Research highlights
► Chlorinated HDL promotes expression of heme oxygenase-1 (HO-1) in endothelial cells. ► Expression involves p42/44 MAPK and activation of transcription factor Egr-1. ► EMSA demonstrates induction of Egr-1 DNA binding activity. ► Immunocytochemistry shows translocation of Egr-1 to the nucleus. ► Silencing of p42/44 MAPK and Egr-1 impairs HO-1 expression to baseline levels.
Keywords: Chlorinated HDL, Hypochlorous acid, Inflammation, Myeloperoxidase, MPO–H2O2–chloride system, Nrf2
Abstract
Modification/chlorination of high-density lipoprotein (HDL) by hypochlorous acid (HOCl), formed by the myeloperoxidase–H2O2–chloride system of activated phagocytes, converts an anti-atherogenic lipoprotein into a pro-inflammatory lipoprotein particle. Chlorinated HDL is present in human lesion material, binds to and is internalized by endothelial cells and impairs expression and activity of endothelial nitric oxide synthase. The present study aimed at clarifying whether exposure of endothelial cells to pro-inflammatory HOCl–HDL impacts on expression of heme oxygenase-1, a potential rescue pathway against endothelial dysfunction. Our findings revealed that HDL modified by HOCl, added as reagent or generated enzymatically, induced phosphorylation of p42/44 mitogen-activated protein kinase, expression of transcription factor early growth response-1 (Egr-1) and enhanced expression of heme oxygenase-1 in human endothelial cells. Upregulation of heme oxygenase-1 could be blocked by an inhibitor upstream of p42/44 mitogen-activated protein kinase and/or knockdown of Egr-1 by RNA-interference. Electrophoretic mobility shift assays demonstrated HOCl–HDL-mediated induction of the Egr-1 DNA binding activity. Immunocytochemical and immunoblotting experiments demonstrated HOCl–HDL-induced translocation of Egr-1 to the nucleus. The present study demonstrates a novel compensatory pathway against adverse effects of HOCl–HDL, providing cytoprotection in a number of pathological conditions including cardiovascular disease.
Introduction
Epidemiological studies have established an inverse correlation between the risk for atherosclerosis, the plasma levels of apolipoprotein A–I (apoA–I)1 and high-density lipoprotein (HDL)–cholesterol. In line, pharmacological strategies show that raising HDL–cholesterol prevents coronary heart disease [1]. Besides an array of potential anti-inflammatory and anti-atherogenic activities [2] the most important vasculoprotective function of apoA–I and HDL is the role in ‘reverse cholesterol transport’ [3]. Accumulating evidence suggested that HDL and apoA–I become dysfunctional in ‘reverse cholesterol transport’ after oxidative modification. The phagocytic enzyme myeloperoxidase (MPO), an established cardiovascular risk factor [4], is considered a prime modulator to form a pro-inflammatory and pro-atherogenic HDL particle. MPO, abundantly present in neutrophils and monocytes uses H2O2 and chloride ions to generate chlorinating oxidants, in particular hypochlorous acid (HOCl), which play an important role in killing microorganisms [5–7].
MPO is enzymatically active in human atheroma [8] and immunoreactivity for the enzyme is prominent in intimal mononuclear cells in all regions of human atheroma [9]. Immunohistochemical studies demonstrated that specific epitopes derived from HOCl colocalized with apoA–I [10,11]. Findings that apoA–I represents a selective target for MPO-catalyzed oxidation [12] support observations on functional impairment of modified HDL in vivo and in vitro [13,14].
Attachment of MPO to and colocalization of HOCl-modified epitopes with apoA–I on endothelial cells [10,14] suggested adverse cellular effects of chlorinated HDL. We previously reported that HOCl–HDL interacts with human umbilical venous endothelial cells [15] thereby impairing their vasculoprotective function [16]. However, at sites of vascular inflammation endothelial cells may exert adaptive responses against cell damage and denudation that promote plaque erosion and thrombosis in vivo. Induction of inducible heme oxygenase-1 (HO-1) by a broad spectrum of agents represents an important cytoprotective pathway to alleviate endothelial dysfunction in response to inflammatory insults.
The cytoprotective effects of the stress-response protein HO-1 are reported to be due to its ability to degrade the pro-oxidative heme to anti-oxidative bile pigments [17] and to generate carbon monoxide that exhibits anti-proliferative, anti-inflammatory and vasodilatory properties [18]. It was shown previously that HO-1 accumulation is accompanied by increased bilirubin levels in rabbit atherosclerotic lesions [19]. Induction of HO-1 expression reversed the progression of vulnerable plaques into more stable lesions [20,21], whereas the lack of HO-1 led to an exacerbation of plaque formation [22].
Nuclear factor E2-related factor 2 (Nrf2), a basic leucine zipper transcription factor, binds to the cis-acting antioxidant response element (ARE) of stress responsive genes including HO-1 [23]. It was shown previously that oxidized low-density lipoprotein promotes induction of HO-1 in human smooth muscle cells via activation of protein kinase C, stress-activated protein kinase/c-jun-NH2-terminal kinase, p38 and p42/44 mitogen-activated protein kinases (MAPKs), and Nrf2 [24].
The zinc-finger transcription factor early growth response factor-1 (Egr-1) belongs to the family of immediate-early genes that binds to GC-rich sequences in the promoter region of target genes involved in the development of atherosclerosis [25–27]. Egr-1 is highly expressed in human and mouse atherosclerotic lesions [28] and is induced by modified low-density lipoprotein in human monocytes [29]. Previous reports show that phosphorylation of p42/44 MAPK leads to induction of Egr-1 expression [30,31] and that transcriptional activation of HO-1 by Egr-1 is induced by several triggers present in atherosclerotic vascular disease [32,33].
Oxidative stress causes enhanced endothelial cell injury in human and animal HO-1 deficiency [22,34]. As staining for MPO and HOCl-modified epitopes was present within and around endothelial cells in both human and animal lesions [35,36] where HO-1 is also highly expressed [37] the present study aimed at investigating signalling pathways leading to HO-1 expression in human endothelial cells in response to HOCl–HDL.
Materials and methods
Cell culture
Human EA.hy926 endothelial cells were grown in DMEM (Gibco Invitrogen, Lofer, Austria) with 4.5 g/l glucose, 3.97 mM l-glutamine and 1 mM sodium pyruvate supplemented with 10% (v:v) FCS, 1% penicillin–streptomycin, 1× HAT supplement at 37 °C in a humidified atmosphere of 5% CO2.
Isolation and modification of human lipoproteins
Low-density lipoprotein (LDL, d = 1.035–1.065 g/ml) was isolated from plasma of healthy, normolipidemic subjects by ultracentrifugation as described [38,39]. The protein of the final LDL preparation consisted of 96–98% apoB-100 as measured immunochemically. HDL (subclass 3, density = 1.125–1.21 g/ml) was further isolated by discontinuous density ultracentrifugation [10] and stored at 4 °C until usage. The protein concentration was determined by the Lowry method, using bovine serum albumin (BSA) as a standard. Native LDL and HDL were desalted using PD-10 desalting columns (GE Healthcare, Vienna, Austria) prior to modification. HOCl–LDL was prepared as described [40]. Briefly, one mg LDL protein per ml PBS (10mM, pH 7.4) was incubated with HOCl-solution (60 min, 4 °C). For LDL incubated in between 0.2 and 1.6 mM final HOCl concentration this resulted in an oxidant:lipoprotein molar ratio in between 100:1 and 800:1. Modification of HDL by reagent HOCl was performed as described [10,41]. Briefly, 2.5 mg of HDL/ml of PBS (10 mM, pH 7.4) were modified by the single addition of HOCl (see above). For HDL (and BSA) incubated in between 0.2 and 1.6 mM final HOCl concentration this resulted in an oxidant:(lipo)protein molar ratio in between 25:1 and 200:1 [15,40]. Alternatively, HDL was modified by the MPO–H2O2–chloride system [41] (approx. oxidant:lipoprotein molar ratio of 25:1). Briefly, 1 mg of HDL/ml of PBS was modified at pH 7.4 (37 °C) by alternate additions of H2O2 [41] and MPO (final concentrations: 440 μM H2O2, 8 μg/ml MPO (Planta Natural Products, Vienna, Austria)); in some experiments ascorbate was added (final concentration 14 μM; assuming a quantitative conversion of H2O2, an oxidant:lipoprotein molar ratio up to 50:1 can be expected). The modified (lipo)protein-preparations were passed over a PD10 column to remove unreacted HOCl and used immediately for cell culture experiments.
Cell culture experiments
Cells were seeded into respective cell culture dishes (Greiner Bio-One, Frickenhausen, Germany) and grown until reaching confluence. The medium was changed to complete medium without FCS and HAT and after 16 h incubation time, native or modified HDL, LDL or BSA was directly added into the medium to start the experiment. In some experiments, the cells were preincubated for 30 min with the respective signal transduction inhibitors for p42/44 MAPK kinase (PD98059, 25 μM) and for p38 MAPK (SB203580, 10 μM) (Merck Biosciences, Darmstadt, Germany). Experiments were terminated by aspiration of the medium and washing the cells twice with cold PBS.
Western blot analysis
Cells were lysed on ice in 100 μl lysis buffer (50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 10 mM Na4P2O7, 2 mM Na3VO4, 10 mM NaF, 1% (v:v) Triton X-100, 10% (v:v) glycerol and Complete Mini protease inhibitor cocktail tablets (Roche Applied Science, Vienna, Austria); pH 7.4) for 10 min. Cell lysates were scraped and centrifuged to remove cell debris. Protein concentration was determined according to Lowry. Three times reducing sample buffer (8% (w:v) SDS, 18% (v:v) glycerine, 114 mM Tris (pH 6.8), 0.04% (w:v) bromphenol-blue and 5% (v:v) mercaptoethanol) was added to equal amounts (50–100 μg) of total cell lysates, and heated for 5 min at 95 °C. Samples were subjected to electrophoresis on NuPAGE 4–12% gradient Bis–Tris Gels (Invitrogen, Lofer, Austria) and proteins were blotted to nitrocellulose. Membranes were blocked with 5% (w:v) non-fat milk (30 min) and afterwards incubated with the primary antibody at 4 °C overnight. The following primary antibodies were used: Mouse monoclonal β-actin (C4) antibody, rabbit polyclonal lamin A/C (H-110) antibody, rabbit polyclonal Egr-1 (C19) antibody and rabbit polyclonal Nrf2 (C-20) antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA); mouse monoclonal phospho-p44/42 MAPK (Erk1/2) antibody and rabbit polyclonal phospho-p38 MAPK antibody (Cell Signaling Technology, Inc., Danvers, MA, USA); rabbit polyclonal HO-1 (Hsp32) antibody (Assay Designs, Inc., Ann Arbor, MI, USA). Peroxidase-conjugated goat anti-rabbit IgG (Pierce Biotechnology, Inc., Rockford, IL, USA) and peroxidase-conjugated goat anti-mouse IgG (Rockland Immunochemicals, Inc., Gilbertsville, PA, USA) were used as secondary antibodies and added at 25 °C for 1.5 h. Immunoreactive bands were visualized by Super Signal West Pico Chemiluminescent substrate.
RNA isolation and real-time RT-PCR
Total RNA from endothelial cells was isolated using QIAshredder and RNeasy Mini Kit (QIAGEN, Hilden, Germany). One microgram of RNA was subjected to DNaseI digestion and reverse transcription (Invitrogen, Lofer, Austria) using random hexamer primers (Applied Biosystems Inc., Foster City, CA, USA). Six nanograms of cDNA was used as template for each Real-time PCR. Gene expression was determined by quantitative Real-time PCR using the LightCycler 480 system (Roche Diagnostics, Vienna, Austria) and the QIAGEN QuantiFast SYBR Green PCR Kit. QIAGEN QuantiTect Primer Assays were used to determine gene expression of Egr-1 (Hs_Egr1_2_SG), HO-1 (Hs_HMOX1_1_SG), and glyceraldehyde-3-phosphate dehydrogenase (Hs_GAPDH_2_SG), respectively. For detection of Nrf2 gene expression the following primer pair was used: forward 5′-TAC TCC CAG GTT GCC CAC A-3′ and reverse 5′-CAT CTA CAA ACG GGA ATG TCT GC-3′. Fold change in gene expression levels of the target gene compared to the housekeeping gene GAPDH was calculated by the method.
Electrophoretic mobility shift assay (EMSA)
Cells were scraped in PBS, centrifuged at 500g (4 °C, 3 min) and nuclear proteins were isolated using NE-PER extraction reagents as recommended by the manufacturer (Pierce Biotechnology, Inc., Rockford, IL, USA). Nuclear protein concentration was determined using the BCA™ Protein Assay kit (Pierce Biotechnology, Inc.). Oligonucleotide sequences of the Egr-1 consensus-binding site, reported by Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA) and synthesized by TibMolBiol (Berlin, Germany), were 5′-GGA TCC AGC GGG GGC GAG CGG GGG CGA-3′ and 3′-CCT AGG TCG CCC CCG CTC GCC CCC GCT-5′. Complementary oligonucleotides were annealed and radioactive labelled using T4 Polynucleotide kinase and [γ-32P]ATP. After removal of unincorporated nucleotides by Micro Bio-Spin-6 Chromatography Columns (Bio-Rad Laboratories, Inc., Vienna, Austria), 5 μg of nuclear protein extracts were incubated with 10 mM Tris pH 7.5, 50 mM NaCl, 1 mM DTT, 0.1 mM EDTA, 5% glycerol (v:v), 0.25 μg Poly[d(I-C)] and 5 μg BSA at 25 °C for 10 min. Labelled oligonucleotides were added to each sample and EMSA binding reaction took place at 25 °C for 20 min. A 20-fold molar excess of non-radiolabelled oligonucleotides was added to the samples prior to the addition of labelled probe for competition experiments. Glycerol was added to a final concentration of 20% (v:v) and samples were subjected to polyacrylamide gel electrophoresis (3.9% polyacrylamide gel; acrylamide:bisacrylamide, 29:1) in 0.25% (v:v) Tris boric acid buffer at 25 °C and 120 V for 3.5 h. The gel was dried under vacuum at 80 °C for 1.5 h and then exposed to Hyperfilm™ MP (GE Healthcare, Vienna, Austria) for 12–24 h.
Isolation of cytosolic and nuclear proteins
For translocation experiments, nuclear and cytosolic proteins were isolated using a modification of the method of Dignam et al. [42,43]. Briefly, cells were washed twice with cold PBS and once with cold buffer A (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT and Complete Mini protease inhibitor cocktail tablets). Cells were lysed in buffer A including 0.1% (v:v) Nonidet P-40 for 10 min on ice, centrifuged at 4500g (4 °C, 10 min); the supernatant, containing the cytosolic fraction, was stored at −70 °C. The nuclear pellet was washed once in PBS, once in buffer A and lysed in buffer C (20 mM Hepes pH 7.9, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 25% (v:v) glycerol and Complete Mini protease inhibitor cocktail tablets) for 30 min on ice. Samples were centrifuged at 21,000g (4 °C, 20 min); the supernatant, containing the nuclear fraction, was stored at −70 °C.
Immunofluorescence and confocal laser scanning microscopy
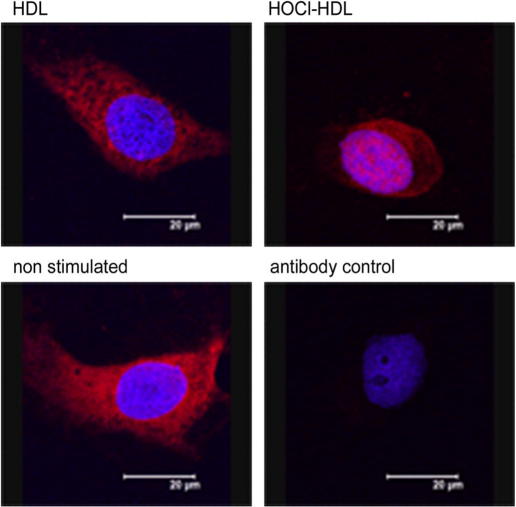
Cells were cultured on Lab-Tek chamber slides (Thermo Fisher Scientific, Rochester, NY, USA) and incubated in the absence (non-stimulated) or presence of 100 μg/ml lipoprotein (HDL or HOCl–HDL (200:1)) for 1 h. Afterwards the slides were washed with PBS, fixed with 3.7% (v:v) formaldehyde for 20 min and re-hydrated in PBS. After incubation with cold acetone for 3 min, slides were again re-hydrated in PBS and blocked with DakoCytomation Antibody Diluent with Background Reducing Components (Dako, Inc., Carpinteria, CA, USA) for 45 min. Slides were then incubated with a polyclonal rabbit anti-Egr-1 (C19) antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA, diluted with antibody diluent 1:50) for 45 min, washed with PBS and incubated with a Cy-3-labelled goat anti-rabbit antibody (1:250, Jackson ImmunoResearch Labs, Inc., West Grove, PA, USA) for 45 min. After rinsing in PBS, Hoechst 33258 was added to the slides for 15 min to counter stain nuclei. Slides were washed with PBS and mounted with Moviol (Merck Chemicals, Ltd., Nottingham, UK). Slides were analyzed on a confocal laser-scanning microscope (Leica SP2, Leica Lasertechnik GmbH, Heidelberg, Germany) using the 405 nm laser line for the excitation of Hoechst and the 543 nm line for Cy-3. Detection settings were: 420–470 nm for Hoechst Stain and 590–670 nm for Cy-3.
siRNA transfection
Cells were cultured in six-well plates and grown until reaching 50% confluence. siRNA transfection using Lipofectamine RNAiMAX or Oligofectamine (Invitrogen, Lofer, Austria) and 50 or 100 nM of siRNA, respectively, was performed according to the manufacturer’s suggestions. Cells were transfected either with Egr-1 siRNA or with scrambled control siRNA (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) for 6 h. Medium was changed to complete medium without FCS and HAT 30 h after transfection and 16 h prior to incubation with 0.1 mg/ml native HDL or HOCl–HDL for indicated time points.
Statistical analysis
Data are expressed as means ± SD for real-time PCR and densitometric evaluations of immunoreactive bands. One-way ANOVA including Bonferroni post-test was performed by using Prism 5 software (Graph pad Software, La Jolla, CA, USA). Means were considered as significantly different at ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
Results
HOCl–HDL-mediates activation of p42/44 and p38 MAPK
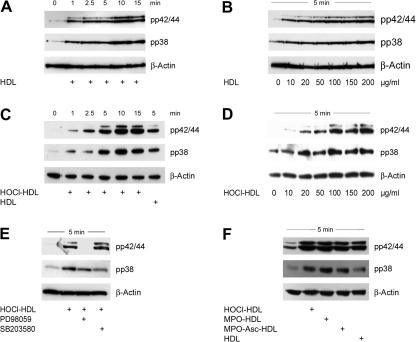
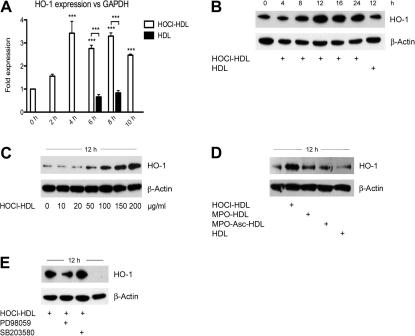
Native HDL has been reported to promote activation of p38 MAPK in human umbilical venous endothelial cells [44]. We here show that native HDL is able to induce phosphorylation of p42/44 and p38 MAPKs in a time- and concentration-dependent manner in human EA.hy926 endothelial cells (Fig. 1A and B).
Fig. 1.
Western blot analysis of native and modified HDL-induced MAPK activation in the absence or presence of MAPK inhibitors in endothelial cells. (A and C) Cells were incubated with 100 μg/ml HDL or HOCl–HDL (oxidant:lipoprotein molar ratio 200:1) for the indicated times or (B and D) with indicated concentrations of HDL or HOCl–HDL for 5 min. (E) Cells were preincubated for 30 min with 25 μM PD98059 or 10 μM SB203580 prior to stimulation with 100 μg/ml HOCl–HDL for 5 min. (F) Cells were incubated with 100 μg/ml native HDL or HDL modified by reagent HOCl (HOCl–HDL) or by the MPO–H2O2-system in the absence (MPO–HDL) or presence of ascorbate (MPO–Asc–HDL) for 5 min. (A–F) Cells were lysed and equal amounts of proteins were subjected to Western blot analysis for pp42/44 and pp38 MAPK. After stripping membranes, β-actin was used as a loading control. In all Western blots, lane 1 represents non-stimulated controls, i.e. cells in the absence of lipoproteins. One representative experiment out of three is shown.
We further tested the capacity of chlorinated HDL to activate these MAPKs. HOCl–HDL induced phosphorylation of p42/44 and p38 MAPKs also in a time- and concentration-dependent manner (Fig. 1C and D); maximum phosphorylation for both MAPKs was observed between 5 and 10 min after stimulation (Fig. 1C) at a protein concentration of 100 μg protein (Fig. 1D, Supplement Fig. ID). Phosphorylation of p42/44 by HOCl–HDL after 5 min was more pronounced when compared to native HDL (Supplement Fig. IC). Incubation of cells with PD98059 (an inhibitor for p42/44 MAPK kinase) for 30 min prior to stimulation with HOCl–HDL completely blocked phosphorylation of p42/44 MAPK (Fig. 1E). In contrast, SB203580 did not impair immunoreactive pp38 MAPK but enhanced p42/44 MAPK phosphorylation (Supplement Fig. IE).
Next, modification of HDL was performed enzymatically as occurring under in vivo conditions. Modification of HDL with HOCl generated by the MPO–H2O2–chloride system in the absence or presence of ascorbate (an enhancer of chlorination reaction of MPO [45]) also led to pronounced phosphorylation of p42/44 and p38 MAPKs as shown for HDL modified by reagent HOCl (Fig. 1F).
HOCl–HDL mediated activation of transcription factor Egr-1 via pp42/44 MAPK signalling
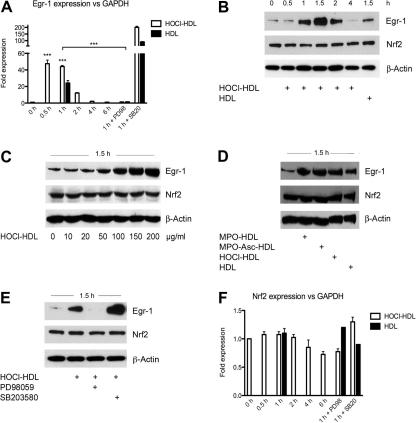
Next, we tested whether HOCl–HDL is able to induce known transcriptional activators of the HO-1 gene, i.e. Egr-1 and Nrf2. Fig. 2A shows time-dependent transcriptional activation of Egr-1 in response to HOCl–HDL. HOCl–HDL rapidly activated transcription of Egr-1 with maximum levels observed at 30 min. The response to native HDL after 1 h was 50% lower compared to HOCl–HDL. The p42/44 MAPK kinase inhibitor PD98059 completely prevented transcriptional activation of Egr-1 while the p38 MAPK antagonist increased transcriptional activation of Egr-1 in cells incubated with HOCl–HDL or HDL for 1 h (Fig. 2A). On the protein level, HOCl–HDL potently induced expression of Egr-1 in a time- and concentration-dependent manner (Fig. 2B and C); maximum of Egr-1 induction was in between 1 and 1.5 h after stimulation (Fig. 2B); native HDL only slightly induced Egr-1 on the protein level (Fig. 2B; for densitometric evaluation see Supplement Fig. IIB and C). As shown in Fig. 1F, HDL modified by HOCl generated via the MPO–H2O2–chloride system in the absence or presence of ascorbate had the same effect on Egr-1 protein expression as HDL modified by reagent HOCl (Fig. 2D); the effect of native HDL on Egr-1 mRNA and protein expression was less pronounced (Fig. 2A, B and D).
Fig. 2.
Real-time RT-PCR and Western blot analysis for lipoprotein-induced Egr-1 and Nrf2 expression in endothelial cells in the absence or presence of MAPK inhibitors. (A) Cells were preincubated for 30 min with 25 μM PD98059 or 10 μM SB20358 prior to stimulation with 100 μg/ml lipoprotein for the indicated times. Equal amounts of RNA were subjected to Real-time RT-PCR. Egr-1 mRNA expression level was normalized to GAPDH (∗∗∗p < 0.001). (B) Cells were incubated with 100 μg/ml lipoprotein for the indicated times. (C) Cells were incubated with indicated concentrations of HOCl–HDL for 1.5 h. (D) Cells were incubated with 100 μg/ml lipoprotein for 1.5 h. (E) Cells were preincubated for 30 min with 25 μM PD98059 or 10 μM SB203580 prior to stimulation with 100 μg/ml HOCl–HDL for 1.5 h. (B–E) Cells were lysed and equal amounts of proteins were subjected to Western blot analysis for Egr-1 and Nrf2. After stripping membranes, β-actin was used as a loading control. (F) Cells were preincubated for 30 min with 25 μM PD98059 or 10 μM SB20358 prior to stimulation with 100 μg/ml lipoprotein for the indicated times. Equal amounts of RNA were subjected to real-time RT-PCR. Nrf2 expression level was normalized to GAPDH. (B–E) Lane 1 represents non-stimulated controls, i.e. cells in the absence of lipoproteins. (A–F) One representative experiment (performed in triplicate A, F) out of three is shown.
To clarify whether MAPK activation is directly involved in Egr-1 protein expression, cells were preincubated with PD98059 or SB203580 before stimulation with HOCl–HDL. In line with data from Fig. 2A, the p42/44 MAPK kinase inhibitor completely blocked expression of Egr-1 protein in response to HOCl–HDL (Fig. 2E). Consistent with Egr-1 expression on mRNA level (Fig. 2A), preincubation of cells with SB203580 prior to stimulation with HOCl–HDL potentiated expression of Egr-1 also on the protein level (Fig. 2E). In contrast to Egr-1, Nrf2 expression on mRNA (Fig. 2F) and protein level (Fig. 2B–E) was unaffected by HOCl- or MPO-modified HDL preparations.
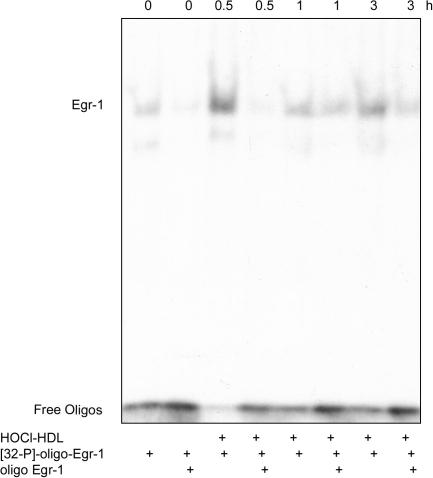
To follow whether HOCl–HDL-mediated stimulation induces Egr-1 DNA binding activity, nuclear proteins were subjected to EMSA using a specific probe for Egr-1. Time-dependent stimulation of cells with HOCl–HDL led to a maximum in Egr-1 binding after 0.5 h. Competition experiments with an excess of the non-radiolabelled Egr-1 probe confirmed the EMSA band as specific for Egr-1 (Fig. 3). This demonstrates that HOCl–HDL enhanced the binding of Egr-1 to its consensus-binding site in the promoter region of target genes.
Fig. 3.
HOCl–HDL-induced Egr-1 DNA binding activity in endothelial cells. EMSA of HOCl–HDL-induced Egr-1 DNA binding activity. Cells were stimulated with 100 μg/ml HOCl–HDL for the indicated times, nuclear extracts were isolated and incubated with radioactively labelled oligonucleotides of the Egr-1 consensus-binding site ([32P]-oligo-Egr-1). For competition experiments extracts were preincubated with a 20-fold molar excess of unlabeled oligonucleotides (oligo Egr-1) specific for Egr-1. Lanes 1 and 2 represents non-stimulated controls, i.e. cells in the absence of lipoproteins. One experiment out of three is shown.
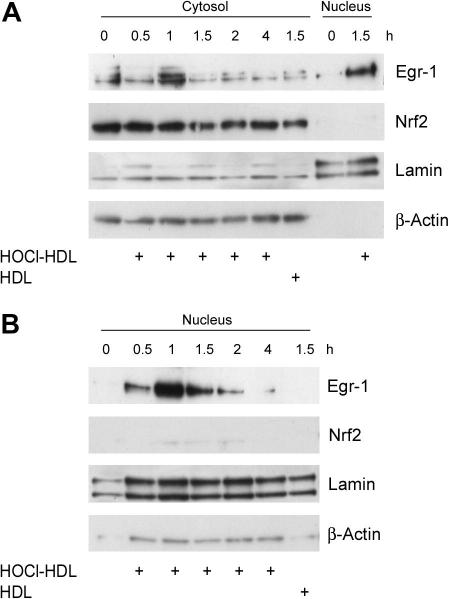
Time-course experiments of Egr-1 expression in the cytosolic protein fraction revealed maximum expression in HOCl–HDL-treated cells approx. after 1 h (Fig. 4A). At this (Fig. 4B) and later time points (1.5 h, Fig. 4A), pronounced Egr-1 expression became apparent in the nuclear fraction. Egr-1 expression by native HDL was less pronounced in the nuclear fraction compared to HOCl–HDL (Fig. 4B). Time-course experiments demonstrated that HOCl–HDL did not affect expression of Nrf2 in the cytosol nor promote nuclear translocation of Nrf2 (Fig. 4A). Immunofluorescence microscopy confirmed HOCl–HDL-induced nuclear translocation of Egr-1 in endothelial cells (Fig. 5). In non-stimulated cells or cells incubated with native HDL staining for Egr-1 was almost exclusively present in the cytosol (Fig. 5).
Fig. 4.
HOCl–HDL-induced Egr-1 nuclear translocation in endothelial cells. (A and B) Cells were incubated with 100 μg/ml lipoprotein for the indicated times. Cells were lysed, nuclear and cytosolic extracts were isolated and equal amounts of proteins were subjected to Western blot analysis for Egr-1 and Nrf2. After stripping membranes, β-actin or lamin was used as cytosolic or nuclear loading control. (A) Lanes 1 and 8 represents non-stimulated controls. (B) Lane 1 represents non-stimulated controls. (A and B) One experiment out of three is shown.
Fig. 5.
Immunocytochemistry of HOCl–HDL-induced nuclear translocation of Egr-1. Cells were cultured in the absence (non-stimulated) or presence of 100 μg/ml lipoprotein for 1 h. Cells were stained for Egr-1 (rabbit anti-Egr-1 antibody followed by Cy-3-labelled goat anti-rabbit antibody, red) and Hoechst (blue). Control experiments (antibody control) were performed by omitting the primary antibody. Slides were then analyzed on a confocal laser-scanning microscope. The bar represents 20 μm. One representative experiment out of three is shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
HOCl–HDL-mediated induction of HO-1 via pp42/44 MAPK signalling
Next, we tested whether HOCl–HDL promotes induction of HO-1. HOCl–HDL promoted high expression of HO-1 on mRNA level (maximum in between 4 and 8 h); native HDL had no effect on HO-1 expression (Fig. 6A). Western blot experiments revealed that HOCl–HDL induced expression of HO-1 in a time- and concentration-dependent manner (Fig. 6B and C) with a maximum HO-1 expression after 12 h (Fig. 6B). Induction of HO-1 was similar in cells in the absence or presence of native HDL (Fig. 6B). HDL modified by the MPO–H2O2–chloride system had a less pronounced effect on HO-1 expression (Fig. 6D). To directly address involvement of MAPK activation in HO-1 expression, cells were preincubated with respective MAPK (kinase) inhibitors. Only PD98059 impaired expression of immunoreactive HO-1 (Fig. 6E). Although the p38 MAPK inhibitor SB203580 promoted activation of p42/44 MAPK and expression of Egr-1, HO-1 expression was not affected.
Fig. 6.
Real-time RT-PCR and Western blot analysis of lipoprotein-induced HO-1 expression in endothelial cells. (A) Cells were stimulated with 100 μg/ml lipoprotein for the indicated times. Equal amounts of RNA were subjected to Real-time RT-PCR. HO-1 expression was normalized to GAPDH (∗∗∗p < 0.001). (B) Cells were incubated with 100 μg/ml lipoprotein for the indicated times. (C) Cells were incubated with indicated concentrations of HOCl–HDL for 12 h. (D) Cells were incubated with 100 μg/ml lipoprotein for 12 h. (E) Cells were preincubated for 30 min with 25 μM PD98059 or 10 μM SB203580 prior to stimulation with 100 μg/ml HOCl–HDL for 12 h. Lane 4 represents non-stimulated controls, i.e. cells in the absence of lipoproteins. (B–D) Lane 1 represents non-stimulated controls. (B–E) Cells were lysed and equal amounts of proteins were subjected to Western blot analysis for HO-1. After stripping membranes, β-actin was used as a loading control. (A–E) One representative experiment (performed in triplicate A) out of three is shown.
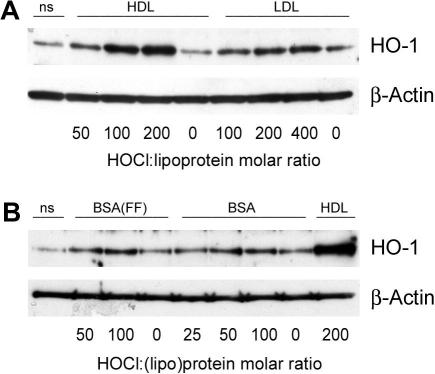
Fig. 7A demonstrates that HO-1 expression increases with increasing degree of HDL modification. To reveal whether up-regulation of HO-1 by HOCl–HDL is specific for HDL, the cells were incubated with HOCl–LDL (Fig. 7A) or HOCl–BSA (Fig. 7B). Although an increasing oxidant:(lipo)protein molar ratio of LDL and BSA tended to increase HO-1 expression, HOCl–LDL and HOCl–BSA were less effective inducers of HO-1 upregulation when compared to HOCl–HDL. No difference in HO-1 expression was observed when BSA or fatty acid-free BSA was used in cell culture experiments.
Fig. 7.
Western blot analysis of (lipo)protein-induced HO-1 expression in endothelial cells. (A) Cells were incubated with 100 μg protein/ml native and modified lipoproteins (at indicated oxidant:lipoprotein molar ratio, 50:1 to 400:1) or (B) 100 μg protein/ml native and modified BSA (at indicated oxidant:protein molar ratio, 25:1 to 200:1, FF = fatty-acid free) for 12 h. HOCl–HDL (200:1) was used as a positive control. Cells were lysed and equal amounts of proteins were subjected to Western blot analysis for HO-1. After stripping membranes, β-actin was used as a loading control. Lane 1 represents non-stimulated controls (ns), i.e. cells in the absence of lipoproteins. (A/B) One representative experiment out of three is shown.
Egr-1 is involved in HOCl–HDL-mediated activation of HO-1
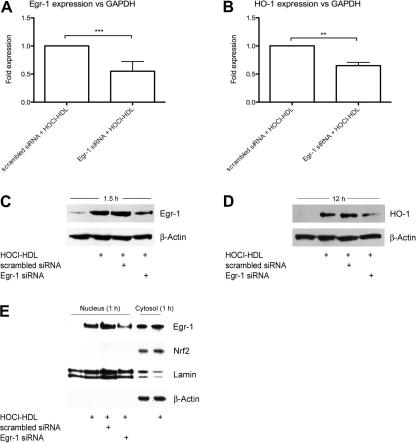
To reveal that Egr-1 is directly involved in HOCl–HDL-mediated activation of HO-1, Egr-1 was silenced by RNA interference. Due to the low basal Egr-1 mRNA and protein levels the silencing efficiency of RNA interference was analyzed in HOCl–HDL-treated cells. Therefore, experiments revealed that under our experimental conditions transfection of Egr-1 specific siRNA led to a reduced silencing efficiency. Fig. 8A/B showed that a specific siRNA directed against Egr-1 inhibits HOCl–HDL-mediated expression of Egr-1 and HO-1 on mRNA level in between 40% and 50%. Western blot experiments confirmed knockdown of Egr-1 (1.5 h) and HO-1 protein (12 h) up to 60% (Fig. 8C/D); the scrambled control siRNA had no effect on HOCl–HDL-mediated induction of Egr-1 and HO-1.
Fig. 8.
Real-time RT-PCR and Western blot analysis of HOCl–HDL-stimulated Egr-1 and HO-1 expression in endothelial cells treated with siRNA against Egr-1. (A/B) Cells were transfected with 50 nM siRNA against Egr-1 or with scrambled control siRNA. Forty-eight hours after transfection, cells were stimulated with 100 μg/ml HOCl–HDL for 1 h to determine Egr-1 mRNA expression (A) and for 4 h to assess HO-1 mRNA expression (B). Equal amounts of RNA were subjected to Real-time RT-PCR. Egr-1 and HO-1 expression were normalized to GAPDH (∗∗∗p < 0.001, ∗∗p < 0.01). (C and D) Cells were transfected with 50 nM siRNA against Egr-1 or with scrambled control siRNA and after 48 h cells were stimulated with 100 μg/ml HOCl–HDL for 1.5 h to assess Egr-1 (C) and for 12 h to determine HO-1 (D) protein expression. Cells were lysed and equal amounts of proteins were subjected to Western blot analysis. β-Actin was used as a loading control. Lane 1 represents non-stimulated controls, i.e. cells in the absence of lipoproteins. (E) Western blot analysis of HOCl–HDL-induced nuclear translocation of Egr-1 in cells treated with siRNA against Egr-1. Cells were transfected with 100 nM siRNA against Egr-1 or with scrambled control siRNA and after 48 h cells were stimulated with 100 μg/ml HOCl–HDL for 1 h. Cells were lysed, nuclear and cytosolic extracts were isolated and equal amounts of proteins were subjected to Western blot analysis for Egr-1 and HO-1. After stripping membranes, β-actin was used as a cytosolic and lamin as a nuclear loading control. Lanes 1 and 5 represents non-stimulated controls. (A–E) One representative experiment (performed in triplicate A/B) out of three is shown.
Fig. 8E showed the presence of immunoreactive Egr-1 in the cytosolic fraction. HOCl–HDL only slightly enhanced immunoreactive Egr-1 in the cytosolic fraction compared to non-stimulated cells (lanes 5 and 6); however, in parallel, HOCl–HDL increased immunoreactive Egr-1 in the nucleus (lane 2). Silencing of Egr-1 resulted in a lower nuclear Egr-1 protein content as compared to cells treated with scrambled control siRNA (Fig. 8E, lanes 3 and 4).
Expression of Nrf2 remained constant in the cytosol in the absence or presence of HOCl–HDL and was not detected in the nucleus (Fig. 8E). These findings indicate that HOCl–HDL mediates HO-1 expression via activation of transcription factor Egr-1.
Discussion
Accumulating reports confirmed that MPO-mediated loss of the atheroprotective functional properties of HDL might provide a novel mechanism linking inflammation and oxidative stress to the pathogenesis of atherosclerosis [14,46,47]. HDL isolated from human atheroma is modified by MPO-derived oxidants and even acts as an in vivo carrier of MPO [13]. Most importantly, chloramines, formed during HOCl-treatment of apoA–I lysine residues enhance binding of MPO to modified/chlorinated HDL [48] thus favouring amplification of the modification cycle. In addition to decreased cholesterol acceptor activities, HDL modified by HOCl (added as reagent or generated by the MPO–H2O2–chloride system) is subject to efficient uptake and degradation by macrophages [41,49], an event leading to foam cell formation in vitro and in vivo.
Wang and co-workers [37] previously demonstrated high HO-1 expression in macrophages, foam cells and endothelial cells in human and animal atherosclerotic lesions. Furthermore, selective overexpression of HO-1 decreased lesion size in apoE−/− mice [21,50]. Ishikawa and colleagues [51,52] showed that inducers of HO-1 induce atheroprotective pathways e.g. diminished lesion size in apoE−/− mice and Watanabe heritable hyperlipidemic rabbits. In line, double knockout mice deficient in HO-1 and apoE develop larger and more advanced lesions than apoE−/− littermates when fed a hypercholesterolemic diet [22].
Induction of HO-1 expression functions as a protective feedback mechanism to prevent endothelial cell dysfunction in response to inflammatory assaults. Results of the present study indicate that upregulation of HO-1 in response to pro-inflammatory chlorinated HDL might represent a hitherto unrecognized protection pathway in endothelial cells. We propose that the underlying signalling machinery that leads to HO-1 transcription might function as a first line of defence against HOCl–HDL-mediated cell dysfunction.
Low concentrations of HOCl induce the apoptotic machinery in endothelial cells [53] while high concentrations promote a significant rise in HO-1 via the Nrf2/ARE pathway to counteract mitochondrial dysfunction and cell death [54]. Nrf2 has recently been reported to regulate the induction of HO-1 in response to various forms of cellular stress, including hemodynamics and endoplasmic reticulum stress [50]. Moreover, fibroblasts from Nrf2−/− mice express reduced levels of HO-1 mRNA [55]. Results of the present study identified an alternative pathway for HO-1 induction by HOCl–HDL via intermediate activation of Egr-1. A striking observation was that the commonly used pharmacological inhibitor SB203580 did not inhibit lipoprotein-induced p38 MAPK phosphorylation (Fig. 1E). However, this is in line with findings that SB203580 does not block all p38 MAPK isoforms [56] but preferentially exerts inhibition downstream of pp38 MAPK signalling [57]. Another observation was that the pyridinyl imidazole p38 MAPK inhibitor SB203580 even enhanced p42/44 MAPK activation in the presence of HOCl–HDL (Fig. 1E). This one-way communication between p38 and p42/44 MAPKs has previously been addressed in other cellular systems [58–61]. SB203580 inhibits p38 MAPK α-isoform signalling leading to increased phosphorylation of p42/44 MAPK [59,60]. As Egr-1 expression is p42/44 MAPK-dependent under our experimental conditions, enhanced expression of Egr-1 on mRNA and protein levels in cells treated with HOCl–HDL and SB203580 may be expected (Fig 2A/E). Recent findings revealed that SB203580-mediated expression of Egr-1 in the absence or presence of other agonists is completely blocked by addition of p42/44 MAPK inhibitors [61]. Egr-1 transcription is activated by different subgroups of MAPKs [30,31,62,63]. We here show that activation of p42/44 by HOCl–HDL is an initial event in this cascade. HOCl–HDL binds to scavenger receptor SR-BI [10,15] and lipoprotein binding may promote phosphorylation of p42/44 MAPK [64]. Therefore, binding of HOCl–HDL to SR-BI might be an initiating event in this cascade.
Egr-1 acts as transcriptional regulator of several genes implicated in the pathogenesis of vascular disease [63]. Indeed, high-level expression of Egr-1 and Egr-1 inducible genes have been observed in human and murine lesion material [28]. Other data obtained with fibroblasts provide evidence that Egr-1 might directly activate HO-1 [32,33]. We here show that treatment of endothelial cells with chlorinated HDL involves activation of p42/44 MAPK, subsequent expression and nuclear translocation of Egr-1 and direct activation of HO-1. Multiple approaches were used to validate this signalling pathway(s): first, expression of Egr-1 and HO-1 in response to HOCl–HDL was investigated on mRNA and protein level; second, a well characterized inhibitor revealed that Egr-1 and HO-1-mediated activation is p42/44 MAPK dependent; third, involvement of Egr-1 in HO-1 expression and its nuclear localization was confirmed by EMSA, immunoblotting, immunofluorescence, and Egr-1 RNA interference, respectively. Most importantly, Egr-1 activation was observed in response to HDL modified by HOCl that was added as reagent or generated enzymatically by the MPO–H2O2–chloride system; the HOCl:HDL molar ratio used during the present study (25:1 to 50:1 for MPO-modified HDL) most probably reflects physiological conditions. Assuming a HOCl concentration up to 340 μM [65] under acute inflammation and a HDL3 plasma concentration of 6–12 μM [41] this would result in a HOCl:lipoprotein molar ratio under in vivo conditions as mentioned above.
Treatment of human umbilical venous endothelial cells with reagent HOCl or HOCl-modified lipoproteins leads to reduced endothelial nitric oxide synthase (eNOS) dimer stability [66] and reduced expression and activity of eNOS [16,67] apparently due to eNOS delocalization from the plasma membrane. Heiss and co-workers [68] demonstrated that eNOS downregulation in endothelial cells parallels HO-1 induction with subsequent local and transient heme deficiency. Most importantly, murine Egr-1 has recently been identified to promote transcriptional repression of δ-aminolevulinic acid synthase 1, the first and rate-limiting enzyme in heme biosynthesis [69]. Also taurine, when chlorinated by the MPO–H2O2–chloride system, induces HO-1 expression and inhibits NO synthesis [70]; however, this activation process occurring in macrophages involves the Nrf2 but not the Egr-1/HO-1 axis [71].
HO-1 is a cytoprotective enzyme and its induction commonly occurs in the setting of increased cellular response to maintain physiological haemostasis [50,56]. More recently it has been reported that also in cultured smooth muscle cells, endoplasmic reticulum stress increased HO-1 mRNA and protein via the ARE, and this was associated with cell survival. However, these studies mostly include activation of HO-1 via the Nrf2-axis [72]. HO-1 regulation responds to a large and broad spectrum of chemical stresses, including agents that cause oxidative stress or diminish oxygen and thiol-reactive agents. A number of in vitro studies have shown that hydroperoxides, oxidized phospholipids, H2O2, and HOCl, respectively, may induce HO-1 [50,54].
We now show that chlorinated HDL is able to promote expression of HO-1 via a selective signalling cascade involving p42/44 activation and nuclear translocation of Egr-1 (Figs. 3–8) although the possibility of alternative activation pathways cannot be excluded. The ability of endothelial cells to respond adequately against adverse effects of chlorinated HDL suggests a novel, Egr-1- but not Nrf2-mediated compensatory pathway, providing cytoprotection in a number of pathological conditions including cardiovascular disease. Whether the lipid moiety of modified HDL or chloramines are responsible for this effect is currently under investigation.
Acknowledgments
The authors are grateful to Dr. M. Vadon (Graz, Austria) for providing human plasma. The expert technical assistance of M. Sundl is appreciated. C.R. is funded by the PhD Program Molecular Medicine of the Medical University of Graz. This work was supported by the Austrian Science Fund (FWF, P19074-B05, F3007).
Footnotes
Presented in part as a poster at the 6th International Human Peroxidase Meeting 2009 in Chapel Hill, NC, USA.
Abbreviations used: apoA–I, apolipoprotein A–I; ARE, antioxidant response element; BSA, bovine serum albumin; Egr-1, early growth response-1; EMSA, electrophoretic mobility shift assay; eNOS, endothelial nitric oxide synthase; HDL, high-density lipoprotein; HO-1, heme oxygenase-1; HOCl, hypochlorous acid; LDL, low-density lipoprotein; MAPK, mitogen-activated protein kinase; MPO, myeloperoxidase; Nrf2, nuclear factor E2-related factor 2.
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.abb.2011.02.016.
Appendix A. Supplementary data
References
- 1.Linsel-Nitschke P., Tall A.R. Nat. Rev. Drug Discov. 2005;4:193–205. doi: 10.1038/nrd1658. [DOI] [PubMed] [Google Scholar]
- 2.Kontush A., Chapman M.J. Pharmacol. Rev. 2006;58:342–374. doi: 10.1124/pr.58.3.1. [DOI] [PubMed] [Google Scholar]
- 3.von Eckardstein A., Nofer J.R., Assmann G. Arterioscler. Thromb. Vasc. Biol. 2001;21:13–27. doi: 10.1161/01.atv.21.1.13. [DOI] [PubMed] [Google Scholar]
- 4.Zhang R., Brennan M.L., Fu X., Aviles R.J., Pearce G.L., Penn M.S., Topol E.J., Sprecher D.L., Hazen S.L. JAMA. 2001;286:2136–2142. doi: 10.1001/jama.286.17.2136. [DOI] [PubMed] [Google Scholar]
- 5.Winterbourn C.C., Kettle A.J. Free Radic. Biol. Med. 2000;29:403–409. doi: 10.1016/s0891-5849(00)00204-5. [DOI] [PubMed] [Google Scholar]
- 6.Davies M.J., Hawkins C.L., Pattison D.I., Rees M.D. Antioxid. Redox Signal. 2008;10:1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- 7.Arnhold J., Flemmig J. Arch. Biochem. Biophys. 2010;500:92–106. doi: 10.1016/j.abb.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Daugherty A., Dunn J.L., Rateri D.L., Heinecke J.W. J. Clin. Invest. 1994;94:437–444. doi: 10.1172/JCI117342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennathur S., Bergt C., Shao B., Byun J., Kassim S.Y., Singh P., Green P.S., McDonald T.O., Brunzell J., Chait A., Oram J.F., O’Brien K., Geary R.L., Heinecke J.W. J. Biol. Chem. 2004;279:42977–42983. doi: 10.1074/jbc.M406762200. [DOI] [PubMed] [Google Scholar]
- 10.Marsche G., Hammer A., Oskolkova O., Kozarsky K.F., Sattler W., Malle E. J. Biol. Chem. 2002;277:32172–32179. doi: 10.1074/jbc.M200503200. [DOI] [PubMed] [Google Scholar]
- 11.Bergt C., Pennathur S., Fu X., Byun J., O’Brien K., McDonald T.O., Singh P., Anantharamaiah G.M., Chait A., Brunzell J., Geary R.L., Oram J.F., Heinecke J.W. Proc. Natl. Acad. Sci. USA. 2004;101:13032–13037. doi: 10.1073/pnas.0405292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergt C., Oettl K., Keller W., Andreae F., Leis H.J., Malle E., Sattler W. Biochem. J. 2000;346(Pt 2):345–354. [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng L., Nukuna B., Brennan M.L., Sun M., Goormastic M., Settle M., Schmitt D., Fu X., Thomson L., Fox P.L., Ischiropoulos H., Smith J.D., Kinter M., Hazen S.L. J. Clin. Invest. 2004;114:529–541. doi: 10.1172/JCI21109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malle E., Marsche G., Panzenboeck U., Sattler W. Arch. Biochem. Biophys. 2006;445:245–255. doi: 10.1016/j.abb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 15.Marsche G., Levak-Frank S., Quehenberger O., Heller R., Sattler W., Malle E. FASEB J. 2001;15:1095–1097. doi: 10.1096/fj.00-0532fje. [DOI] [PubMed] [Google Scholar]
- 16.Marsche G., Heller R., Fauler G., Kovacevic A., Nuszkowski A., Graier W., Sattler W., Malle E. Arterioscler. Thromb. Vasc. Biol. 2004;24:2302–2306. doi: 10.1161/01.ATV.0000148703.43429.25. [DOI] [PubMed] [Google Scholar]
- 17.Stocker R., Yamamoto Y., McDonagh A.F., Glazer A.N., Ames B.N. Science. 1987;235:1043–1046. doi: 10.1126/science.3029864. [DOI] [PubMed] [Google Scholar]
- 18.Morita T. Arterioscler. Thromb. Vasc. Biol. 2005;25:1786–1795. doi: 10.1161/01.ATV.0000178169.95781.49. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama M., Takahashi K., Komaru T., Fukuchi M., Shioiri H., Sato K., Kitamuro T., Shirato K., Yamaguchi T., Suematsu M., Shibahara S. Arterioscler. Thromb. Vasc. Biol. 2001;21:1373–1377. doi: 10.1161/hq0801.093592. [DOI] [PubMed] [Google Scholar]
- 20.Cheng C., Noordeloos A.M., Jeney V., Soares M.P., Moll F., Pasterkamp G., Serruys P.W., Duckers H.J. Circulation. 2009;119:3017–3027. doi: 10.1161/CIRCULATIONAHA.108.808618. [DOI] [PubMed] [Google Scholar]
- 21.Juan S.H., Lee T.S., Tseng K.W., Liou J.Y., Shyue S.K., Wu K.K., Chau L.Y. Circulation. 2001;104:1519–1525. doi: 10.1161/hc3801.095663. [DOI] [PubMed] [Google Scholar]
- 22.Yet S.F., Layne M.D., Liu X., Chen Y.H., Ith B., Sibinga N.E., Perrella M.A. FASEB J. 2003;17:1759–1761. doi: 10.1096/fj.03-0187fje. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen T., Sherratt P.J., Pickett C.B. Annu. Rev. Pharmacol. Toxicol. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- 24.Anwar A.A., Li F.Y., Leake D.S., Ishii T., Mann G.E., Siow R.C. Free Radic. Biol. Med. 2005;39:227–236. doi: 10.1016/j.freeradbiomed.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Sukhatme V.P., Cao X.M., Chang L.C., Tsai-Morris C.H., Stamenkovich D., Ferreira P.C., Cohen D.R., Edwards S.A., Shows T.B., Curran T. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 26.Blaschke F., Bruemmer D., Law R.E. Rev. Endocr. Metab. Disord. 2004;5:249–254. doi: 10.1023/B:REMD.0000032413.88756.ee. [DOI] [PubMed] [Google Scholar]
- 27.Khachigian L.M. Circ Res. 2006;98:186–191. doi: 10.1161/01.RES.0000200177.53882.c3. [DOI] [PubMed] [Google Scholar]
- 28.McCaffrey T.A., Fu C., Du B., Eksinar S., Kent K.C., Bush H., Jr., Kreiger K., Rosengart T., Cybulsky M.I., Silverman E.S., Collins T. J. Clin. Invest. 2000;105:653–662. doi: 10.1172/JCI8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoyanova E., Tesch A., Armstrong V.W., Wieland E. Clin. Biochem. 2001;34:483–490. doi: 10.1016/s0009-9120(01)00258-2. [DOI] [PubMed] [Google Scholar]
- 30.Guha M., O’Connell M.A., Pawlinski R., Hollis A., McGovern P., Yan S.F., Stern D., Mackman N. Blood. 2001;98:1429–1439. doi: 10.1182/blood.v98.5.1429. [DOI] [PubMed] [Google Scholar]
- 31.Day F.L., Rafty L.A., Chesterman C.N., Khachigian L.M. J. Biol. Chem. 1999;274:23726–23733. doi: 10.1074/jbc.274.34.23726. [DOI] [PubMed] [Google Scholar]
- 32.Chen H., Wang L., Gong T., Yu Y., Zhu C., Li F., Li C. Biochem. Biophys. Res. Commun. 2010;396:388–393. doi: 10.1016/j.bbrc.2010.04.102. [DOI] [PubMed] [Google Scholar]
- 33.Yang G., Nguyen X., Ou J., Rekulapelli P., Stevenson D.K., Dennery P.A. Blood. 2001;97:1306–1313. doi: 10.1182/blood.v97.5.1306. [DOI] [PubMed] [Google Scholar]
- 34.Yachie A., Niida Y., Wada T., Igarashi N., Kaneda H., Toma T., Ohta K., Kasahara Y., Koizumi S. J. Clin. Invest. 1999;103:129–135. doi: 10.1172/JCI4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malle E., Waeg G., Schreiber R., Grone E.F., Sattler W., Grone H.J. Eur. J. Biochem. 2000;267:4495–4503. doi: 10.1046/j.1432-1327.2000.01498.x. [DOI] [PubMed] [Google Scholar]
- 36.Malle E., Wag G., Thiery J., Sattler W., Grone H.J. Biochem. Biophys. Res. Commun. 2001;289:894–900. doi: 10.1006/bbrc.2001.6074. [DOI] [PubMed] [Google Scholar]
- 37.Wang L.J., Lee T.S., Lee F.Y., Pai R.C., Chau L.Y. Am. J. Pathol. 1998;152:711–720. [PMC free article] [PubMed] [Google Scholar]
- 38.Malle E., Ibovnik A., Steinmetz A., Kostner G.M., Sattler W. Arterioscler. Thromb. 1994;14:345–352. doi: 10.1161/01.atv.14.3.345. [DOI] [PubMed] [Google Scholar]
- 39.Malle E., Ibovnik A., Leis H.J., Kostner G.M., Verhallen P.F., Sattler W. Arterioscler. Thromb. Vasc. Biol. 1995;15:377–384. doi: 10.1161/01.atv.15.3.377. [DOI] [PubMed] [Google Scholar]
- 40.Malle E., Hazell L., Stocker R., Sattler W., Esterbauer H., Waeg G. Arterioscler. Thromb. Vasc. Biol. 1995;15:982–989. doi: 10.1161/01.atv.15.7.982. [DOI] [PubMed] [Google Scholar]
- 41.Panzenboeck U., Raitmayer S., Reicher H., Lindner H., Glatter O., Malle E., Sattler W. J. Biol. Chem. 1997;272:29711–29720. doi: 10.1074/jbc.272.47.29711. [DOI] [PubMed] [Google Scholar]
- 42.Dignam J.D., Lebovitz R.M., Roeder R.G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solan N.J., Miyoshi H., Carmona E.M., Bren G.D., Paya C.V. J. Biol. Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 44.Norata G.D., Callegari E., Inoue H., Catapano A.L. Arterioscler. Thromb. Vasc. Biol. 2004;24:871–877. doi: 10.1161/01.ATV.zhq0504.1403. [DOI] [PubMed] [Google Scholar]
- 45.Marquez L.A., Dunford H.B., Van Wart H. J. Biol. Chem. 1990;265:5666–5670. [PubMed] [Google Scholar]
- 46.Nicholls S.J., Zheng L., Hazen S.L. Trends Cardiovasc. Med. 2005;15:212–219. doi: 10.1016/j.tcm.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Shao B., Oda M.N., Oram J.F., Heinecke J.W. Curr. Opin. Cardiol. 2006;21:322–328. doi: 10.1097/01.hco.0000231402.87232.aa. [DOI] [PubMed] [Google Scholar]
- 48.Marsche G., Furtmuller P.G., Obinger C., Sattler W., Malle E. Cardiovasc. Res. 2008;79:187–194. doi: 10.1093/cvr/cvn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergt C., Reicher H., Malle E., Sattler W. FEBS Lett. 1999;452:295–300. doi: 10.1016/s0014-5793(99)00677-8. [DOI] [PubMed] [Google Scholar]
- 50.Stocker R., Perrella M.A. Circulation. 2006;114:2178–2189. doi: 10.1161/CIRCULATIONAHA.105.598698. [DOI] [PubMed] [Google Scholar]
- 51.Ishikawa K., Sugawara D., Goto J., Watanabe Y., Kawamura K., Shiomi M., Itabe H., Maruyama Y. Circulation. 2001;104:1831–1836. doi: 10.1161/hc3901.095897. [DOI] [PubMed] [Google Scholar]
- 52.Ishikawa K., Sugawara D., Wang X., Suzuki K., Itabe H., Maruyama Y., Lusis A.J. Circ. Res. 2001;88:506–512. doi: 10.1161/01.res.88.5.506. [DOI] [PubMed] [Google Scholar]
- 53.Vissers M.C., Pullar J.M., Hampton M.B. Biochem. J. 1999;344(Pt 2):443–449. [PMC free article] [PubMed] [Google Scholar]
- 54.Wei Y., Liu X.M., Peyton K.J., Wang H., Johnson F.K., Johnson R.A., Durante W. Am. J. Physiol. Cell. Physiol. 2009;297:C907–915. doi: 10.1152/ajpcell.00536.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leung L., Kwong M., Hou S., Lee C., Chan J.Y. J. Biol. Chem. 2003;278:48021–48029. doi: 10.1074/jbc.M308439200. [DOI] [PubMed] [Google Scholar]
- 56.Ryter S.W., Alam J., Choi A.M. Physiol. Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 57.Kumar S., Jiang M.S., Adams J.L., Lee J.C. Biochem. Biophys. Res. Commun. 1999;263:825–831. doi: 10.1006/bbrc.1999.1454. [DOI] [PubMed] [Google Scholar]
- 58.Birkenkamp K.U., Tuyt L.M., Lummen C., Wierenga A.T., Kruijer W., Vellenga E. Br. J. Pharmacol. 2000;131:99–107. doi: 10.1038/sj.bjp.0703534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H., Shi X., Hampong M., Blanis L., Pelech S. J. Biol. Chem. 2001;276:6905–6908. doi: 10.1074/jbc.C000917200. [DOI] [PubMed] [Google Scholar]
- 60.Westermarck J., Li S.P., Kallunki T., Han J., Kahari V.M. Mol. Cell. Biol. 2001;21:2373–2383. doi: 10.1128/MCB.21.7.2373-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaefer A., Kosa F., Bittorf T., Magocsi M., Rosche A., Ramirez-Chavez Y., Marotzki S., Marquardt H. Cell. Signal. 2004;16:223–234. doi: 10.1016/j.cellsig.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 62.Santiago F.S., Lowe H.C., Day F.L., Chesterman C.N., Khachigian L.M. Am. J. Pathol. 1999;154:937–944. doi: 10.1016/S0002-9440(10)65341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Silverman E.S., Collins T. Am. J. Pathol. 1999;154:665–670. doi: 10.1016/S0002-9440(10)65312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grewal T., de Diego I., Kirchhoff M.F., Tebar F., Heeren J., Rinninger F., Enrich C. J. Biol. Chem. 2003;278:16478–16481. doi: 10.1074/jbc.C300085200. [DOI] [PubMed] [Google Scholar]
- 65.Katrantzis M., Baker M.S., Handley C.J., Lowther D.A. Free Radic. Biol. Med. 1991;10:101–109. doi: 10.1016/0891-5849(91)90003-l. [DOI] [PubMed] [Google Scholar]
- 66.Stocker R., Huang A., Jeranian E., Hou J.Y., Wu T.T., Thomas S.R., Keaney J.F., Jr. Arterioscler. Thromb. Vasc. Biol. 2004;24:2028–2033. doi: 10.1161/01.ATV.0000143388.20994.fa. [DOI] [PubMed] [Google Scholar]
- 67.Nuszkowski A., Grabner R., Marsche G., Unbehaun A., Malle E., Heller R. J. Biol. Chem. 2001;276:14212–14221. doi: 10.1074/jbc.M007659200. [DOI] [PubMed] [Google Scholar]
- 68.Heiss E.H., Schachner D., Werner E.R., Dirsch V.M. J. Biol. Chem. 2009;284:31579–31586. doi: 10.1074/jbc.M109.009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gotoh S., Nakamura T., Kataoka T., Taketani S. Gene. 2011;472:28–36. doi: 10.1016/j.gene.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 70.Park E., Quinn M.R., Wright C.E., Schuller-Levis G. J. Leukoc. Biol. 1993;54:119–124. doi: 10.1002/jlb.54.2.119. [DOI] [PubMed] [Google Scholar]
- 71.Kim C., Jang J.S., Cho M.R., Agarawal S.R., Cha Y.N. Int. Immunopharmacol. 2010;10:440–446. doi: 10.1016/j.intimp.2009.12.018. [DOI] [PubMed] [Google Scholar]
- 72.Liu X.M., Peyton K.J., Ensenat D., Wang H., Hannink M., Alam J., Durante W. Cardiovasc. Res. 2007;75:381–389. doi: 10.1016/j.cardiores.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.