Abstract
Signal recognition particle (SRP) is a cytoplasmic ribonucleoprotein that targets a subset of nascent presecretory proteins to the endoplasmic reticulum membrane. We have considered the SRP cycle from the perspective of molecular evolution, using recently determined sequences of genes or cDNAs encoding homologs of SRP (7SL) RNA, the Srp54 protein (Srp54p), and the alpha subunit of the SRP receptor (SR alpha) from a broad spectrum of organisms, together with the remaining five polypeptides of mammalian SRP. Our analysis provides insight into the significance of structural variation in SRP RNA and identifies novel conserved motifs in protein components of this pathway. The lack of congruence between an established phylogenetic tree and size variation in 7SL homologs implies the occurrence of several independent events that eliminated more than half the sequence content of this RNA during bacterial evolution. The apparently non-essential structures are domain I, a tRNA-like element that is constant in archaea, varies in size among eucaryotes, and is generally missing in bacteria, and domain III, a tightly base-paired hairpin that is present in all eucaryotic and archeal SRP RNAs but is invariably absent in bacteria. Based on both structural and functional considerations, we propose that the conserved core of SRP consists minimally of the 54 kDa signal sequence-binding protein complexed with the loosely base-paired domain IV helix of SRP RNA, and is also likely to contain a homolog of the Srp68 protein. Comparative sequence analysis of the methionine-rich M domains from a diverse array of Srp54p homologs reveals an extended region of amino acid identity that resembles a recently identified RNA recognition motif. Multiple sequence alignment of the G domains of Srp54p and SR alpha homologs indicates that these two polypeptides exhibit significant similarity even outside the four GTPase consensus motifs, including a block of nine contiguous amino acids in a location analogous to the binding site of the guanine nucleotide dissociation stimulator (GDS) for E. coli EF-Tu. The conservation of this sequence, in combination with the results of earlier genetic and biochemical studies of the SRP cycle, leads us to hypothesize that a component of the Srp68/72p heterodimer serves as the GDS for both Srp54p and SR alpha. Using an iterative alignment procedure, we demonstrate similarity between Srp68p and sequence motifs conserved among GDS proteins for small Ras-related GTPases. The conservation of SRP cycle components in organisms from all three major branches of the phylogenetic tree suggests that this pathway for protein export is of ancient evolutionary origin.
Full text
PDF


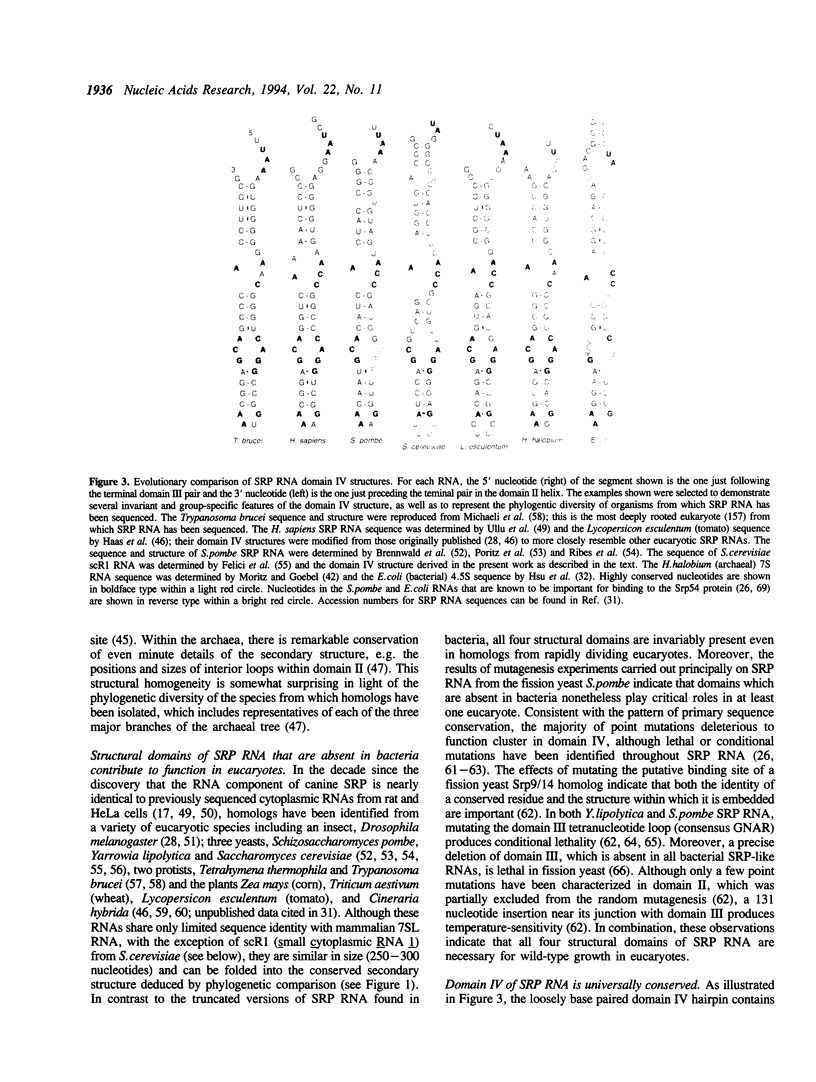










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amaya Y., Nakano A., Ito K., Mori M. Isolation of a yeast gene, SRH1, that encodes a homologue of the 54K subunit of mammalian signal recognition particle. J Biochem. 1990 Mar;107(3):457–463. doi: 10.1093/oxfordjournals.jbchem.a123067. [DOI] [PubMed] [Google Scholar]
- Amaya Y., Nakano A. SRH1 protein, the yeast homologue of the 54 kDa subunit of signal recognition particle, is involved in ER translocation of secretory proteins. FEBS Lett. 1991 Jun 3;283(2):325–328. doi: 10.1016/0014-5793(91)80619-e. [DOI] [PubMed] [Google Scholar]
- Andreazzoli M., Gerbi S. A. Changes in 7SL RNA conformation during the signal recognition particle cycle. EMBO J. 1991 Apr;10(4):767–777. doi: 10.1002/j.1460-2075.1991.tb08008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews D. W., Walter P., Ottensmeyer F. P. Evidence for an extended 7SL RNA structure in the signal recognition particle. EMBO J. 1987 Nov;6(11):3471–3477. doi: 10.1002/j.1460-2075.1987.tb02671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Benson S. A., Hall M. N., Silhavy T. J. Genetic analysis of protein export in Escherichia coli K12. Annu Rev Biochem. 1985;54:101–134. doi: 10.1146/annurev.bi.54.070185.000533. [DOI] [PubMed] [Google Scholar]
- Berchtold H., Reshetnikova L., Reiser C. O., Schirmer N. K., Sprinzl M., Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993 Sep 9;365(6442):126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Poritz M. A., Strub K., Hoben P. J., Brenner S., Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989 Aug 10;340(6233):482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Bernstein H. D., Zopf D., Freymann D. M., Walter P. Functional substitution of the signal recognition particle 54-kDa subunit by its Escherichia coli homolog. Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5229–5233. doi: 10.1073/pnas.90.11.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993 Nov 19;75(4):717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Boguski M. S., McCormick F. Proteins regulating Ras and its relatives. Nature. 1993 Dec 16;366(6456):643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bourgaize D. B., Fournier M. J. Initiation of translation is impaired in E. coli cells deficient in 4.5S RNA. Nature. 1987 Jan 15;325(6101):281–284. doi: 10.1038/325281a0. [DOI] [PubMed] [Google Scholar]
- Bourgaize D. B., Phillips T. A., VanBogelen R. A., Jones P. G., Neidhardt F. C., Fournier M. J. Loss of 4.5S RNA induces the heat shock response and lambda prophage in Escherichia coli. J Bacteriol. 1990 Feb;172(2):1151–1154. doi: 10.1128/jb.172.2.1151-1154.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Brennwald P. J., Siegel V., Walter P., Wise J. A. Sequence and structure of Tetrahymena SRP RNA. Nucleic Acids Res. 1991 Apr 25;19(8):1942–1942. doi: 10.1093/nar/19.8.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Liao X., Holm K., Porter G., Wise J. A. Identification of an essential Schizosaccharomyces pombe RNA homologous to the 7SL component of signal recognition particle. Mol Cell Biol. 1988 Apr;8(4):1580–1590. doi: 10.1128/mcb.8.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S., Fournier M. J. The 4.5 S RNA gene of Escherichia coli is essential for cell growth. J Mol Biol. 1984 Sep 25;178(3):533–550. doi: 10.1016/0022-2836(84)90237-7. [DOI] [PubMed] [Google Scholar]
- Brown S. Genes for 7S RNAs can replace the gene for 4.5S RNA in growth of Escherichia coli. J Bacteriol. 1991 Mar;173(5):1835–1837. doi: 10.1128/jb.173.5.1835-1837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. Mutations in the gene for EF-G reduce the requirement for 4.5S RNA in the growth of E. coli. Cell. 1987 Jun 19;49(6):825–833. doi: 10.1016/0092-8674(87)90620-9. [DOI] [PubMed] [Google Scholar]
- Brown S., Thon G., Tolentino E. Genetic selection and DNA sequences of 4.5S RNA homologs. J Bacteriol. 1989 Dec;171(12):6517–6520. doi: 10.1128/jb.171.12.6517-6520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. Time of action of 4.5 S RNA in Escherichia coli translation. J Mol Biol. 1989 Sep 5;209(1):79–90. doi: 10.1016/0022-2836(89)90171-x. [DOI] [PubMed] [Google Scholar]
- Béjà O., Ullu E., Michaeli S. Identification of a tRNA-like molecule that copurifies with the 7SL RNA of Trypanosoma brucei. Mol Biochem Parasitol. 1993 Feb;57(2):223–229. doi: 10.1016/0166-6851(93)90198-7. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Campos N., Palau J., Zwieb C. Diversity of 7 SL RNA from the signal recognition particle of maize endosperm. Nucleic Acids Res. 1989 Feb 25;17(4):1573–1588. doi: 10.1093/nar/17.4.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter P. B., Hanlon D. W., Ordal G. W. flhF, a Bacillus subtilis flagellar gene that encodes a putative GTP-binding protein. Mol Microbiol. 1992 Sep;6(18):2705–2713. doi: 10.1111/j.1365-2958.1992.tb01447.x. [DOI] [PubMed] [Google Scholar]
- Chant J., Corrado K., Pringle J. R., Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991 Jun 28;65(7):1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Connolly T., Gilmore R. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTP-binding protein. J Cell Biol. 1986 Dec;103(6 Pt 1):2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T., Gilmore R. GTP hydrolysis by complexes of the signal recognition particle and the signal recognition particle receptor. J Cell Biol. 1993 Nov;123(4):799–807. doi: 10.1083/jcb.123.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T., Gilmore R. The signal recognition particle receptor mediates the GTP-dependent displacement of SRP from the signal sequence of the nascent polypeptide. Cell. 1989 May 19;57(4):599–610. doi: 10.1016/0092-8674(89)90129-3. [DOI] [PubMed] [Google Scholar]
- Crowley K. S., Reinhart G. D., Johnson A. E. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993 Jun 18;73(6):1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Philipson L., Mattaj I. W. Ribonucleoprotein particles in cellular processes. J Cell Biol. 1988 May;106(5):1419–1425. doi: 10.1083/jcb.106.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Swanson M. S., Piñol-Roma S. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci. 1988 Mar;13(3):86–91. doi: 10.1016/0968-0004(88)90046-1. [DOI] [PubMed] [Google Scholar]
- Farnsworth C. L., Feig L. A. Dominant inhibitory mutations in the Mg(2+)-binding site of RasH prevent its activation by GTP. Mol Cell Biol. 1991 Oct;11(10):4822–4829. doi: 10.1128/mcb.11.10.4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felici F., Cesareni G., Hughes J. M. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol Cell Biol. 1989 Aug;9(8):3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn G. C., Chappell T. G., Rothman J. E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989 Jul 28;245(4916):385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Franklin A. E., Hoffman N. E. Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J Biol Chem. 1993 Oct 15;268(29):22175–22180. [PubMed] [Google Scholar]
- Gatignol A., Buckler C., Jeang K. T. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol Cell Biol. 1993 Apr;13(4):2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Blobel G. Transient involvement of signal recognition particle and its receptor in the microsomal membrane prior to protein translocation. Cell. 1983 Dec;35(3 Pt 2):677–685. doi: 10.1016/0092-8674(83)90100-9. [DOI] [PubMed] [Google Scholar]
- Gilmore R., Blobel G., Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982 Nov;95(2 Pt 1):463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R. Protein translocation across the endoplasmic reticulum: a tunnel with toll booths at entry and exit. Cell. 1993 Nov 19;75(4):589–592. doi: 10.1016/0092-8674(93)90476-7. [DOI] [PubMed] [Google Scholar]
- Gropp R., Gropp F., Betlach M. C. Association of the halobacterial 7S RNA to the polysome correlates with expression of the membrane protein bacterioopsin. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1204–1208. doi: 10.1073/pnas.89.4.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E. D., Di Carlo M., Zopf D., Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984 Oct;3(10):2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991 Jul 12;253(5016):157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Görlich D., Hartmann E., Prehn S., Rapoport T. A. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992 May 7;357(6373):47–52. doi: 10.1038/357047a0. [DOI] [PubMed] [Google Scholar]
- Haas B., Klanner A., Ramm K., Sänger H. L. The 7S RNA from tomato leaf tissue resembles a signal recognition particle RNA and exhibits a remarkable sequence complementarity to viroids. EMBO J. 1988 Dec 20;7(13):4063–4074. doi: 10.1002/j.1460-2075.1988.tb03300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann B. C., Poritz M. A., Walter P. Saccharomyces cerevisiae and Schizosaccharomyces pombe contain a homologue to the 54-kD subunit of the signal recognition particle that in S. cerevisiae is essential for growth. J Cell Biol. 1989 Dec;109(6 Pt 2):3223–3230. doi: 10.1083/jcb.109.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hann B. C., Stirling C. J., Walter P. SEC65 gene product is a subunit of the yeast signal recognition particle required for its integrity. Nature. 1992 Apr 9;356(6369):532–533. doi: 10.1038/356532a0. [DOI] [PubMed] [Google Scholar]
- Hann B. C., Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991 Oct 4;67(1):131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- He F., Beckerich J. M., Gaillardin C. A mutant of 7SL RNA in Yarrowia lipolytica affecting the synthesis of a secreted protein. J Biol Chem. 1992 Jan 25;267(3):1932–1937. [PubMed] [Google Scholar]
- He F., Beckerich J. M., Ribes V., Tollervey D., Gaillardin C. M. Two genes encode 7SL RNAs in the yeast Yarrowia lipolytica. Curr Genet. 1989 Dec;16(5-6):347–350. doi: 10.1007/BF00340713. [DOI] [PubMed] [Google Scholar]
- He F., Yaver D., Beckerich J. M., Ogrydziak D., Gaillardin C. The yeast Yarrowia lipolytica has two, functional, signal recognition particle 7S RNA genes. Curr Genet. 1990 Apr;17(4):289–292. doi: 10.1007/BF00314874. [DOI] [PubMed] [Google Scholar]
- Herskovits J. S., Burgess C. C., Obar R. A., Vallee R. B. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993 Aug;122(3):565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz J., Flint N., Stanley K., Frank R., Dobberstein B. The 68 kDa protein of signal recognition particle contains a glycine-rich region also found in certain RNA-binding proteins. FEBS Lett. 1990 Dec 10;276(1-2):103–107. doi: 10.1016/0014-5793(90)80518-n. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- High S., Dobberstein B. The signal sequence interacts with the methionine-rich domain of the 54-kD protein of signal recognition particle. J Cell Biol. 1991 Apr;113(2):229–233. doi: 10.1083/jcb.113.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Nakamura K., Nishiguchi M., Yamane K. Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J Bacteriol. 1993 Aug;175(15):4885–4894. doi: 10.1128/jb.175.15.4885-4894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L. M., Zagorski J., Fournier M. J. Cloning and sequence analysis of the Escherichia coli 4.5 S RNA gene. J Mol Biol. 1984 Sep 25;178(3):509–531. doi: 10.1016/0022-2836(84)90236-5. [DOI] [PubMed] [Google Scholar]
- Janiak F., Walter P., Johnson A. E. Fluorescence-detected assembly of the signal recognition particle: binding of the two SRP protein heterodimers to SRP RNA is noncooperative. Biochemistry. 1992 Jun 30;31(25):5830–5840. doi: 10.1021/bi00140a019. [DOI] [PubMed] [Google Scholar]
- Kaine B. P., Merkel V. L. Isolation and characterization of the 7S RNA gene from Methanococcus voltae. J Bacteriol. 1989 Aug;171(8):4261–4266. doi: 10.1128/jb.171.8.4261-4266.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaine B. P. Structure of the archaebacterial 7S RNA molecule. Mol Gen Genet. 1990 May;221(3):315–321. doi: 10.1007/BF00259394. [DOI] [PubMed] [Google Scholar]
- Kohno K., Normington K., Sambrook J., Gething M. J., Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993 Feb;13(2):877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg U. C., Walter P., Johnson A. E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia T. V., Wiedmann M., Girshovich A. S., Bochkareva E. S., Bielka H., Rapoport T. A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986 Apr 17;320(6063):634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- Lai C. C., Boguski M., Broek D., Powers S. Influence of guanine nucleotides on complex formation between Ras and CDC25 proteins. Mol Cell Biol. 1993 Mar;13(3):1345–1352. doi: 10.1128/mcb.13.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991 Jan 25;19(2):209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffer L., Garcia P. D., Harkins R. N., Coussens L., Ullrich A., Walter P. Topology of signal recognition particle receptor in endoplasmic reticulum membrane. 1985 Nov 28-Dec 4Nature. 318(6044):334–338. doi: 10.1038/318334a0. [DOI] [PubMed] [Google Scholar]
- Li W. Y., Reddy R., Henning D., Epstein P., Busch H. Nucleotide sequence of 7 S RNA. Homology to Alu DNA and La 4.5 S RNA. J Biol Chem. 1982 May 10;257(9):5136–5142. [PubMed] [Google Scholar]
- Liao X. B., Brennwald P., Wise J. A. Genetic analysis of Schizosaccharomyces pombe 7SL RNA: a structural motif that includes a conserved tetranucleotide loop is important for function. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4137–4141. doi: 10.1073/pnas.86.11.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Selinger D., Althoff S., Chiang A., Hamilton D., Ma M., Wise J. A. Random mutagenesis of Schizosaccharomyces pombe SRP RNA: lethal and conditional lesions cluster in presumptive protein binding sites. Nucleic Acids Res. 1992 Apr 11;20(7):1607–1615. doi: 10.1093/nar/20.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach K., Zwieb C., Webb J. R., Marshallsay C., Hoben P. J., Walter P., Dobberstein B. Isolation and characterization of a cDNA clone encoding the 19 kDa protein of signal recognition particle (SRP): expression and binding to 7SL RNA. Nucleic Acids Res. 1988 Oct 25;16(20):9431–9442. doi: 10.1093/nar/16.20.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luehrsen K. R., Nicholson D. E., Jr, Fox G. E. Widespread distribution of a 7S RNA in archaebacteria. Curr Microbiol. 1985;12:69–72. doi: 10.1007/BF01567394. [DOI] [PubMed] [Google Scholar]
- Luirink J., High S., Wood H., Giner A., Tollervey D., Dobberstein B. Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature. 1992 Oct 22;359(6397):741–743. doi: 10.1038/359741a0. [DOI] [PubMed] [Google Scholar]
- Lütcke H., High S., Römisch K., Ashford A. J., Dobberstein B. The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J. 1992 Apr;11(4):1543–1551. doi: 10.1002/j.1460-2075.1992.tb05199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H., Prehn S., Ashford A. J., Remus M., Frank R., Dobberstein B. Assembly of the 68- and 72-kD proteins of signal recognition particle with 7S RNA. J Cell Biol. 1993 Jun;121(5):977–985. doi: 10.1083/jcb.121.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshallsay C., Prehn S., Zwieb C. cDNA cloning of the wheat germ SRP 7S RNAs. Nucleic Acids Res. 1989 Feb 25;17(4):1771–1771. doi: 10.1093/nar/17.4.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D. I., Krause E., Dobberstein B. Secretory protein translocation across membranes-the role of the "docking protein'. Nature. 1982 Jun 24;297(5868):647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Michaeli S., Podell D., Agabian N., Ullu E. The 7SL RNA homologue of Trypanosoma brucei is closely related to mammalian 7SL RNA. Mol Biochem Parasitol. 1992 Mar;51(1):55–64. doi: 10.1016/0166-6851(92)90200-4. [DOI] [PubMed] [Google Scholar]
- Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990 Feb 23;247(4945):939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Miller J. D., Wilhelm H., Gierasch L., Gilmore R., Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature. 1993 Nov 25;366(6453):351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- Moritz A., Goebel W. Characterization of the 7S RNA and its gene from halobacteria. Nucleic Acids Res. 1985 Oct 11;13(19):6969–6979. doi: 10.1093/nar/13.19.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Oubridge C., Jessen T. H., Li J., Evans P. R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990 Dec 6;348(6301):515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Imai Y., Nakamura A., Yamane K. Small cytoplasmic RNA of Bacillus subtilis: functional relationship with human signal recognition particle 7S RNA and Escherichia coli 4.5S RNA. J Bacteriol. 1992 Apr;174(7):2185–2192. doi: 10.1128/jb.174.7.2185-2192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., Walter P. Protein targeting to and translocation across the membrane of the endoplasmic reticulum. Curr Opin Cell Biol. 1992 Aug;4(4):573–580. doi: 10.1016/0955-0674(92)90074-m. [DOI] [PubMed] [Google Scholar]
- Ogg S. C., Poritz M. A., Walter P. Signal recognition particle receptor is important for cell growth and protein secretion in Saccharomyces cerevisiae. Mol Biol Cell. 1992 Aug;3(8):895–911. doi: 10.1091/mbc.3.8.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Overbeek R., Larsen N., Marsh T. L., McCaughey M. J., Maciukenas M. A., Kuan W. M., Macke T. J., Xing Y., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1992 May 11;20 (Suppl):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Smith D. Ribonuclease P: function and variation. J Biol Chem. 1990 Mar 5;265(7):3587–3590. [PubMed] [Google Scholar]
- Phillips G. J., Silhavy T. J. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992 Oct 22;359(6397):744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Bernstein H. D., Strub K., Zopf D., Wilhelm H., Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990 Nov 23;250(4984):1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Poritz M. A., Siegel V., Hansen W., Walter P. Small ribonucleoproteins in Schizosaccharomyces pombe and Yarrowia lipolytica homologous to signal recognition particle. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4315–4319. doi: 10.1073/pnas.85.12.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz M. A., Strub K., Walter P. Human SRP RNA and E. coli 4.5S RNA contain a highly homologous structural domain. Cell. 1988 Oct 7;55(1):4–6. doi: 10.1016/0092-8674(88)90003-7. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993 Mar;57(1):50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez C., Matheson A. T. A gene in the archaebacterium Sulfolobus solfataricus that codes for a protein equivalent to the alpha subunits of the signal recognition particle receptor in eukaryotes. Mol Microbiol. 1991 Jul;5(7):1687–1693. doi: 10.1111/j.1365-2958.1991.tb01916.x. [DOI] [PubMed] [Google Scholar]
- Rapoport T. A. Protein transport across the endoplasmic reticulum membrane: facts, models, mysteries. FASEB J. 1991 Oct;5(13):2792–2798. doi: 10.1096/fasebj.5.13.1916103. [DOI] [PubMed] [Google Scholar]
- Rapoport T. A. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992 Nov 6;258(5084):931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- Ribes V., Dehoux P., Tollervey D. 7SL RNA from Schizosaccharomyces pombe is encoded by a single copy essential gene. EMBO J. 1988 Jan;7(1):231–237. doi: 10.1002/j.1460-2075.1988.tb02804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V., Römisch K., Giner A., Dobberstein B., Tollervey D. E. coli 4.5S RNA is part of a ribonucleoprotein particle that has properties related to signal recognition particle. Cell. 1990 Nov 2;63(3):591–600. doi: 10.1016/0092-8674(90)90454-m. [DOI] [PubMed] [Google Scholar]
- Rothman J. H., Raymond C. K., Gilbert T., O'Hara P. J., Stevens T. H. A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell. 1990 Jun 15;61(6):1063–1074. doi: 10.1016/0092-8674(90)90070-u. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb J., Herz J., Prehn S., Frank R., Vingron M., Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989 Aug 10;340(6233):478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb J., Lingelbach K., Gausepohl H., Dobberstein B. The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J Cell Biol. 1990 Nov;111(5 Pt 1):1793–1802. doi: 10.1083/jcb.111.5.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T. A Mycoplasma protein homologous to mammalian SRP54 recognizes a highly conserved domain of SRP RNA. Nucleic Acids Res. 1992 Nov 11;20(21):5763–5770. doi: 10.1093/nar/20.21.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson T., Guindy Y. Nucleotide sequence of a Mycoplasma mycoides RNA which is homologous to E. coli 4.5S RNA. Nucleic Acids Res. 1990 Aug 25;18(16):4938–4938. doi: 10.1093/nar/18.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. L., Schekman R. Polypeptide translocation across the endoplasmic reticulum membrane. J Biol Chem. 1992 Jul 15;267(20):13791–13794. [PubMed] [Google Scholar]
- Schlenstedt G., Gudmundsson G. H., Boman H. G., Zimmermann R. A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and post-translationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J Biol Chem. 1990 Aug 15;265(23):13960–13968. [PubMed] [Google Scholar]
- Selinger D., Brennwald P., Liao X., Wise J. A. Identification of RNA sequences and structural elements required for assembly of fission yeast SRP54 protein with signal recognition particle RNA. Mol Cell Biol. 1993 Mar;13(3):1353–1362. doi: 10.1128/mcb.13.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger D., Liao X., Wise J. A. Functional interchangeability of the structurally similar tetranucleotide loops GAAA and UUCG in fission yeast signal recognition particle RNA. Proc Natl Acad Sci U S A. 1993 Jun 15;90(12):5409–5413. doi: 10.1073/pnas.90.12.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein-RNA "footprinting". Proc Natl Acad Sci U S A. 1988 Mar;85(6):1801–1805. doi: 10.1073/pnas.85.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988 Jan 15;52(1):39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- Siegel V., Walter P. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J Cell Biol. 1985 Jun;100(6):1913–1921. doi: 10.1083/jcb.100.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature. 1986 Mar 6;320(6057):81–84. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- Simoneau P., Hu P. C. The gene for a 4.5S RNA homolog from Mycoplasma pneumoniae: genetic selection, sequence, and transcription analysis. J Bacteriol. 1992 Jan;174(2):627–629. doi: 10.1128/jb.174.2.627-629.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L., Gunderson J. H., Elwood H. J., Alonso R. A., Peattie D. A. Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science. 1989 Jan 6;243(4887):75–77. doi: 10.1126/science.2911720. [DOI] [PubMed] [Google Scholar]
- Stirling C. J., Hewitt E. W. The S. cerevisiae SEC65 gene encodes a component of yeast signal recognition particle with homology to human SRP19. Nature. 1992 Apr 9;356(6369):534–537. doi: 10.1038/356534a0. [DOI] [PubMed] [Google Scholar]
- Stirling C. J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992 Feb;3(2):129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Moss J., Walter P. Binding sites of the 9- and 14-kilodalton heterodimeric protein subunit of the signal recognition particle (SRP) are contained exclusively in the Alu domain of SRP RNA and contain a sequence motif that is conserved in evolution. Mol Cell Biol. 1991 Aug;11(8):3949–3959. doi: 10.1128/mcb.11.8.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Walter P. Assembly of the Alu domain of the signal recognition particle (SRP): dimerization of the two protein components is required for efficient binding to SRP RNA. Mol Cell Biol. 1990 Feb;10(2):777–784. doi: 10.1128/mcb.10.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Walter P. Isolation of a cDNA clone of the 14-kDa subunit of the signal recognition particle by cross-hybridization of differently primed polymerase chain reactions. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9747–9751. doi: 10.1073/pnas.86.24.9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck J. C., Lempicki R. A., Toschka H. Y., Erdmann V. A., Fournier M. J. Escherichia coli 4.5S RNA gene function can be complemented by heterologous bacterial RNA genes. J Bacteriol. 1990 Mar;172(3):1284–1288. doi: 10.1128/jb.172.3.1284-1288.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck J. C., Toschka H. Y., Specht T., Erdmann V. A. Common structural features between eukaryotic 7SL RNAs, eubacterial 4.5S RNA and scRNA and archaebacterial 7S RNA. Nucleic Acids Res. 1988 Aug 11;16(15):7740–7740. doi: 10.1093/nar/16.15.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struck J. C., Vogel D. W., Ulbrich N., Erdmann V. A. The Bacillus subtilis scRNA is related to the 4.5S RNA from Escherichia coli. Nucleic Acids Res. 1988 Mar 25;16(6):2719–2719. doi: 10.1093/nar/16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha M. K., Dupuy B., Saurin W., So M., Marchal C. Control of pilus expression in Neisseria gonorrhoeae as an original system in the family of two-component regulators. Mol Microbiol. 1991 Jan;5(1):137–148. doi: 10.1111/j.1365-2958.1991.tb01834.x. [DOI] [PubMed] [Google Scholar]
- Toschka H. Y., Struck J. C., Erdmann V. A. The 4.5S RNA gene from Pseudomonas aeruginosa. Nucleic Acids Res. 1989 Jan 11;17(1):31–36. doi: 10.1093/nar/17.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Wainright P. O., Hinkle G., Sogin M. L., Stickel S. K. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993 Apr 16;260(5106):340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Disassembly and reconstitution of signal recognition particle. Cell. 1983 Sep;34(2):525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982 Oct 21;299(5885):691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981 Nov;91(2 Pt 1):557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol. 1981 Nov;91(2 Pt 1):551–556. doi: 10.1083/jcb.91.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Lingappa V. R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S. L., Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J Cell Biol. 1989 Dec;109(6 Pt 1):2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H., Luirink J., Tollervey D. Evolutionary conserved nucleotides within the E.coli 4.5S RNA are required for association with P48 in vitro and for optimal function in vivo. Nucleic Acids Res. 1992 Nov 25;20(22):5919–5925. doi: 10.1093/nar/20.22.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaver D. S., Matoba S., Ogrydziak D. M. A mutation in the signal recognition particle 7S RNA of the yeast Yarrowia lipolytica preferentially affects synthesis of the alkaline extracellular protease: in vivo evidence for translational arrest. J Cell Biol. 1992 Feb;116(3):605–616. doi: 10.1083/jcb.116.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., O'Brien J. M., Ouellette L. A., Church W. R., Johnson D. I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1991 Jul;11(7):3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D., Bernstein H. D., Johnson A. E., Walter P. The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J. 1990 Dec;9(13):4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf D., Bernstein H. D., Walter P. GTPase domain of the 54-kD subunit of the mammalian signal recognition particle is required for protein translocation but not for signal sequence binding. J Cell Biol. 1993 Mar;120(5):1113–1121. doi: 10.1083/jcb.120.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. Interaction of protein SRP19 with signal recognition particle RNA lacking individual RNA-helices. Nucleic Acids Res. 1991 Jun 11;19(11):2955–2960. doi: 10.1093/nar/19.11.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. The secondary structure of the 7SL RNA in the signal recognition particle: functional implications. Nucleic Acids Res. 1985 Sep 11;13(17):6105–6124. doi: 10.1093/nar/13.17.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Ullu E. Identification of dynamic sequences in the central domain of 7SL RNA. Nucleic Acids Res. 1986 Jun 11;14(11):4639–4657. doi: 10.1093/nar/14.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek A. M., Redelmeier T. E., Damke H., Tisdale E. J., Meyerowitz E. M., Schmid S. L. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993 Aug;122(3):553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]