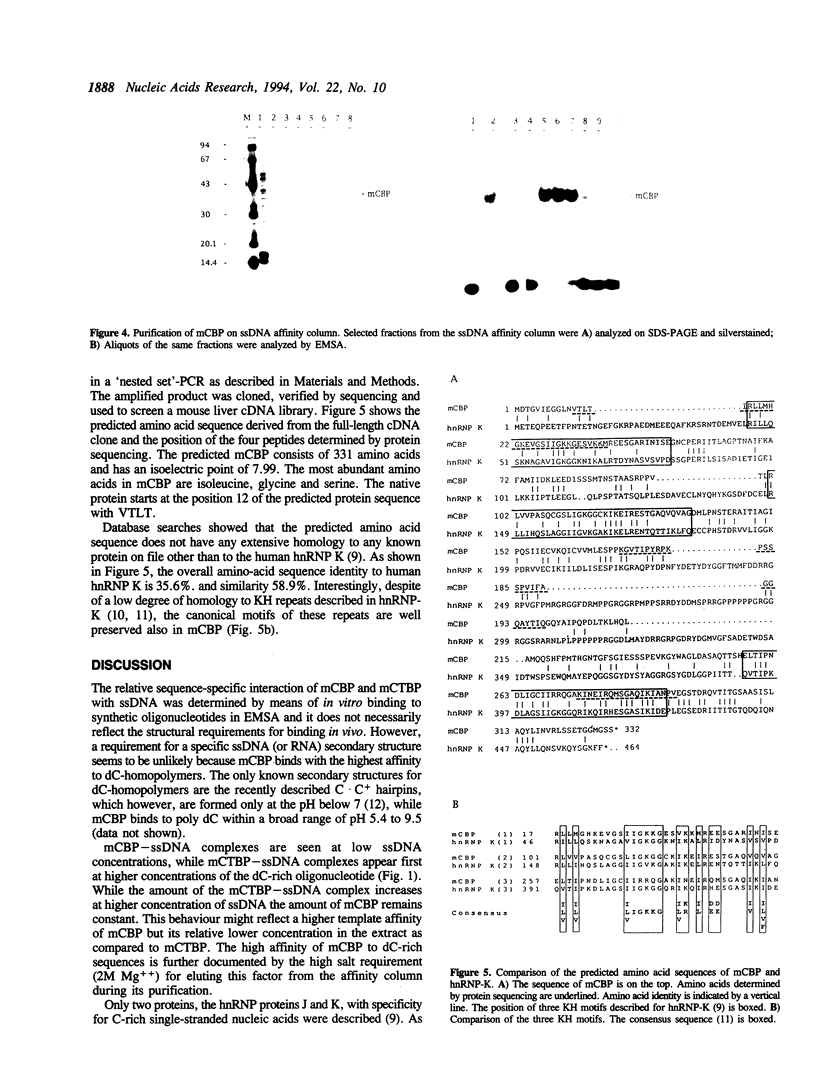
Two single-stranded nucleic acid binding proteins mCBP and mCTBP were identified by means of their binding to a potential recombination hotspot in LTRs of mouse retro-transposons. Both are nuclear proteins of 35 and 55 kDa respectively. mCBP binds preferentially to oligo dC, mCTBP to oligo dCdT. mCBP was purified and its cDNA was isolated and sequenced.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed S., Henderson E. Formation of novel hairpin structures by telomeric C-strand oligonucleotides. Nucleic Acids Res. 1992 Feb 11;20(3):507–511. doi: 10.1093/nar/20.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase J. W., Williams K. R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Matunis M. J., Piñol-Roma S., Burd C. G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Edelmann W., Kröger B., Goller M., Horak I. A recombination hotspot in the LTR of a mouse retrotransposon identified in an in vitro system. Cell. 1989 Jun 16;57(6):937–946. doi: 10.1016/0092-8674(89)90332-2. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y., Kohwi-Shigematsu T. Altered gene expression correlates with DNA structure. Genes Dev. 1991 Dec;5(12B):2547–2554. doi: 10.1101/gad.5.12b.2547. [DOI] [PubMed] [Google Scholar]
- Kohwi Y., Panchenko Y. Transcription-dependent recombination induced by triple-helix formation. Genes Dev. 1993 Sep;7(9):1766–1778. doi: 10.1101/gad.7.9.1766. [DOI] [PubMed] [Google Scholar]
- Matunis M. J., Michael W. M., Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992 Jan;12(1):164–171. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H., Matunis M. J., Michael W. M., Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993 Mar 11;21(5):1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto M., Tomonaga T., Matunis M., Avigan M., Krutzsch H., Dreyfuss G., Levens D. Specific binding of heterogeneous ribonucleoprotein particle protein K to the human c-myc promoter, in vitro. J Biol Chem. 1993 Aug 25;268(24):18249–18258. [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. The structure of DNA. Cold Spring Harb Symp Quant Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- Wirth T., Glöggler K., Baumruker T., Schmidt M., Horak I. Family of middle repetitive DNA sequences in the mouse genome with structural features of solitary retroviral long terminal repeats. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3327–3330. doi: 10.1073/pnas.80.11.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagil G. Paranemic structures of DNA and their role in DNA unwinding. Crit Rev Biochem Mol Biol. 1991;26(5-6):475–559. doi: 10.3109/10409239109086791. [DOI] [PubMed] [Google Scholar]