Abstract
The de novo design of protein-binding peptides is challenging, because it requires identifying both a sequence and a backbone conformation favorable for binding. We used a computational strategy that iterates between structure and sequence optimization to redesign the C-terminal portion of the RGS14 GoLoco motif peptide so that it adopts a new conformation when bound to Gαi1. An X-ray crystal structure of the redesigned complex closely matches the computational model, with a backbone RMSD of 1.1 Å.
Computational protein design tests our understanding of protein energetics and allows the creation of new functional proteins1–3. In the area of protein interface design, computer-based methods have been used to stabilize protein-protein interactions, redesign protein-binding specificities and design new interactions from scratch4,5. In these studies, the designs were based on high-resolution structures and did not involve large perturbations to the backbone conformation of either interacting partner. However, in many naturally occurring interactions there are large conformational changes that accompany binding. Such changes particularly occur in peptide-protein interactions where the peptide usually populates an ensemble of conformations in the unbound state but adopts a single conformation when bound to the target protein. To design a new protein-binding peptide, it is necessary to identify both an amino acid sequence and backbone conformation that are favorable for binding.
A variety of approaches have been described for coupling structure and sequence optimization6–14. Here, we iterate between sequence design and structure refinement with the Rosetta molecular modeling program15,16 to design the bound conformation of a protein-binding peptide. Our model system is the GoLoco motif from the G-protein regulator and H-Ras effector17 RGS14, which binds to the heterotrimeric G-protein alpha subunit Gαi1 in its inactive, GDP-complexed state18. The 36-residue GoLoco peptide interacts with both the Ras-like and all-helical domains of Gαi1·GDP (Figure S1), burying 1900 Å2 of surface area (PDB ID 2OM2)19. In this study, we focus on the last 12 residues of the RGS14 GoLoco motif, which bind with an irregular secondary structure to a hydrophobic groove between the αA and αB helices of the Gαi1 all-helical domain (Figure 1). Removing these residues from the GoLoco motif weakens the binding affinity for Gαi1·GDP from 95 nM to ~20 μM (Figure S2c). We used our flexible-backbone design protocol to replace the last 12 residues of the RGS14 GoLoco motif peptide with a new 16-residue sequence designed to adopt an α-helix when bound to Gαi1.
Figure 1.
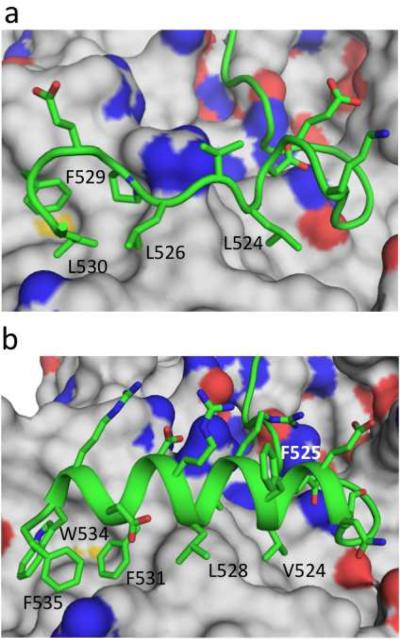
Redesigning the RGS14 GoLoco motif. (a) Crystal structure of the wildtype RGS14 GoLoco motif peptide bound to Gαi1. (b) Model of the redesigned GoLoco motif, GLhelix-4, bound to Gαi1. Both the wildtype GoLoco motif and GLhelix-4 are shown in cartoon representation with selected side chains displayed and labeled. Gαi1 is in surface representation with side-chain nitrogens colored blue and side-chain oxygens colored red.
First, the backbone conformation of the GoLoco motif C-terminus was rebuilt using a fragment assembly protocol with helical fragments from the Protein Data Bank20. A low-resolution score function was used to prevent steric overlap with Gαi1 and to favor the burial of hydrophobic residues15. The initial sequence of the helix was set to have a hydrophobic face in order to favor docking into the hydrophobic groove between the αA and αB helices of Gαi1. Independent fragment assembly trajectories were used to produce 2000 starting structures for high-resolution sequence design and backbone refinement. In all of the starting structures the new helix made contact with Gαi1, but the orientation and distance of the helix relative to Gαi1 varied between models (Figure S3).
Each of the starting structures was optimized with four rounds of sequence design followed by backbone and side chain minimization with a high-resolution score function that evaluates van der Waals contacts, hydrogen bonding, desolvation energies, backbone and side-chain torsion potentials and residue type-based reference energies10. Sequence optimization was performed using a simulated annealing protocol with a backbone dependent rotamer library21. Backbone and side-chain torsion angle minimization used a gradient-based, quasi-Newtonian method15. During this process the amino acid sequence of Gαi1 was held fixed, but side-chain and backbone torsion angles in the Gαi1 αA and αB helices were allowed to vary. In general, after four rounds of sequence design followed by backbone refinement, the energies of the models stopped decreasing, suggesting that most sequence/structure combinations were trapped in a local minima on the energy landscape.
The resulting models were filtered to remove designs that introduced unsatisfied, buried hydrogen-bonding partners. Of the remaining designs, those with the lowest Rosetta energy and the highest quality packing (SASApack score22) (Figure S4) were visually inspected. Several designs contained hydrophobic residues in positions that were primarily solvent-exposed. In most cases, these residues were replaced by performing fixed backbone design, only allowing polar amino acids. In some of the best scoring models, we also saw small cavities that could potentially be filled if neighboring GoLoco motif side chains were enlarged. However, when we forced a mutation to a larger amino acid, we introduced clashes that raised the overall energy of the model. This result suggested that the protein should be relaxed before evaluating the favorability of the mutation. Our standard protocol performs mutation and backbone relaxation as separate steps and does not easily allow for coupled changes that may be needed to move from small to large side chains. Previously, we developed a strategy for identifying affinity enhancing point mutations that cycles through every position at an interface and evaluates all possible point mutations19. Neighboring side-chain and backbone torsion angles are allowed to relax before evaluating the energy of the mutation. We used this protocol to scan our top scoring GoLoco motif designs for mutations that could further improve packing at the GoLoco motif-Gαi1 interface. In many cases no mutations were identified, but in our most successful design, GLhelix-4, an important mutation was identified, His 531 to Phe (Figure 1).
Four GoLoco motif peptide designs were selected for experimental characterization, GLhelix-1, GLhelix-2, GLhelix-3 and GLhelix-4 (Figure S2a, Table S1). To measure binding affinities for Gαi1·GDP, the GLhelix peptides were labeled with fluorescein, and fluorescence polarization was monitored as a function of Gαi1·GDP concentration. Interestingly, two of the designs, GLhelix-1 and GLhelix-2, bound more weakly to Gαi1·GDP than the truncated GoLoco motif peptide with its C-terminal residues removed (Figure S2b–c). This result suggests that these two peptides may be forming intra- or intermolecular interactions that compete with binding to Gαi1. Consistent with this interpretation, dye-labeled GLhelix-1 and GLhelix-2 have higher intrinsic fluorescence polarization values than GLhelix-3 and GLhelix-4. GLhelix-4 binds Gαi1·GDP more tightly (810 nM) than the truncated GoLoco motif (~20 μM), indicating that the designed residues are forming favorable interactions with Gαi1 (Figure 1b and Figure S2b–c).
To determine whether GLhelix-4 adopts the designed conformation when bound to Gαi1 we elucidated the crystal structure of the complex (PDB ID 2XNS, Table S2). Our structure was determined using diffraction data to a resolution of 3.4 Å (R factor = 22.3, Rfree = 24.4). The redesigned portion of the RGS14 GoLoco motif is well-defined in the electron density and clearly adopts an α-helical backbone (Figure 2). Strong electron density is observed for the aromatic side chains Phe 525, Phe 531 and Trp 534 on the designed helix. Except for the last two residues of the peptide, the computational design model closely matches the crystal structure (Figure 3). The backbone RMSD between peptide residues 520–533 of the designed model and the equivalent residues in the crystal structure is 1.1 Å when aligning the complexes with the helical domain of Gαi1. As designed, the side chains of Val 524 and Phe 531 pack on either side of Phe 108 from Gαi1.
Figure 2.

Crystal Structure of GLhelix-4 bound to Gai1·GDP. The unbiased 2Fo-Fc electron density (blue mesh, contoured to σ=1.5, see Supplementary Methods), indicates a GoLoco motif peptide C-terminal α-helix (salmon) bound to the all-helical domain of Gαi1 (gray surface). The well-defined aromatic side chains Phe 525, Phe 531, and Trp 534 establish the correct helical orientation and register.
Figure 3.

Model (light and dark green) and crystal structure (salmon and maroon) of GLhelix-4 bound to Gαi1. The structures were superimposed by minimizing the root mean square deviation between backbone heavy atoms in the all helical domain of Gαi1. The peptide side chains of Val 524 and Phe 531 pack on either side of Phe 108 from Gai1, as predicted. Only the orientation of Trp 534 and the absence of a definable residue 535 in the crystal structure deviate significantly from the predicted model.
The very C-terminus of the peptide is located close to a crystallographic symmetry axis, and thus, the corresponding electron density for Phe 535 overlaps with its symmetry-related counterpart. The deviation of the observed peptide structure from the computational model may be due in part to crystal-packing effects. In the crystal, the preceding Trp 534 packs against the neighboring GLhelix-4 peptide (Figure S5). Further evidence that Trp 534 is not making strong interactions with Gαi1 comes from mutagenesis studies. Mutating Trp 534 to an alanine has a modest effect on binding affinity (Kd = 1.4 μM compared to 810 nM). In contrast, mutating Phe 531 to an alanine weakens binding over 10-fold (Figure S6).
In conclusion, our results demonstrate that sequence optimization combined with backbone modeling can be used to rationally design the bound conformation of a protein-binding peptide. Such capabilities represent important steps towards the full de novo design of peptide-protein interactions. In this study, we designed an α-helical peptide to bind to a hydrophobic groove and therefore did not need to consider hydrogen bonds across the interface. One of the challenges for future designs is the creation of interfaces that are more polar in character and involve hydrogen-bond networks with both side chain and backbone groups from the peptide.
Supplementary Material
Acknowledgements
Work in the Kuhlman lab was funded by NIH grant R01 GM073960 and the W.M. Keck Foundation. Work in the Siderovski lab was funded by NIH grants R01 GM074268 (to D.P.S.) and T32 GM008719 (to D.E.B.). We are grateful for the resources of the SER-CAT 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory, IL.
Footnotes
Supporting Information Available: Supplementary figures, experimental procecdures, and details of the X-ray analysis.
References
- (1).Van der Sloot AM, Kiel C, Serrano L, Stricher F. Protein Eng Des Sel. 2009;22:537–42. doi: 10.1093/protein/gzp032. [DOI] [PubMed] [Google Scholar]
- (2).Kang SG, Saven JG. Curr Opin Chem Biol. 2007;11:329–34. doi: 10.1016/j.cbpa.2007.05.006. [DOI] [PubMed] [Google Scholar]
- (3).Lippow SM, Tidor B. Curr Opin Biotechnol. 2007;18:305–11. doi: 10.1016/j.copbio.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Karanicolas J, Kuhlman B. Curr Opin Struct Biol. 2009;19:458–63. doi: 10.1016/j.sbi.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mandell DJ, Kortemme T. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- (6).Fu X, Apgar JR, Keating AE. J Mol Biol. 2007;371:1099–117. doi: 10.1016/j.jmb.2007.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Humphris EL, Kortemme T. Structure. 2008;16:1777–88. doi: 10.1016/j.str.2008.09.012. [DOI] [PubMed] [Google Scholar]
- (8).Mandell DJ, Kortemme T. Curr Opin Biotechnol. 2009;20:420–8. doi: 10.1016/j.copbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- (9).Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. Science. 1998;282:1462–7. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- (10).Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- (11).Georgiev I, Donald BR. Bioinformatics. 2007;23:i185–94. doi: 10.1093/bioinformatics/btm197. [DOI] [PubMed] [Google Scholar]
- (12).Georgiev I, Keedy D, Richardson JS, Richardson DC, Donald BR. Bioinformatics. 2008;24:i196–204. doi: 10.1093/bioinformatics/btn169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Desjarlais JR, Handel TM. J Mol Biol. 1999;290:305–18. doi: 10.1006/jmbi.1999.2866. [DOI] [PubMed] [Google Scholar]
- (14).Su A, Mayo SL. Protein Sci. 1997;6:1701–7. doi: 10.1002/pro.5560060810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Rohl CA, Strauss CE, Misura KM, Baker D. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- (16).Das R, Baker D. Annu Rev Biochem. 2008;77:363–82. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- (17).Willard FS, Willard MD, Kimple AJ, Soundararajan M, Oestreich EA, Li X, Sowa NA, Kimple RJ, Doyle DA, Der CJ, Zylka MJ, Snider WD, Siderovski DP. PLoS One. 2009;4:e4884. doi: 10.1371/journal.pone.0004884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kimple RJ, Kimple ME, Betts L, Sondek J, Siderovski DP. Nature. 2002;416:878–81. doi: 10.1038/416878a. [DOI] [PubMed] [Google Scholar]
- (19).Sammond DW, Eletr ZM, Purbeck C, Kimple RJ, Siderovski DP, Kuhlman B. J Mol Biol. 2007;371:1392–404. doi: 10.1016/j.jmb.2007.05.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Simons KT, Kooperberg C, Huang E, Baker D. J Mol Biol. 1997;268:209–25. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- (21).Kuhlman B, Baker D. Proc Natl Acad Sci U S A. 2000;97:10383–8. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Sood VD, Baker D. J Mol Biol. 2006;357:917–27. doi: 10.1016/j.jmb.2006.01.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.