Abstract
Background & Aims
In approximately 70% of patients with hepatocellular carcinoma (HCC) treated by resection or ablation, disease recurs within 5 years. Although gene expression signatures have been associated with outcome, there is no method to predict recurrence based on combined clinical, pathology, and genomic data (from tumor and cirrhotic tissue). We evaluated gene expression signatures associated with outcome in a large cohort of patients with early-stage (BCLC 0/A), single-nodule HCC and heterogeneity of signatures within tumor tissues.
Methods
We assessed 287 HCC patients undergoing resection and tested genome-wide expression platforms using tumor (n=287) and adjacent non-tumor, cirrhotic tissue (n=226). We evaluated gene expression signatures with reported prognostic ability generated from tumor or cirrhotic tissue in 18 and 4 reports, respectively. In 15 additional patients, we profiled samples from the center and periphery of the tumor, to determine stability of signatures. Data analysis included Cox modeling and random survival forests to identify independent predictors of tumor recurrence.
Results
Gene expression signatures that were associated with aggressive HCC were clustered, as well as those associated with tumors of progenitor cell origin and those from non-tumor, adjacent, cirrhotic tissues. On multivariate analysis, the tumor-associated signature “G3-proliferation” (hazard ratio [HR]=1.75, P=0.003) and an adjacent “poor-survival” signature (HR=1.74, P=0.004) were independent predictors of HCC recurrence, along with satellites (HR=1.66, P=0.04). Samples from different sites in the same tumor nodule were reproducibly classified.
Conclusions
We developed a composite prognostic model for HCC recurrence, based on gene expression patterns in tumor and adjacent tissues. These signatures predict early and overall recurrence in patients with HCC, and complement findings from clinical and pathology analyses.
Keywords: liver cancer, relapse, prognosis, microarray
INTRODUCTION
Hepatocellular carcinoma (HCC) is among the top-five cancer killers worldwide. In the United States, during the 1990-2005 interval, death rate due to liver cancer increased both in males and females1. Roughly 30% of newly diagnosed patients will be eligible for potential curative therapies (e.g. liver transplantation, surgical resection and percutaneous ablation) in the West2. Liver resection, the curative therapy most frequently applied, provides 5-year survival rates of 70%. Ideal candidates are patients with single nodules, well preserved liver function, absence of portal hypertension, without symptoms or extra-hepatic spread3. In these patients, however, survival is often jeopardized by tumor recurrence.
Robust estimates suggest that close to 70% of patients will relapse within 5 years following surgery3. Typically, recurrence rate in HCC follows a 2 peak distribution: the first peak, usually within 2 years after resection, is mostly related to true metastatic spread (i.e., early recurrence); whereas the second peak mainly results from de novo tumors, as a consequence of the carcinogenic cirrhotic field (i.e., late recurrence)4. Vascular invasion (both macroscopic and microscopic) is the strongest predictor of recurrence although other variables, such as tumor size, number of nodules, alpha-fetoprotein (AFP) levels, degree of differentiation and satellites have also been found associated to recurrence3. Unfortunately, microvascular invasion and satellites can only be assessed with the full pathological specimen, a fact that reduces the odds for an accurate preoperative prediction of HCC recurrence. Besides cancer, another life-threatening condition (i.e., cirrhosis) is present in more than 80% of patients with HCC, what renders prognosis prediction a major challenge. Some clinical-based staging systems, specially the widely accepted Barcelona-Clinic Liver Cancer (BCLC) algorithm5, address both conditions, establishing a road-map for routine clinical decision-making. Nonetheless, it still lacks molecular information, which can complement the portrait of prognosis in complex solid neoplasms.
Genomic profiling has a great potential as diagnostic and prognostic tool in molecular medicine. Recent technological advances have even demonstrated its performance in formalin-fixed paraffin-embedded (FFPE) archived samples6, what will further increase its applicability. Gene signatures have allowed accurate prediction of prognosis and response to therapy in oncology7, 8, clearing the path for personalized cancer medicine9. Likewise, many studies have addressed prognosis prediction in HCC using array-based gene expression profiling, obtained from tumor or non-tumoral adjacent cirrhotic tissue. However, some of these signatures were frequently ill-defined, being generated in patients at different stages and with distinct etiologies for their underlying liver damage. Hence, the concordance of these signatures in a patient-by-patient basis is still unknown. In addition, it is of particular interest their prognosis performance in patients with early HCC, ideal candidates for liver resection.
Here, we simultaneously analyze 22 gene signatures with reported prognostic power in a cohort of 287 patients with early stage HCC (BCLC 0/A) treated by surgical resection. In this setting, we propose a composite genomic-based prognostic model incorporating molecular information from the tumor and the adjacent cirrhotic tissue. In addition, to ensure that a signature reflects the profile of a given tumor, we simultaneously tested samples from different sites of the same tumor nodule to evaluate the stability of genomic predictions.
METHODS
Patients and genomic profiling
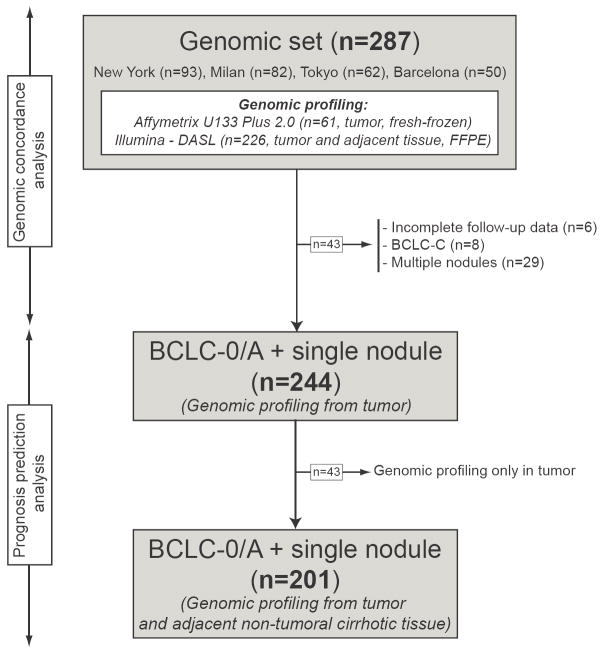
We included 287 HCC patients treated with surgical resection in four institutions, three from the HCC Genomic Consortium: Mount Sinai School of Medicine, New York (n=93), IRCCS Istituto Nazionale Tumori, Milan (n=82), Hospital Clinic, Barcelona (n=50); and the Toranomon Hospital in Tokyo (n=62). All study protocols were previously approved by their respective Institutional Review Boards6, 10. Sample processing, RNA extraction, and hybridization procedures are thoroughly described elsewhere6, 10. Profiling was conducted using 3 high-throughput genomic platforms: Affymetrix U133 Plus 2.0 (Gene Expression Omnibus (GEO) database ID GPL570), 6k Transcriptionally Informative Gene Panel for DASL (Illumina, GEO-GPL5474) and whole-genome DASL (Illumina, GEO-GPL8432). Gene expression data were already available in 123 tumors (i.e., 61 fresh-frozen and 62 FFPE tissues) and in 226 adjacent non-tumoral cirrhotic FFPE tissues. For the purpose of this study, we did additional DASL genomic profiling of 164 tumor samples. Microarray data of these newly profiled samples are available in GEO under accession number GSE20140. In summary, among the 287 patients included in this study, 244 patients had single nodules and genomic data from the tumor, among which 201 had gene expression data from the tumor and non-tumoral adjacent cirrhotic tissue (Figure 1).
Figure 1. Flow chart of the study.
We collected genomic information from 287 HCC patients treated with surgical resection in 4 different institutions. All patients had tumors profiled using genome-wide platforms, whereas in 226 there was also genomic data available from the non-tumoral adjacent cirrhotic tissue. For prognosis studies, we focused on those patients with single nodule, early stage (BCLC 0/A) HCC (see methods for details).
Table 1 summarizes the 22 gene signatures obtained from the tumor (18) or adjacent non-tumoral cirrhotic tissue (4), which reported prognostic value for either recurrence or survival in HCC patients undergoing surgical resection 11-14. We required that the genes included in the signature were clearly described in their respective reports. Predefined prognosis analysis was restricted to patients with early HCC, meaning stages 0 or A of the BCLC classification5. BCLC 0 included patients with a single tumor less than 2 cm without microvascular invasion or satellites. Additionally, since only the largest nodule was profiled, we excluded patients with multiple tumors to avoid the possibility that prognosis would be determined by tumor nodules not profiled (i.e., intra-individual genomic heterogeneity).
Table 1.
Gene signatures included in the study
| Author | Year | Signature Name (original description) | Signature names (MSIGBD§) | Patients with the signature (%)† | ||
|---|---|---|---|---|---|---|
| Signatures generated in tumor | 1 | Ye23 | 2003 | Signature of metastatic HCC | YE_LIVER_CANCER_INTRAHEPATIC_METS_UP | 14 (4.8) |
| 2 | Iizuka35 | 2003 | Signature of early intrahepatic recurrence | IIZUKA_LIVER_CANCER_EARLY_RECURRENCE_DN | NA* | |
| 3 | Lee24 | 2004 | Cluster A | LEE_LIVER_CANCER_POOR-SURVIVAL_UP, _DN | 91 (31.7) | |
| 4 | Kurokawa36 | 2004 | Signature of tumor recurrence | KUROKAWA_LIVER_CANCER_EARLY_RECURRENCE_UP, _DN | NA* | |
| 5 | Kaposi-Novak26 | 2006 | MET signature | NOVAK_LIVER_CANCER_MET_UP, _DN | 45 (15.6) | |
| 6 | Boyault12 | 2007 | G3 class | BOYAULT_LIVER_CANCER_SUBCLASS_G3_UP, _DN | 90 (31.3) | |
| 7 | Chiang10 | 2008 | Proliferation class | CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_UP, _DN | 112 (39) | |
| 8 | Villanueva | 2008 | CK19 human signature | VILLANUEVA_LIVER_CANCER_KRT19_UP, _DN | 98 (34.1) | |
| 9 | Minguez | 2008 | Vascular invasion signature | MINGUIZ_LIVER_CANCER_VASCULAR_INVASION_UP, _DN | 72 (25.1) | |
| 10 | Coulouarn25 | 2008 | Late TGFB | COULOUARN_LIVER_CANCER_TGF_BETA_LATE_VS_EARLY_UP, _DN | 70 (24.3) | |
| 11 | Woo29 | 2008 | Signature of recurrence | WOO_LIVER_CANCER_RECURRENCE_UP, _DN | 84 (29.2) | |
| 12 | Yamashita28 | 2008 | EpCAM signature | YAMASHITA_LIVER_CANCER_EPCAM_UP, _DN | 35 (12.1) | |
| 13 | Cairo14 | 2008 | Hepatoblastoma_C2 class | CAIRO_LIVER_CANCER_HEPATOBLAST_C2_POOR-PROGNOSIS_UP, _DN | 20 (6.9) | |
| 14 | Yoshioka37 | 2009 | Signature of early recurrence | YOSHIOKA_LIVER_CANCER_EARLY_RECURRENCE_UP, _DN | NA* | |
| 15 | Hoshida27 | 2009 | Class S1 (WNT-TGFB) | HOSHIDA_LIVER_CANCER_META_ANALYSIS_SUBCLASS | 80 (27.8) | |
| 16 | Hoshida27 | 2009 | Class S2 (AKT-MYC) | HOSHIDA_LIVER_CANCER_META_ANALYSIS_SUBCLASS | 44 (15.3) | |
| 17 | Andersen11 | 2010 | CK19 rat signature | ANDERSEN_LIVER_CANCER_KRT19_UP, _DN | 95 (33.1) | |
| 18 | Woo38 | 2010 | Cholangiocarcinoma-like signature | WOO_LIVER_CANCER_CHOLANGIOCA_LIKE_UP, _DN | 115 (40.1) | |
| Signatures generated in adjacent non-tumor tissue | 19 | Hoshida6 | 2008 | Poor survival signature | HOSHIDA_LIVER_CANCER_POOR-SURVIVAL_UP, _DN | 72 (31.8)ø |
| 20 | Hoshida6 | 2008 | Late recurrence signature | HOSHIDA_LIVER_CANCER_LATE_RECURRENCE_UP, _DN | 89 (39.3)ø | |
| 21 | Budhu13 | 2006 | Metastasis inclined microenvironment | BUDHU_LIVER_CANCER_METASTASIS_UP, _DN | NA* | |
| 22 | Okamato39 | 2006 | Signature of multicentric HCC | OKAMOTO_LIVER_CANCER_MULTICENTRIC_OCCURRENCE_UP, _DN | NA* | |
Patients with a predicted poor prognosis signature according to each of the gene signatures evaluated (FDR<0.05, see methods for details)
Nearest-template prediction method failed to classify any sample with the signature, based on a FDR<0,05
Prediction done on 226 patients (tissue from adjacent cirrhotic tissue available)
CK19 gene signature
All these 22 signatures have been previously reported, except for our CK19 human signature that was also considered for the analysis. This signature was generated by supervised analysis of the gene expression data of 77 fresh frozen HCC samples profiled with Affymetrix U133 Plus 2.010. We considered cytokeratin-19 (CK19) staining status as a surrogate marker of progenitor cell origin15. Immunohistochemistry was performed on FFPE slides using a predefined protocol16 with a 1:50 dilution of CK19 antibody (Dako North America, Carpinteria, CA). As previously reported, samples were considered positive when at least 5% of tumor cells were stained for CK1915. Validation of the performance of the signature to predict CK19 positive staining was evaluated in a different set of 34 FFPE HCC samples profiled with DASL-Illumina and stained for CK19 using the same protocol.
Genomic heterogeneity assessment
In order to assess whether a given genomic signature is consistent throughout different sites of a tumor, we analyzed in a separate cohort of 15 HCC patients (not included in the prognosis study) treated with surgical resection. In these tumors, genomic profiling of fresh frozen tissue was conducted at the center and periphery of the nodule. Minimum size of the tumors tested was of 4 cm in diameter, and tissue was collected with a distance of at least 2 cm between center and periphery samples, being the periphery sample at less than 1 cm from the tumor-cirrhosis interface. Sample processing, RNA extraction, and hybridization procedures are thoroughly described elsewhere6, 10. Profiling of these samples was conducted using Illumina HT-12 arrays.
Statistical analysis
Study design followed general recommendations included in the REMARK statement for reporting tumor marker prognostic studies17. All signatures included, except for our CK19 human and the vascular invasion signature (manuscript under review) were already reported, being deposited in the Molecular Signature Database (www.broadinstitute.org/gsea/msigdb18, Table 1), and predictions were made using nearest-template prediction method (NTP, extensively reviewed19) as implemented in the NTP module of the Gene Pattern software. Briefly, a template of poor prognosis was assigned a value of 1 or -1 to over- or under-expressed genes in patients with poor prognosis. Proximity of the expression pattern of the signature genes to the template was calculated using cosine distance. If the distance was smaller, a prediction of “poor prognosis” was assigned to the sample. Significance for the prediction was assessed by computing a nominal p-value estimated based on a null distribution for the distance to the templates generated by randomly resampling the same number of genes from the microarray dataset for the sample 1000 times. False discovery rate (FDR) was used to correct for multiple hypothesis20. Samples predicted as having “poor prognosis” with FDR<0.05 were compared to the rest of the samples in the following analysis. The prediction analysis was performed separately for each dataset/platform. Genes in each signature were converted into gene symbols provided by NCBI’s Entrez Gene database before subjected to NTP. Signature genes mapped onto the test datasets used in this study are summarized in Supplementary Table 1.
Robustness of the gene-expression signature-based prediction was evaluated as follows. For each dataset, a subset of the samples (90%) was randomly resampled 100 times, and the prediction was performed for each subset. The G3 signature from the tumor and the poor-survival signature from adjacent non-tumor liver tissues were assessed with the prediction confidence of FDR<0.05. For each sample, proportion of poor-prognosis prediction was calculated and compared with original prediction made in entire dataset.
Correlation among predictions was calculated with the Fisher Exact Test, using FDR correction for multiple testing20. Two-way contingency tables and Cramer’s V statistic were computed to quantify the strength of the association between signatures as previously described21. Cramer’s V statistic values range from 0 to 1, indicating no relation or identical, respectively. Values between 0.36 and 0.49 indicate substantial correlation and values higher than 0.50 indicate strong correlation.
Kaplan-Meier plots and Cox regression were used to assess association with outcome of each poor-outcome prediction and clinical variable. The primary end-point of the study is overall recurrence, but early recurrence – defined as that occurring within the first 2 years upon resection4 – was also assessed. Samples from the Japanese cohort (n=62) were previously used to train signatures identifying survival and late recurrence (i.e., poor-survival and late-recurrence signatures from the adjacent tissue6). Hence, survival was only a secondary end-point in our study, and the late-recurrence signature was not included in the prognosis prediction analysis. Several clinical variables reported as predictors of tumor recurrence in different studies were also assessed in the multivariable analysis (Supplementary table 2): sample origin, gender, age, etiology of liver disease, tumor size, serum albumin, serum bilirubin, platelet count, serum AFP, vascular invasion, satellites, degree of differentiation and BCLC stage. Those variables (clinical or genomic) showing a p-value less than 0.05 in the univariate Cox regression analysis were separately evaluated in a multivariate Cox model aimed to identify independent predictors of early and overall recurrence. Missing values were less than 5% for each variable included in the analysis. We performed a different approach to identify variables associated with early/overall recurrence by using random survival forests (RSF). RSF is an extension of random forests (RF) developed by Ishwaran et al. for right-censored data22, able to improve ensemble learning by injecting randomization into the base learning process. RSF provides ‘variable importance’ (VIMP) values for all candidate predictors of early/overall recurrence. Higher VIMP values indicate variables with predictive ability whereas zero or negative values identify non-predictive variables to be filtered. A forest of 4000 trees was grown using ‘logrankscore’ splitting, and VIMP for each variable was recorded. The analysis was repeated 100 times independently and VIMP averaged over the runs. Analyses were performed using the GenePattern analytical toolkit (www.broad.mit.edu/cancer/software/genepattern/), the R statistical package (www.r-project.org) and the SPSS software® (version 16).
RESULTS
Genomic landscape of early hepatocellular carcinoma
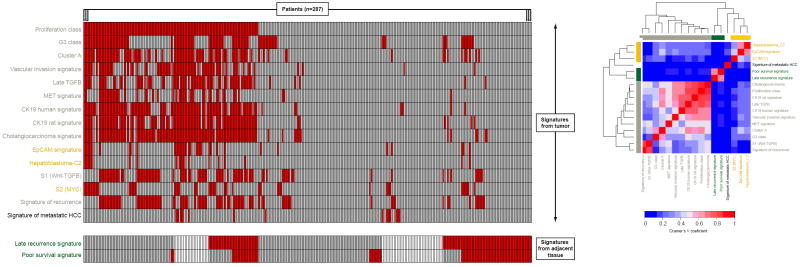
Among the 22 signatures evaluated, 17 (i.e., 15 from the tumor and 2 form the adjacent non-tumoral cirrhotic tissue) were able to confidently allocate patients (FDR<0.05) within their predicted poor outcome subclass, as opposed to the remaining 5 signatures (Table 1). Figure 2 summarizes the prediction obtained for each of the 287 patients. The proliferation signature10 was the most prevalent prediction in our dataset (39%, 112/287), whereas the signature of metastatic HCC23 was identified only in 4.8% (14/287) of patients. We then sought to evaluate the prediction similarity of these 17 signatures. Pair-wise comparisons showed a significant overlap between most poor-outcome signatures derived from the tumor (P<0.001), except for the metastatic HCC signature23. Signature clustering based on Cramer’s V coefficients indicated substantial relation among 3 groups of signatures (Figure 2): (1) signatures identifying biological processes related to increased cell proliferation, progression in cell cycle and activation of specific pathways, such as proliferation10, G312, cluster A24, vascular invasion signature, late TGFB25, MET signature26, S127; (2) those generated in the adjacent tissue, such as poor-survival and late recurrence signatures6 and (3) those suggesting a probable progenitor cell tumor origin, (e.g., hepatoblastoma_C214, EpCAM28 and S227). Interestingly, 25% (72/287) of tumor samples harbored concurrently 6 or more poor-outcome signatures. When etiologic differences were considered, patients with hepatitis B related liver damage were significantly enriched in the proliferation10 (32/110 vs 27/172, P=0.01) and CK19 human (30/97 vs 29/185, P=0.003) signatures.
Figure 2. Concordance of poor-outcome signatures in the whole set (n=287).
Figure includes only those signatures that confidently identified patients within their respective poor-outcome class (FDR<0.05). Left panel: Each column represents prediction for each patient. Red bars mean positive prediction for the signature, gray bars mean absence of prediction, and white bars that there was no genomic data available for that sample. Signatures are organized according to the type of tissue where they were generated and evaluated (tumor in the top and non-tumoral cirrhotic adjacent in the bottom). Right panel: Heatmap of Cramer’s V coefficients corresponding to signature pair wise comparisons. Signatures are clustered according to their degree of overlap (signatures generated from adjacent tissue are shown in green).
Interestingly, poor-outcome signatures from the tumor were not associated with those obtained from non-tumor adjacent tissue on a patient-by-patient basis (Figure 2). Therefore, it seemed that tumor and cirrhotic profiling could capture complementary biological signals essential in prognosis prediction. By contrast, we also identified a subgroup of patients (19%, 43/226) lacking any of the predicted poor-outcome signatures, neither from the tumor nor from the cirrhotic adjacent tissue. As will be discussed below, this ‘genomically indolent’ subgroup held some features compatible with a less aggressive disease.
Combined genomic profiling from tumor (G3) and adjacent tissue (poor-survival) predict recurrence in single-nodule early-stage hepatocellular carcinoma
Predefined prognosis analysis was restricted to patients at early stages (BCLC 0 or A) with single nodules. Table 1 summarizes the clinical data of the 244 patients included in this analysis. Most patients were cirrhotic males (73.3%), with compensated disease and well-preserved liver function (Child-Pugh class A: 97%), and hepatitis C-related liver disease (62%). Vascular invasion and satellites were present in 35.3% and 14.9% of patients, respectively, and 26.7% had high AFP levels (>100 mg/dL).
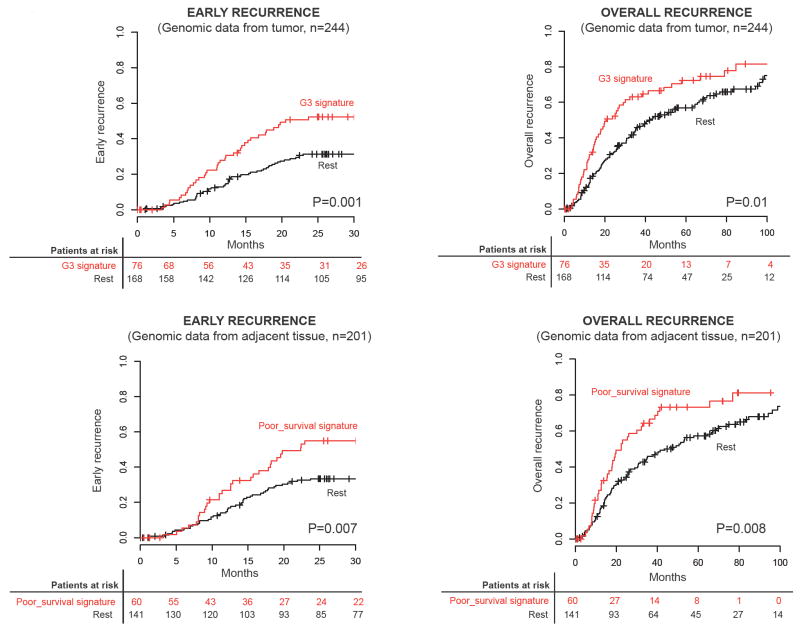
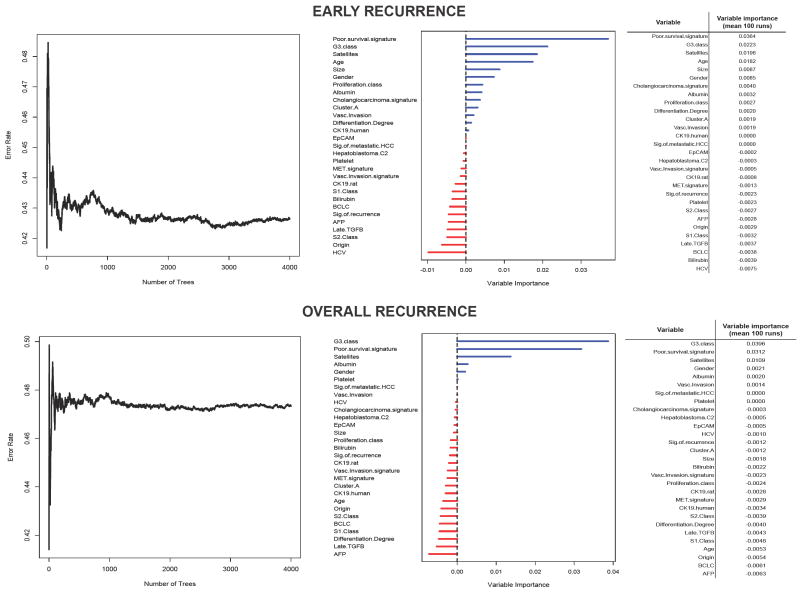
Univariate Cox analysis identified tumor size, vascular invasion, satellites, G312 signature (from tumor, Figure 3) and poor-survival6 signature (from adjacent tissue, Figure 3) as significantly associated with early recurrence, whereas only the latter three were significantly correlated with overall recurrence (Table 3). In multivariate analysis, satellites (HR: 1.80, 95% CI: 1.02-3.18, P=0.04), G312 (HR: 1.71, 95% CI: 1.04-2.81, P=0.03) and poor-survival6 (HR: 1.92, 95% CI: 1.20-3.06, P=0.006) signatures were the only independent predictors of early tumor recurrence, as well as overall recurrence (Table 3). Similar results were obtained using random survival forest. For early recurrence, variables top-ranked after 100 runs were poor-survival signature, G3 signature, satellites and age, whereas for overall recurrence were G3 signature, poor-survival signature and satellites (Fig 4). There were 17 patients (8.5%) with concomitant prediction of the signatures with best prediction performance (i.e., G312 from tumor and poor-survival6 from adjacent tissue). When compared to those lacking both signatures, these patients had HR for early and overall recurrence of 4 and 3.2, respectively. Although due to study design survival was not considered as a primary endpoint for genomic analysis, when non-genomic variables were tested, tumor size, serum albumin, vascular invasion and BCLC stage were significantly associated with overall survival.
Figure 3. Kaplan-Meier estimates of early and overall recurrence according to G3 (tumor) and poor-survival (adjacent non-tumoral cirrhotic tissue) signature status.
Top panels show results of patients with genomic data available from the tumor (n=244), whereas lower panels show results of those with genomic information from tumor and adjacent tissue (n=201). P values were obtained from the log-rank test. (+) denotes observations that were censored owing to loss of follow-up or the date the last contact.
Table 3.
Univariate and multivariate analysis of clinical variables and gene signatures for overall (A) and early (B) recurrence.
| A | OVERALL RECURRENCE
|
|||||||
|---|---|---|---|---|---|---|---|---|
| UNIVARIATE ANALYSIS | MULTIVARIATE ANALYSIS | |||||||
| Variables | n | P-value | HR | 95% CI | n | P-value | HR | 95% CI |
| Origin (Japan) | 244 | 0.766 | 1.05 | 0.75-1.49 | ||||
| Gender (Male) | 244 | 0.520 | 1.13 | 0.78-1.62 | ||||
| Age (>60) | 241 | 0.556 | 0.91 | 0.66-1.25 | ||||
| Etiology (HCV) | 242 | 0.240 | 1.22 | 0.88-1.70 | ||||
| Size (>2 cm) | 244 | 0.471 | 1.14 | 0.80-1.63 | ||||
| Albumin (<3.5 g/L) | 237 | 0.090 | 1.49 | 0.94-2.37 | ||||
| Bilirubin (>1 mg/dL) | 240 | 0.341 | 0.85 | 0.61-1.19 | ||||
| Platelet count (<100,000/mm3) | 240 | 0.423 | 1.17 | 0.80-1.72 | ||||
| AFP (>100 mg/dL) | 234 | 0.330 | 0.830 | 0.57-1.20 | ||||
| Vascular invasion | 239 | 0.321 | 1.18 | 0.85-1.65 | ||||
| Satellites | 241 | 0.016 | 1.70 | 1.10-2.62 | 198 | 0.041 | 1.66 | 1.02-2.71 |
| Degree of differentiation (moderately or poor) | 241 | 0.548 | 1.12 | 0.77-1.62 | ||||
| BCLC stage (A) | 244 | 0.509 | 1.15 | 0.76-1.76 | ||||
| Signature of metastatic HCC | 244 | 0.383 | 0.64 | 0.24-1.74 | ||||
| G3 signature | 244 | 0.011 | 1.55 | 1.11-2.16 | 198 | 0.003 | 1.75 | 1.20-2.53 |
| Proliferation class | 244 | 0.665 | 1.07 | 0.78-1.49 | ||||
| CK19 signature | 244 | 0.971 | 1.01 | 0.72-1.41 | ||||
| Ck19 rat signature | 244 | 0.828 | 1.04 | 0.74-1.47 | ||||
| Vascular invasion signature | 244 | 0.921 | 0.98 | 0.66-1.45 | ||||
| Late TGFB | 244 | 0.517 | 0.88 | 0.60-1.29 | ||||
| Cluster A | 244 | 0.496 | 1.12 | 0.80-1.58 | ||||
| MET signature | 244 | 0.891 | 1.03 | 0.66-1.61 | ||||
| HCC recurrence signature | 244 | 0.490 | 0.88 | 0.62-1.26 | ||||
| EPCAM signature | 244 | 0.506 | 0.83 | 0.49-1.42 | ||||
| Hepatoblastoma C2 | 244 | 0.661 | 0.87 | 0.47-1.61 | ||||
| Class S1 | 244 | 0.883 | 1.03 | 0.72-1.47 | ||||
| Class S2 | 244 | 0.791 | 1.06 | 0.69-1.63 | ||||
| Colangiocarcinoma signature | 244 | 0.485 | 0.88 | 0.64-1.23 | ||||
| Poor survival signature* | 201 | 0.007 | 1.67 | 1.15-2.44 | 198 | 0.004 | 1.74 | 1.19-2.55 |
|
| ||||||||
| B |
EARLY RECURRENCE
|
|||||||
| UNIVARIATE ANALYSIS | MULTIVARIATE ANALYSIS | |||||||
|
| ||||||||
| Variables | n | P-value | HR | 95% CI | n | P-value | HR | 95% CI |
|
| ||||||||
| Origin (Japan) | 244 | 0.325 | 1.26 | 0.80-1.98 | ||||
| Gender (Male) | 244 | 0.522 | 1.17 | 0.72-1.92 | ||||
| Age (>60) | 241 | 0.412 | 0.84 | 0.54-1.28 | ||||
| Etiology (HCV) | 242 | 0.809 | 0.95 | 0.62-1.46 | ||||
| Size (>2 cm) | 244 | 0.037 | 1.78 | 1.03-3.06 | 193 | 0.109 | 1.62 | 0.89-2.92 |
| Albumin (<3.5 g/L) | 237 | 0.085 | 1.65 | 0.93-2.94 | ||||
| Bilirubin (>1 mg/dL) | 240 | 0.595 | 1.13 | 0.73-1.75 | ||||
| Platelet count (<100,000/mm3) | 240 | 0.939 | 1.02 | 0.61-1.72 | ||||
| AFP (>100 mg/dL) | 234 | 0.607 | 1.13 | 0.70-1.81 | ||||
| Vascular invasion | 239 | 0.039 | 1.57 | 1.02-2.40 | 193 | 0.950 | 1.01 | 0.59-1.73 |
| Satellites | 241 | 0.012 | 1.91 | 1.15-3.18 | 193 | 0.041 | 1.80 | 1.02-3.18 |
| Degree of differentiation (moderately or poor) | 241 | 0.059 | 1.71 | 0.98-2.99 | ||||
| BCLC stage (A) | 244 | 0.24 | 1.46 | 0.78-2.27 | ||||
| Signature of metastatic HCC | 244 | 0.572 | 0.67 | 0.16-2.71 | ||||
| G3 signature | 244 | 0.001 | 1.99 | 1.30-3.05 | 193 | 0.033 | 1.71 | 1.04-2.81 |
| Proliferation class | 244 | 0.073 | 1.47 | 0.96-2.25 | ||||
| CK19 signature | 244 | 0.135 | 1.39 | 0.90-2.14 | ||||
| CK19 rat signature | 244 | 0.242 | 1.30 | 0.84-2.04 | ||||
| Vascular invasion signature | 244 | 0.38 | 1.25 | 0.76-2.04 | ||||
| Late TGFB | 244 | 0.193 | 1.37 | 0.85-2.19 | ||||
| Cluster A | 244 | 0.142 | 1.39 | 0.90-2.14 | ||||
| MET signature | 244 | 0.865 | 0.95 | 0.52-1.74 | ||||
| HCC recurrence signature | 244 | 0.534 | 0.86 | 0.52-1.40 | ||||
| EPCAM signature | 244 | 0.247 | 0.61 | 0.27-1.41 | ||||
| Hepatoblastoma C2 | 244 | 0.889 | 0.94 | 0.41-2.16 | ||||
| Class S1 | 244 | 0.84 | 1.05 | 0.65-1.69 | ||||
| Class S2 | 244 | 0.347 | 1.30 | 0.75-2.23 | ||||
| Colangiocarcinoma signature | 244 | 0.441 | 1.18 | 0.77-1.81 | ||||
| Poor survival signature* | 201 | 0.007 | 1.88 | 1.18-2.98 | 193 | 0.006 | 1.92 | 1.20-3.06 |
Signature generated in adjacent non-tumoral cirrhotic tissue
Figure 4. Random survival forests for early (top panel) and overall (lower panel) recurrence.
Left figures show the error rate according to the number of trees generated and right panel shows VIMP values for each variable evaluated. Adjacent tables show the average VIMP values for each variable after 100 runs.
We finally evaluated the clinical characteristics of those patients lacking any of the 22 predicted poor-outcome signatures (19.4%, 39/201), the ‘genomically indolent’ subgroup. These patients presented well-differentiated tumors (21/45 vs 16/154, P<0.001), less vascular invasion (6/67 vs 32/129, P=0.007) and lower rates of early recurrence (P=0.03).
Genomic stability of tumor genomic prediction
Internal stability of the tumor G3 and adjacent poor-survival signature predictions were assessed using bootstrap prediction analysis. We observed highly stable predictions for both signatures since more than 90% of samples constantly received prediction of poor prognosis in more than 90% of the bootstrap trials (Supplementary Figure 1).
Since evaluation of vascular invasion and satellites is currently not feasible prior surgery, we consider the scenario where genomic profiling of the tumor and adjacent tissue would be obtained preoperatively from fine-needle biopsies. We previously demonstrated the feasibility of conducting whole-genome expression study form this limited amount of biological material6. Accordingly, stability of the gene signature predictions at different tumor sites is of utmost importance when considering clinical deployment of genomic-based prognostic tools. Hence, we profiled 15 additional tumors obtaining tissue from two separate nodule locations: center and periphery. Assuming certain degree of intratumoral molecular heterogeneity, we hypothesize that in early stage HCC, this would not significantly impact genomic predictions, especially for those gene signatures harboring a significant number of genes. Among those 15 tumors, signature predictions were identical in the center and periphery of the tumors in more than 80% of cases for all signatures except for the signature of recurrence29, which had a concordance rate of 73% (11/15).
Limited prognostic impact of tumor lineage in early hepatocellular carcinoma
Previous studies have suggested that HCCs with supposedly progenitor cell origin had significant worse prognosis28, 30. To evaluate this hypothesis, we first generated a CK19 signature using tumor CK19 staining as a surrogate marker of progenitor cell origin15. Only 11.6% (9/77) of samples were positively stained for CK19. Most (8/9, 88.8%) belonged to the proliferation subclass of our reported molecular classification10. We validate the performance of the signature to predict CK19 staining in a different set of 34 FFPE HCC samples. Even though all four CK19 stained samples were identified by the signature, it was overestimating staining frequency (4/16 vs 0/18, P=0.04). When applied to our study group, 34% (98/287) of tumors showed confident prediction for the CK19 human signature. As expected, these samples were enriched in the proliferation subclass (P<0.001). This human-based CK19 signature was highly correlated with the recently reported CK19 signature generated in a rat model of HCC11 (Cramer V coefficient 0.7, P<0.001), and was also significantly associated with the other two progenitor-derived signatures, EpCAM28 and Hepatoblastoma-C214 (P<0.001). Despite the low number of patients predicted in these two signatures; 12.1% (35/278) for EpCAM and 6.9% (20/278) for Hepatoblastoma-C2, they also showed a high degree of overlap (Cramer’s V statistic 0.65, P<0.001).
Regarding clinical and pathological correlations, CK19-human and CK19-rat signatures were associated with vascular invasion (P=0.003 and P=0.001, respectively) and poorly differentiated tumors (P<0.001 both). Similarly, EpCAM and Hepatoblastoma-C2 signatures were also overrepresented in patients with vascular invasion (P<0.001) and poorly differentiated tumors (P=0.04), respectively. Although preliminary data suggested that CK19-human signature was associated with early recurrence, it was not confirmed in this set of BCLC0/A single-nodule HCCs. Likewise, neither EpCAM nor Hepatoblastoma-C2 signatures show a significant role as prognosis predictors in surgically resected single-nodule early HCC. A subgroup analysis was done to analyze their performance in patients mimicking the scenario where these signatures were generated (i.e., more advanced disease: single tumors larger than 3 cm with vascular invasion, n=59). All these signatures from supposedly derived progenitor-derived tumors were able to significantly predict early tumor recurrence in this subgroup: CK19-human (P=0.01), CK19-rat (P=0.01), EpCAM (P=0.003) and Hepatoblasoma-C2 (P<0.001), along with AFP levels (P=0.04).
DISCUSSION
Accurate prognosis prediction is crucial in modern oncology. In HCC, clinical-based staging algorithms (e.g., BCLC) provide a useful framework for decision-making31. This study introduces an integrated prognostic model combining genomic and clinico-pathological data to improve outcome prediction in patients with single-nodule early HCC. In addition, we show that genomic profiles of both the tumor and the adjacent tissue are complementary in refining the prognosis of individuals undergoing surgical resection for a liver cancer. However, the results also indicate that at early stages genomic signatures reflecting potential progenitor cell origin of the tumor are not associated to worse outcome, as stated in previous publications. These novel data can improve our understanding of the pathogenesis of the disease, potentially allowing to refine clinical staging systems and model trial design in terms of stratification of patients prior randomization.
Prediction of recurrence in patients undergoing resection represents a clinical challenge. The dual mode of recurrence depends on both tumor aggressiveness and the liver carcinogenic field. Tumor aggressiveness has been defined by clinical (e.g., size, AFP levels) and pathological data (e.g., vascular invasion, satellites). Conversely, the risk of developing de novo tumors has been more difficult to predict. During the last decade several gene signatures have been reported in HCC reflecting pathway activation, molecular subclasses or outcome prediction. The number and heterogeneity of the signatures reported has created certain confusion in the field in understanding their prognostic relevance. In addition, as opposed to most solid tumors, some of the prognostic signatures have been obtained from adjacent, non-tumoral tissues. Thus, the current study primarily aimed to unravel the prognostic weight of the reported gene signatures in HCC along with clinical variables in a large cohort of early single-nodule HCCs. For that purpose we also confirmed that gene signature prediction was stable when evaluated at distant sites of the same nodule. In our pilot study conducted with 15 solitary tumors beyond 4 cm, the same prediction for the tumor signatures evaluated, either present or absent, was obtained in the center and periphery of the tumor in more than 80% of cases for most signatures. Sufficient prediction consistency at distant tumor portions is a prerequisite for considering preoperative profiling using tumor tissue fine-needle biopsies, specially in small tumors.
We then assessed whether the signatures reflected different biological processes, and thus had distinct molecular significance. Three subgroups of signatures clustered apart: one subgroup defining proliferation traits, another subgroup defining a potential progenitor cell origin and a third reflecting the non-tumoral at-risk carcinogenic field. Among the first group agglutinating most of the already reported poor-outcome signatures derived from the tumor (Figure 1), the G3 signature12 showed the strongest association to tumor recurrence, mostly by detecting risk of early recurrence. Despite the close association between these signatures, the number of predicted patients in our dataset varied among them. This could be due to the differences in the biological background of the samples used to generate each one (e.g., underlying liver disease, stage), not completely reproduced in our dataset. This should also be taken into account when evaluating the prognostic performance of the G3 signature, meaning that a different signature clustered within this ‘proliferation trait’ group could have a better prognostic prediction in a different dataset. Even though G3 signature was originally enriched in TP53 mutations12, when we integrated TP53 mutation information status (n=8110), we only find a non-significant trend towards enriched TP53 mutations in tumors with this signature (data not shown).
A striking result of the study was the lack of prognostic power of signatures allegedly identifying a progenitor cell origin: EpCAM28, Hepatoblasoma-C214, CK19-rat11 and the CK19-human signature generated ad hoc. Their prognostic performance in our study was suboptimal and none was independently associated with either recurrence or survival, as opposed to what has been reported so far. Unlike this study, the performance of these signatures has been previously tested in cohorts of patients at more advanced stages. It remains unknown whether specific signatures better define different evolutionary stages of the tumor progression. We can hypothesize that their prognostic accuracy might improve in advanced tumors where these signatures were originally generated. Nonetheless, our results in early stages challenge the concept of poor outcome related to cell of origin.
The third group of signatures reflected molecular aberrations in the non-tumoral environment (so called ‘field effect’6), which govern liver dysfunction and the risk of HCC development and dissemination6, 13. The relevance of the ‘field effect’ is crucial and, hence, it should be incorporated in any genomic prognostic algorithm. We have reported a gene signature from the adjacent tissue able to accurately predict survival in patients with HCC treated with surgical resection6. This poor-survival signature also predicted liver decompensation, risk of HCC development and death in a cohort of cirrhotic patients followed by a median period of 10 years (manuscript under review). Taken as a whole, these data suggest that the signature captures most biological events conferring aggressive behavior, both in terms of progression of liver dysfunction and facilitating tumor development or dissemination. In the present study, the poor-prognosis signature was an independent predictor of recurrence, and did not correlate with the tumoral proliferative G3, indicating that they might capture different events relevant to disease progression.
A population of tumors lacks any of the reported poor-outcome signatures from the tumor or the adjacent tissue. This ‘genomicaly indolent’ subgroup showed clinical and pathological data compatible with a less aggressive disease (e.g., lower AFP levels, well-differentiated tumors, less vascular invasion), and decreased early recurrence rates. The prognostic significance of such silent group of tumors needs further exploration because current approach failed to capture a clear impact in overall recurrence or survival. There is a medical need to define HCC subclasses with benign outcome, for instance in the setting of liver transplant, where numerous centers are trying to expand the worldwide-accepted Milan criteria for HCC candidates for transplantation based on clinical parameters (e.g., tumor size, number of nodules), obtaining suboptimal results32.
The implications of the current research are two-fold. First, the study presents a unified approach to demonstrate the significance and complementary nature of genomic profiling in outcome prediction in HCC using tumoral (G3-proliferation signature) and non-tumoral tissues (poor-prognosis signature). The fact that these signatures retain independent prognostic significance tested along with clinical and pathological variables (e.g., satellites) reflects advancement in the understanding of the biological events relevant for outcome prediction33. Second, the composite model presented should be confirmed in prospective evaluations since it might have importance in molecular classification of tumors and stratification of patients within clinical investigations testing chemopreventive strategies. Sorafenib, the first systemic therapy that showed unequivocal benefits in patients with advanced HCC34, is currently under evaluation in this setting in a large phase III clinical trial. It is expected that other molecular therapies will soon enter the field. Pre-surgical assessment of recurrence risk using genomic profiling in such early tumors should become instrumental in future trials. It will allow risk stratification, personalized surveillance, and eventually customized neo or adjuvant interventions in patients with early HCC candidates for resection.
Supplementary Material
Table 2.
Clinical data of patients included in prognosis analysis (n=244).
| Variable | New York (%) | Milan (%) | Tokyo (%) | Barcelona (%) | Total (%) |
|---|---|---|---|---|---|
| n | 76 (31.1) | 64 (26.2) | 62 (25.4) | 42 (17.1) | 244 (100.0) |
| Age (>60) | 37 (48.6) | 58 (90.6) | 21 (35.6) | 34 (80.1) | 150 (62.4) |
| Gender (male) | 61 (81.2) | 38 (59.3) | 46 (74.2) | 34 (80.1) | 179 (73.3) |
| Child-Pugh (Class A) | 76 (100.0) | 64 (100) | 56 (90.3) | 38 (95.1) | 226 (96.6) |
| Etiology | |||||
| HCV | 32 (42.1) | 45 (70.3) | 44 (79.9) | 29 (72.5) | 150 (61.9) |
| HBV | 32 (42.1) | 8 (12.5) | 13 (20.9) | 3 (7.5) | 56 (23.1) |
| Alcohol | 0 (0) | 3 (4.6) | 0 (0) | 5 (12.5) | 8 (3.3) |
| Other | 12 (15.7) | 8 (12.5) | 5 (8.0) | 3 (7.5) | 28 (11.6) |
| Tumor size | |||||
| <2 cm | 13 (17.1) | 15 (23.4) | 26 (41.9) | 7 (16.6) | 61 (25) |
| 2-3 cm | 14 (18.4) | 18 (28.1) | 22 (35.4) | 14 (33.3) | 68 (27.9) |
| >3 cm | 49 (64.4) | 31 (48.4) | 14 (22.5) | 21 (50) | 115 (47.1) |
| Degree of differentiation | |||||
| Well | 20 (26.3) | 17 (26.7) | 10 (16.1) | 13 (33.3) | 60 (24.9) |
| Moderate | 44 (57.8) | 33 (51.7) | 40 (64.5) | 23 (58.9) | 140 (58.1) |
| Poor | 12 (15.8) | 14 (21.9) | 12 (19.3) | 3 (7.6) | 41 (17) |
| Satellites | |||||
| Absent | 62 (82.7) | 51 (79.6) | 59 (95.2) | 25 (56.5) | 205 (85.1) |
| Present | 13 (17.3) | 13 (20.3) | 3 (4.8) | 12 (32.4) | 36 (14.9) |
| Vascular invasion | |||||
| Absent | 39 (51.3) | 45 (70.3) | 45 (72.5) | 25 (67.5) | 154 (64.4) |
| Microscopic | 37 (48.7) | 19 (29.7) | 17 (27.4) | 12 (32.4) | 85 (35.5) |
| BCLC stage | |||||
| 0 | 2 (2.6) | 10 (15.6) | 22 (35.4) | 5 (11.9) | 39 (16) |
| A | 74 (97.4) | 54 (84.3) | 40 (64.5) | 37 (88.1) | 205 (84) |
| Albumin (mg/dL) | |||||
| <3.5 | 7 (9.4) | 7 (10.9) | 13 (20.9) | 2 (5.4) | 29 (12.2) |
| Bilirrubin (mg/dL) | |||||
| >1 | 14 (18.9) | 24 (37.5) | 35 (56.4) | 12 (30) | 85 (35.4%) |
| Platelet count (×103/ mm3) | |||||
| <100,000 | 8 (10.6) | 15 (23.4) | 20 (32.2) | 6 (15.4) | 49 (20.4) |
| Alfa-fetoprotein (mg/dL) | |||||
| >100 | 26 (35.6) | 10 (15.8) | 20 (32.2) | 7 (18.4) | 63 (26.7) |
| Events | |||||
|
| |||||
| Follow-up (months)* | 47 (24-69.8) | 56 (32.9-70.2) | 99 (56.0-119.2) | 50 (21.1-71.9) | 58 (29.2-82.5) |
| Deaths | 31 (40.7) | 22 (34.4) | 23 (37.1) | 6 (14.2) | 82 (33.6) |
| Overall recurrence | 45 (59.2) | 36 (56.2) | 49 (79.0) | 25 (59.2) | 155 (63.2) |
| Early recurrence (<2 years) | 26 (34.2) | 20 (31.2) | 27 (43.5) | 14 (33.3) | 87 (35.6) |
Data expressed as median (range Q25-Q75)
Acknowledgments
Grant Support: AV is a recipient of a Sheila Sherlock fellowship (European Association for the Study of the Liver). CA is supported by a grant from Instituto de Salud Carlos III (ISCIII/FIS FI09/00605). HC is supported by a grant from Spanish National Health Institute (SAF-2007-61898). AL is supported by a grant from the U.S. National Institutes of Health (A.L.: 5R01CA121941). JB is supported by a grant from Instituto de Salud Carlos III (ISCIII/FIS PI 05-0150). JML is supported by grants from the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (J.M.L: 1R01DK076986-01), European Comission-FP7 Framework (HEPTROMIC, Proposal No: 259744), The Samuel Waxman Cancer Research Foundation and the Spanish National Health Institute (J.M.L: SAF-2007-61898 & SAF-2010-16055). The study was supported by the Landon Foundation - American Association for Cancer Research Innovator Award for International Collaboration in Cancer Research.
Abbreviations
- AFP
alpha-fetoprotein
- BCLC
Barcelona-Clinic Liver Cancer
- FDR
false discovery rate
- FFPE
Formalin-fixed paraffin-embedded
- HCC
hepatocellular carcinoma
- HR
hazard ratio
Footnotes
- Conception and design of the study: AV, JML.
- Generation, collection, assembly, analysis and/or interpretation of data: AV, YH, CB, VT, DS, CA, HC, AL, MK, HK, ST, JB, PN, SR, VM, MS, JML.
- Drafting or revision of the manuscript: AV, YH, AL, HC, VT, CA, JB, VM, JML.
- Approval of the final version of the manuscript: AV, YH, CB, VT, DS, CA, HC, AL, MK, HK, ST, JB, PN, SR, VM, MS, JML.
Financial disclosures: There are no financial disclosures relevant to this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–17. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Schwartz M, Mazzaferro V. Resection and liver transplantation for hepatocellular carcinoma. Semin Liver Dis. 2005;25:181–200. doi: 10.1055/s-2005-871198. [DOI] [PubMed] [Google Scholar]
- 4.Imamura H, Matsuyama Y, Tanaka E, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–7. doi: 10.1016/s0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Di Bisceglie AM, Bruix J, et al. Design and endpoints of clinical trials in hepatocellular carcinoma. J Natl Cancer Inst. 2008;100:698–711. doi: 10.1093/jnci/djn134. [DOI] [PubMed] [Google Scholar]
- 6.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faivre S, Raymond E, Boucher E, et al. Safety and efficacy of sunitinib in patients with advanced hepatocellular carcinoma: an open-label, multicentre, phase II study. Lancet Oncol. 2009;10:794–800. doi: 10.1016/S1470-2045(09)70171-8. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann WK, de Vos S, Elashoff D, et al. Relation between resistance of Philadelphia-chromosome-positive acute lymphoblastic leukaemia to the tyrosine kinase inhibitor STI571 and gene-expression profiles: a gene-expression study. Lancet. 2002;359:481–6. doi: 10.1016/S0140-6736(02)07678-X. [DOI] [PubMed] [Google Scholar]
- 9.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–70. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 10.Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–88. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen JB, Loi R, Perra A, et al. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology. 2009 doi: 10.1002/hep.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 13.Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Cairo S, Armengol C, De Reyniès A, et al. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–84. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Komuta M, Spee B, Vander Borght S, et al. Clinicopathological study on cholangiolocellular carcinoma suggesting hepatic progenitor cell origin. Hepatology. 2008;47:1544–56. doi: 10.1002/hep.22238. [DOI] [PubMed] [Google Scholar]
- 16.Villanueva A, Chiang DY, Newell P, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–83. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; 1983:e1–11. [Google Scholar]
- 17.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97:1180–4. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoshida Y. Nearest template prediction: a single-sample-based flexible class prediction with confidence assessment. PLoS ONE. 2010;5:e15543. doi: 10.1371/journal.pone.0015543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 21.Fan C, Oh DS, Wessels L, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–9. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 22.Ishwaran H, Kogalur UB. Consistency of Random Survival Forests. Stat Probab Lett. 2010;80:1056–1064. doi: 10.1016/j.spl.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye QH, Qin LX, Forgues M, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–23. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 24.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40:667–76. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 25.Coulouarn C, Factor VM, Thorgeirsson SS. Transforming growth factor-beta gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–67. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, et al. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–95. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoshida Y, Nijman SM, Kobayashi M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–92. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–61. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 29.Woo HG, Park ES, Cheon JH, et al. Gene expression-based recurrence prediction of hepatitis B virus-related human hepatocellular carcinoma. Clin Cancer Res. 2008;14:2056–64. doi: 10.1158/1078-0432.CCR-07-1473. [DOI] [PubMed] [Google Scholar]
- 30.Lee JS, Heo J, Libbrecht L, et al. Nat Med. 2006;12:410–6. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 31.Marrero JA, Fontana RJ, Barrat A, et al. Prognosis of hepatocellular carcinoma: comparison of 7 staging systems in an American cohort. Hepatology. 2005;41:707–16. doi: 10.1002/hep.20636. [DOI] [PubMed] [Google Scholar]
- 32.Mazzaferro V, Llovet JM, Miceli R, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10:35–43. doi: 10.1016/S1470-2045(08)70284-5. [DOI] [PubMed] [Google Scholar]
- 33.Villanueva A, Hoshida Y, Toffanin S, et al. New strategies in hepatocellular carcinoma: genomic prognostic markers. Clin Cancer Res. 2010;16:4688–94. doi: 10.1158/1078-0432.CCR-09-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–90. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 35.Iizuka N, Oka M, Yamada-Okabe H, et al. Oligonucleotide microarray for prediction of early intrahepatic recurrence of hepatocellular carcinoma after curative resection. Lancet. 2003;361:923–9. doi: 10.1016/S0140-6736(03)12775-4. [DOI] [PubMed] [Google Scholar]
- 36.Kurokawa Y, Matoba R, Takemasa I, et al. Molecular-based prediction of early recurrence in hepatocellular carcinoma. J Hepatol. 2004;41:284–91. doi: 10.1016/j.jhep.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 37.Yoshioka S, Takemasa I, Nagano H, et al. Molecular prediction of early recurrence after resection of hepatocellular carcinoma. Eur J Cancer. 2009;45:881–9. doi: 10.1016/j.ejca.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Woo HG, Lee JH, Yoon JH, et al. Identification of a cholangiocarcinoma-like gene expression trait in hepatocellular carcinoma. Cancer Res. 2010;70:3034–41. doi: 10.1158/0008-5472.CAN-09-2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okamoto M, Utsunomiya T, Wakiyama S, et al. Specific gene-expression profiles of noncancerous liver tissue predict the risk for multicentric occurrence of hepatocellular carcinoma in hepatitis C virus-positive patients. Ann Surg Oncol. 2006;13:947–54. doi: 10.1245/ASO.2006.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.