This study shows that, in addition to its known role in protein degradation, the anaphase-promoting complex also regulates transcription of a cell cycle gene, Cyclin B1, and that this regulation, which is mediated by microRNA, is important for pollen development.
Abstract
The anaphase-promoting complex/cyclosome (APC/C), an essential ubiquitin protein ligase, regulates mitotic progression and exit by enhancing degradation of cell cycle regulatory proteins, such as CYCB1;1, whose transcripts are upregulated by DUO POLLEN1 (DUO1). DUO1 is required for cell division in male gametophytes and is a target of microRNA 159 (miR159) in Arabidopsis thaliana. Whether APC/C is required for DUO1-dependent CYCB1;1 regulation is unknown. Mutants in both APC8 and APC13 had pleiotrophic phenotypes resembling those of mutants affecting microRNA biogenesis. We show that these apc/c mutants had reduced miR159 levels and increased DUO1 and CYCB1;1 transcript levels and that APC/C is required to recruit RNA polymerase II to MIR159 promoters. Thus, in addition to its role in degrading CYCB1;1, APC/C stimulates production of miR159, which downregulates DUO1 expression, leading to reduced CYCB1;1 transcription. Both MIR159 and APC8–yellow fluorescent protein accumulated in unicellular microspores and bicellular pollen but decreased in tricellular pollen, suggesting that spatial and temporal regulation of miR159 by APC/C ensures mitotic progression. Consistent with this, the percentage of mature pollen with no or single sperm-like cells increased in apc/c mutants and plants overexpressing APC8 partially mimicked the duo1 phenotype. Thus, APC/C is an integrator that regulates both microRNA-mediated transcriptional regulation of CYCB1;1 and degradation of CYCB1;1.
INTRODUCTION
The gametes of flowering plants are formed within haploid gametophytes (McCormick, 2004). In the male gametophyte, each haploid microspore divides asymmetrically to produce a larger vegetative cell that will form the pollen tube and a smaller generative cell, which divides symmetrically to form twin sperm cells. These sperm cells are delivered to the embryo sac via the pollen tube, where they fuse with an egg and a central cell to produce embryo and endosperm, respectively. The molecular mechanisms underlying the production of male gametes remain largely unknown, although a few required genes have been identified (Okada et al., 2005; Rotman et al., 2005; Iwakawa et al., 2006; Mori et al., 2006; Nowack et al., 2006; Kim et al., 2008; Brownfield et al., 2009b; Gusti et al., 2009; Ron et al., 2010). Among these genes, DUO POLLEN1 (DUO1) encodes a male germ cell–specific R2R3 Myb protein (Rotman et al., 2005) that is required for expression of the Arabidopsis thaliana G2/M regulator CyclinB1;1 (CYCB1;1) in the male germline (Brownfield et al., 2009a), suggesting an integrative role for DUO1 in cell specification and cell cycle progression that is necessary for twin sperm cell production. DUO1 mRNA is targeted directly by microRNA159 (miR159) (Palatnik et al., 2007), which leads to its degradation.
Cell proliferation in all eukaryotes depends on the ubiquitin ligase (E3) activity of the anaphase-promoting complex/cyclosome (APC/C) (Peters, 2006). The APC/C in Arabidopsis contains at least 11 subunits (Capron et al., 2003a). Among them, APC3, APC5, APC6, APC7, and APC8 each contain a tetratricopeptide repeat (TPR) protein–protein interaction domain and are expected to function as receptors that interact with regulatory proteins (Lima et al., 2010). Without APC/C, cells cannot separate their sister chromatids in anaphase, cannot exit from mitosis to divide into two daughter cells, and cannot initiate the steps necessary for DNA replication in S phase (Peters, 2006). The APC/C was initially found to be involved in targeted proteolysis of A- and B-type cyclins, thus facilitating exit from mitosis (Glotzer et al., 1991). The genetic inactivation of APC/C has caused lethality in all species in which it has been investigated so far (Yu et al., 1998; Yamashita et al., 1999; Cullen et al., 2000; Bentley et al., 2002; Garbe et al., 2004; Pál et al., 2007; Jin et al., 2010). For example, in Caenorhabditis elegans, an APC8 mutant, mat-3, is a temperature-sensitive embryonic lethal due to defects in germline meiosis and mitosis (Garbe et al., 2004).
In Arabidopsis, single-copy genes encode counterparts of all known vertebrate APC/C subunits, except for APC3/CDC27, which has two genes (Capron et al., 2003a). Plant APC/C is involved in cell cycle regulation. For example, APC2 and APC6 are important for female gametophyte development (Capron et al., 2003b; Kwee and Sundaresan, 2003), as plants with loss-of-function alleles of either of these genes exhibited female gametophytic lethality due to cell cycle arrest at an early stage of embryo sac development. Reciprocal crosses with the apc6/nomega mutant suggested that most defects came from the female, although male transmission was somewhat reduced (Kwee and Sundaresan, 2003). Interestingly, a mir159ab double mutant (Allen et al., 2007) phenocopied the apc13 mutant (Saze and Kakutani, 2007) and the apc6 and apc10 RNA interference lines (Marrocco et al., 2009) that had pleiotropic morphological defects, including small siliques, curled leaves, and abnormal phylotaxy, suggesting that both APC/C and miR159 might be required for similar developmental processes.
To identify new genes in miRNA biogenesis and function pathways, we performed a forward genetic screen. One mutant had pleiotrophic developmental phenotypes expected for mutants in miRNA pathway genes, such as DICER-LIKE1 (DCL1) (Jacobsen et al., 1999), but surprisingly it was a point mutation in APC8. This point mutant, apc8-1, resulted in reduced MIR159 transcription and lessened accumulation of mature miR159. This caused an increase in transcripts of both DUO1 and CYCB1;1 in pollen. We further showed that APC/C is required for RNA polymerase II (Pol II) recruitment to promoters of MIR genes. CYCB1;1–green fluorescent protein (GFP) accumulated in both the vegetative cell and sperm cells of tricellular pollen in apc8-1 but was absent in wild-type tricellular pollen. We also observed an increased incidence of unicellular and bicellular pollen in mature anthers of apc8-1 and increased ratio of bicellular pollen in mature pollen by overexpressing APC8 in male germlines, suggesting that disrupted balance of CYCB1;1 levels prevented efficient transition of male gametophytes through the bicellular and tricellular stages. Similar results were obtained by analyzing a T-DNA insertion allele of APC13, another component of the APC/C. Therefore, we propose a dual role for the APC/C both in coordinating miRNA-mediated transcriptional regulation of CYCB1;1 and in direct protein degradation of CYCB1;1.
RESULTS
Isolation and Characterization of a Weak APC8 Allele
DCL1 plays a critical role in miRNA biogenesis in plants (Jacobsen et al., 1999), and seeds from dcl1 null alleles cannot be obtained due to embryo lethality. However, a weak allele of dcl1, dcl1-14 (in which the T-DNA insertion is located in the promoter region, SALK_056243; see Supplemental Figure 1A online), showed curly leaves, abnormal reproductive development, and reduced miRNA levels (see Supplemental Figures 1B and 1C online) but is fertile. We therefore performed a genetic screen after ethyl methanesulfonate mutagenesis of dcl1-14, aimed at the isolation of mutants with enhanced or suppressed phenotypes. We isolated a mutant with much more severe developmental phenotypes than those of dcl1-14. The pleiotropic phenotypes included distorted leaf shapes, abnormal shoot meristem development, and a delay in the transition from the vegetative to the reproductive stage (Figures 1A and 1B). Moreover, the mutant had severely abnormal flower and silique development, including bushy inflorescences and shorter siliques (Figures 1C and 1D). We showed that these phenotypes were dcl1-14 independent by backcrossing to wild-type plants. Therefore, the mutant without dcl1-14 background was used for our subsequent analyses. Collectively, these pleiotropic developmental defects suggested that the mutated gene had roles at multiple stages of plant development.
Figure 1.
Phenotypes and Complementation of the apc8-1 Mutant.
(A) A representative 5-week-old seedling of apc8-1.
(B) A representative 8-week-old apc8-1 plant.
(C) Close-up of fasciculate siliques in apc8-1.
(D) A representative 12-week-old apc8-1 plant.
(E) Complementation of apc8-1. A representative apc8-1 plant carrying the pAPC8:APC8-YFP transgene.
(F) Partial amino acid sequences of APC8 in various species. Identical residues are white on black background, and conserved residues are white on gray background. Asterisk indicates the Asp (D309) that was mutated to Asn in apc8-1. At, Arabidopsis thaliana; Pt, Populus trichocarpa; Os, Oryza sativa (japonica); Zm, Zea mays; Hs, Homo sapiens; Mm, Mus musculus; Xl, Xenopus laevis; Dm, Drosophila melanogaster; Sp, Schizosaccharomyces pombe.
Bars = 1 cm in (A), (B), (D), and (E), and 1 mm in (C).
[See online article for color version of this figure.]
To further understand the basis of these heritable pleiotropic phenotypes, we examined their inheritance in F2 progeny from a cross of a mutant plant (Columbia [Col]) to a wild-type Landsberg erecta plant. The genotype was determined for 661 F2 plants with obvious mutant phenotypes, which comprised ~10% of the F2 population. Characterization of Col/Landsberg erecta polymorphisms throughout the genome showed that all plants with the mutant phenotype were homozygous for the Col haplotype at one region on chromosome 3, suggesting that a disrupted gene in this region was responsible for the mutant phenotype. This region was narrowed down to an interval between BAC clones F18E5 and T8O5 (five recombinants and two recombinants, respectively, out of 1322 chromosomes examined). We sequenced the genes in this region and found a point mutation (G to A) in the coding region of APC8. This mutation resulted in the conversion of a highly conserved Asp (Asp-309) to an Asn at a position adjacent to the TPR domain (Figure 1F). To confirm that this mutation was responsible for the phenotype, a genomic fragment was fused to yellow fluorescent protein (YFP) and introduced into the mutant. The morphological phenotypes were fully rescued in 23 of 25 plants (Figure 1E). Therefore, we concluded that the phenotypes were due to the mutation in APC8 and designated this allele apc8-1.
Arabidopsis APC8 Can Complement an APC8 Mutant in Schizosaccaromyces pombe
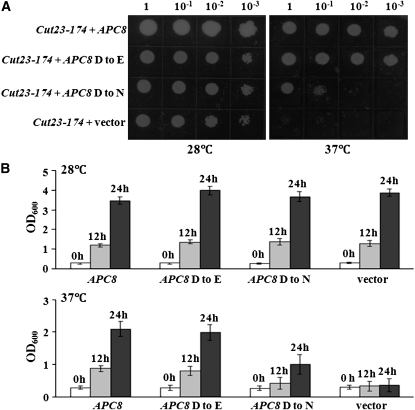
To determine whether Arabidopsis APC8 is a functional ortholog of APC8 found in other eukaryotes, we tested whether Arabidopsis APC8 was able to functionally complement an APC8 mutant in fission yeast. We introduced full-length cDNAs of both APC8 and mutant apc8-1 (APC8 D to N) into a yeast expression vector under the control of the Gal promoter. These constructs were then transformed into the temperature-sensitive cut23-174 mutant, which has a lesion in the APC8 gene in S. pombe. This mutant can grow at 28°C but not at 37°C (Yamashita et al., 1999). Expression of wild-type APC8 rescued the yeast mutant strain at the restrictive temperature, whereas an empty vector or the mutant apc8-1 did not (Figures 2A and 2B). Moreover, a construct in which the Asp was changed to Glu (APC8 D to E) only partially complemented the yeast mutant at 37°C (Figures 2A and 2B), which reinforces the idea that the conserved Asp is important for APC8 function.
Figure 2.
Arabidopsis APC8 Complements the Fission Yeast APC8 Mutant cut23-174.
(A) Growth at the permissive (28°C) or restrictive (37°C) temperature of the cut23-174 yeast strain carrying an empty vector (pYPGE15), APC8 (pYPGE15-APC8), APC8 D to E (pYPGE15-APC8, D was mutated to E), or APC8 D to N (pYPGE15-APC8, cDNA from the apc8-1 mutant).
(B) Quantitative analysis of growth (OD600 measurements) at the permissive (28°C) or restrictive (37°C) temperature of the cut23-174 yeast strain carrying each plasmid as in (A) at 0, 12, and 24 h growth in liquid YEA media. Three colonies for each construct were analyzed, and all measurements represent the average of three biological replicates with error bars representing the se.
Subcellular Localization of APC8 Is Regulated during Male Gametophyte Development
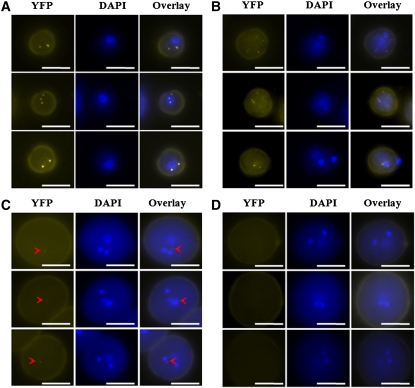
Microarray analyses indicated that APC/C genes were ubiquitously expressed in Arabidopsis tissues (Zimmermann et al., 2004). To validate the expression pattern of APC8, we performed RT-PCR analyses using cDNA samples from different Arabidopsis tissues. APC8 expression was detected in a variety of vegetative and reproductive tissues (see Supplemental Figure 2A online). To substantiate the RT-PCR analyses, we constructed different promoter-reporter constructs. The β-glucuronidase (GUS) reporter was expressed in almost all tissues examined, including leaves, shoot apical meristems, inflorescences, and siliques (see Supplemental Figure 2B online). During male gametophyte development, the GFP reporter was detectable at the unicellular and bicellular stages but appeared less intense at the tricellular stage (see Supplemental Figure 2C online). We therefore examined the subcellular localization of a pAPC8:APC8-YFP fusion protein during male gametophyte development. This protein fusion construct complemented the apc8-1 phenotypes completely (Figure 1E), showing that YFP-tagged APC8 was functional. Foci of APC8-YFP were visible in the nuclei and cytoplasm of unicellular microspores (Figure 3A) and bicellular pollen (Figure 3B), but only a very faint signal was detected in tricellular pollen (Figure 3C), and no signal was detectable in mature pollen (Figure 3D). Moreover, the foci of APC8-YFP varied in different microspores and pollen grains (Figure 3).
Figure 3.
Subcellular Localization of APC8 during Male Gametophyte Development.
Representative fluorescence microscopy images of unicellular pollen (A), bicellular pollen (B), tricellular pollen (C), and mature pollen (D) from at least 10 individual T1 apc8-1 plants harboring the pAPC8:APC8-YFP construct. Three examples for each developmental stage were shown. The left panel for each represents YFP epifluorescence of pAPC8:APC8-YFP. The middle panel for each indicates the developmental stage determined by DAPI staining. The right panel for each shows an overlay of the YFP and DAPI epifluorescence signals. Arrowheads show faint YFP signals. Bars = 10 μm.
[See online article for color version of this figure.]
APC/C Is Required for Efficient Male Transmission
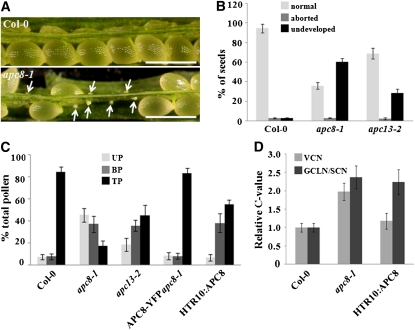
Microscopy analysis showed that the siliques in apc8-1 had many (~66%) apparently undeveloped ovules (Figure 4A), which led to obviously reduced seed set (Figure 4B). The mutant was backcrossed to the wild type, and the F1 was allowed to self-pollinate. Only ~10% of the F2 progeny had the mutant phenotypes, instead of the expected 25%. These results suggested that the mutant had transmission problems. Reciprocal crosses with wild-type plants showed that transmission through the female was mildly affected but was strongly perturbed through the male (Table 1). To rule out the possibility that disturbed male transmission was specific to APC8, we analyzed the T-DNA insertion allele of another APC/C component, APC13 (SALK_027397, here named apc13-2), which was reported to have a bonsai phenotype (Saze and Kakutani, 2007) similar to that of apc8-1. We found slightly reduced seed set in apc13-2 (Figure 4B), and reciprocal crosses with wild-type plants indicated that male transmission was more affected than female transmission (Table 1).
Figure 4.
Seed Set Analysis and Pollen Development of apc8-1.
(A) Dissected mature siliques of wild-type (Col-0, top) and apc8-1 plants (bottom). Undeveloped ovules are indicated with arrows. The undeveloped ovules are tiny and white, whereas developing seeds are large and green. Bar = 500 μm.
(B) Percentage of normal seeds (light gray), aborted seeds (dark gray), and undeveloped ovules (black) from self-pollinated Col-0, apc8-1, and apc13-2 plants. Error bars represent sd from the mean of 10 siliques from each of 20 individual plants.
(C) Distribution of uninucleate microspores (UP; light gray), bicellular pollen (BP; dark gray), and tricellular pollen (TP; black) in mature anthers. Pollen was stained with DAPI, and the percentage of each stage was determined for Col-0, apc8-1, APC8-YFP apc8-1, and HTR10:APC8 (line 8-2). All measurements represent the average of three biological replicates with error bars representing the se. For each independent replicate for each genotype, 600 to 800 pollen grains from 5 to 10 plants were analyzed.
(D) DNA content of vegetative cell nuclei (VCN; light gray), generative cell-like nuclei/sperm cell nuclei (GCLN/SCN; dark gray) in pollen from different genotypes. The relative C values of apc8-1 (n = 108) and HTR10:APC8 (n = 126 from line 8-2) were calculated from DAPI fluorescence values normalized to the mean fluorescence of wild-type cells (n = 98 for vegetative cell nuclei; n = 112 for sperm cell nuclei). n indicates the number of pollen grains that was measured. All measurements represent the average of three biological replicates, with error bars representing the se.
[See online article for color version of this figure.]
Table 1.
Genetic Analysis of apc8-1 and apc13-2
| Parental Genotypes | Genotype of F1 Plants |
Transmission Efficiency | |
| ♀ × ♂ | apc/+ | Wild Type | |
| apc8-1/+ × wild type | 136 | 172 | 136/172 × 100% = 79.1% |
| Wild type × apc8-1/+ | 61 | 238 | 61/238 × 100% = 25.2% |
| apc13-2/+ × wild type | 158 | 187 | 158/187 × 100% = 84.5% |
| Wild type × apc13-2/+ | 109 | 261 | 109/261 × 100% = 41.8% |
Because decreased APC/C function resulted in a reduced seed set phenotype, we next tested whether increased APC/C function had an effect on seed set. We generated transgenic plants overexpressing APC8 under the control of the HISTONE THREE RELATED 10 (HTR10) promoter. HTR10 is specifically expressed in the male germ line, and HTR10 accumulates in generative cell nuclei and subsequently in sperm cell nuclei (Ingouff et al., 2007). We obtained 24 independent T1 lines; T2 plants from five T1 lines with single copy insertions were used for further analyses. The T-DNA cassette carrying APC8 also contained a pollen-specific marker (LAT52-GFP), which we used to confirm homozygous plants. We measured the relative expression levels of APC8 RNA in these plants by quantitative RT-PCR (qPCR) and compared the expression levels with those in wild-type plants (see Supplemental Figure 3A online). We also scored the seed set (see Supplemental Figure 3B online). All T2 progenies tested showed higher (3.4- to 15.2-fold) APC8 expression levels (see Supplemental Figure 3A online). These plants also had, on average, 27% reduced seed set (see Supplemental Figure 3B online), indicating that the higher APC8 levels in the male germline disrupted seed set.
APC/C Is Necessary for Tricellular Pollen Formation
Given the importance of cell cycle regulation by APC/C (Peters, 2006) and the dynamic expression of APC8 during male gametophyte development (Figure 3), we suspected that disrupting APC/C function might lead to disorganized cell divisions during male gametophyte development. Indeed, when mature pollen from apc8-1 plants was stained with 4',6-diamidino-2-phenylindole (DAPI), the proportion of uninucleate cells was 45% and binucleate cells was 37%, whereas in wild-type plants, these proportions were 8 and 9%, respectively (Figures 4C). Introduction of wild-type APC8 into apc8-1 completely rescued these defects of male gametophyte development (Figure 4C), confirming that apc8-1 was responsible for these phenotypes. The defects in pollen maturation in apc13-2 showed a similar trend but were less severe than those in apc8-1 (Figure 4C). A representative HTR10:APC8 transgenic T2 plant (line 8-2) showed increased incidence of bicellular pollen, but no change in the incidence of unicellular pollen (Figure 4C), consistent with the specific expression of HTR10 in the male germline.
If the vegetative cell and the generative cell fail to divide in apc/c mutants and in APC8 overexpression lines, the expectation might be that further DNA replication would not occur. We therefore investigated DNA content in the vegetative cell nuclei and the generative cell-like nuclei in apc8-1 and in the generative cell-like nuclei in an HTR10:APC8 plant (line 8-2). Relative to the nuclear DNA content in the wild type, nuclear DNA content in both the vegetative cell nuclei and the generative cell-like nuclei in apc8-1 was significantly increased, to nearly 2C (Figure 4D), suggesting that in apc8-1, pollen containing only a vegetative cell nucleus and pollen containing a single sperm cell both finish DNA replication but cannot enter the subsequent mitosis. As expected, the nuclear DNA content in the generative cell-like nuclei in HTR10:APC8-expressing plants increased, also to nearly 2C (Figure 4D). To examine whether pollen cell fate was disturbed, we introduced pHTR10:HTR10-mRFP (Ingouff et al., 2007) into apc8-1. This marker was observed in sperm cell nuclei of apc8-1 mature pollen (see Supplemental Figure 3C online), indicating that sperm cells in apc8-1 had not lost their gametic fate. Taken together, our results indicate that APC/C plays an important role in mitotic cell cycle progression during male gametophyte development, promoting tricellular pollen formation.
APC/C Is Required for miR159 Accumulation, and APC/C Promotes Pol II Recruitment to MIR159
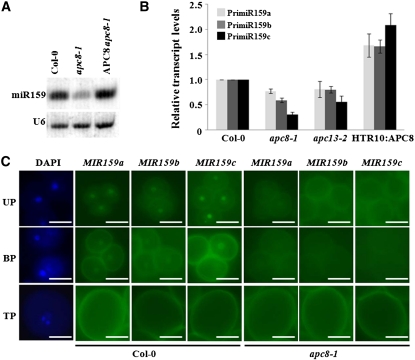
Because apc8-1 was identified while looking for miRNA accumulation mutants, we hoped to determine the molecular mechanism by which APC8 regulates male gametophyte development by examining whether apc8-1 was compromised in miRNA accumulation. Of 10 tested miRNAs, miR159 accumulation was obviously reduced (Figure 5A; see Supplemental Figure 4A online), and this molecular phenotype was rescued by wild-type APC8 (Figure 5A), suggesting that APC8 was involved in the miR159 biogenesis pathway. We also found obvious reductions in accumulation of miR166, miR172, miR173, and miR319, but no detectable changes for miR156, miR164, miR167, miR168, and miR171 (see Supplemental Figure 4A online), suggesting that the role of APC8 in miRNA biogenesis is locus specific. Similar defects in mature miR159 accumulation were observed in apc13-2 (see Supplemental Figure 4A online). miR159 is encoded by three genes (MIR159a, MIR159b, and MIR159c) (Park et al., 2002; Rhoades et al., 2002).
Figure 5.
APC8 Is Required for miR159 Biogenesis during Male Gametophyte Development.
(A) miR159 accumulation in Col-0, apc8-1, and APC8 apc8-1 (the pAPC8:APC8-YFP construct was introduced into apc8-1). Total RNAs were extracted from inflorescences. U6 was the loading control.
(B) Expression of MIR159a (light gray), MIR159b (dark gray), and MIR159c (black) in pollen from Col-0, apc8-1, apc13-2, and HTR10:APC8 (line 8-2). UBIQUITIN5 (UBQ5) was the loading control. All measurements represent the average of three biological replicates with error bars representing the se.
(C) Expression patterns of ProMIR159a, b, c-GFP during pollen development in wild-type and apc8-1 plants. Fifteen individual T1 plants in the wild type or the apc8-1 background were examined. Panels (top to bottom) show three progressive stages: UP, uninucleate microspores; BP, bicellular pollen; TP, tricellular pollen. A representative DAPI-stained image for each stage is shown. Bar = 10 μm.
[See online article for color version of this figure.]
To investigate which step in miR159 biogenesis was affected, we used qPCR to examine whether primary (pri) miRNA transcription of each MIR159 was affected in the pollen of apc/c mutants. We detected reduced pri-miR159 transcription from all three MIR159 genes in apc8-1 and apc13-2, most notably an ~70% decrease for MIR159c in apc8-1 (Figure 5B). As expected, overexpression of APC8 resulted in increased pri-miR159 transcription (Figure 5B). To further confirm these results and to determine the expression patterns of each MIR159 during male gametophyte development, we generated constructs in which the promoter regions of MIR159a, b, or c were fused to GFP. We introduced these constructs into wild-type and apc/c mutants and examined multiple transgenic lines for each construct. In the wild type, MIR159a-GFP, MIR159b-GFP, and MIR159c-GFP were strongly expressed at the unicellular microspore stage, detectable at the bicellular stage, but undetectable at the tricellular stage; their expression was restricted to vegetative cell nuclei (Figure 5C). By contrast, in apc8-1, none of these reporter gene fusions was detectable at any stage of male gametophyte development. Consistent with the much weaker phenotypes in apc13-2 than in apc8-1 described above, we observed only a slight reduction of expression of the three MIR159-GFPs in apc13-2 (see Supplemental Figure 4B online). Since GFP signal intensity was not a quantitative measure of reporter gene expression, we performed qPCR to determine the levels of GFP mRNA in inflorescences. Indeed, GFP mRNA levels were lower in apc/c mutants than in the wild type (see Supplemental Figure 4C online). These results indicate that APC/C is required for miR159 biogenesis during male gametophyte development and that it acts at the transcriptional level.
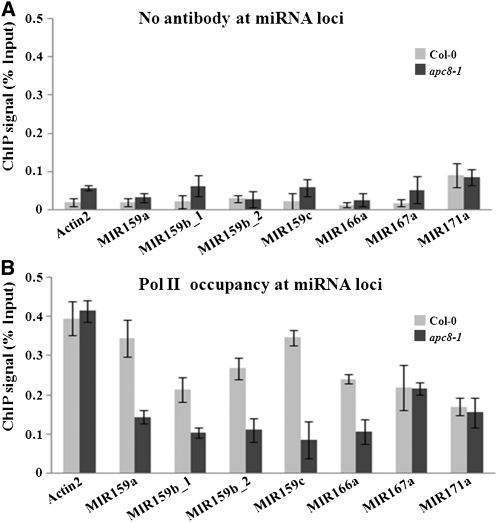
Although we showed that in the apc/c mutants the levels of pri-miR159 and the activities of the MIR159 promoters were compromised, the defects in MIR gene transcription in apc/c mutants could be an indirect effect of reduced expression of certain known genes in the miRNA biogenesis pathway. To evaluate this possibility, we first examined the expression of several miRNA biogenesis pathway genes in apc8-1 by qPCR. None of these genes was affected in apc8-1 (see Supplemental Figure 5A online). Furthermore, the protein levels of these genes were comparable to those in the wild type (see Supplemental Figures 5B and 5C online). Therefore, the reduced pri-miRNA and mature miRNA levels in the apc/c mutants are unlikely due to reduced expression of miRNA biogenesis genes. This result prompted us to examine whether APC/C plays a direct role in the transcription of MIR genes. Pol II is responsible for MIR transcription in animals and plants (Lee et al., 2004; Xie et al., 2005; Kim et al., 2011). Therefore, we next examined Pol II occupancy at promoters of several MIR genes by chromatin immunoprecipitation (ChIP) using an antibody against the largest subunit of Pol II (RPB1). We monitored MIR159a, MIR159b, MIR159c, MIR166a, MIR167a, and MIR171a using Actin2 as a positive control (Figures 6A and 6B). Pol II occupancy was enriched at regions encompassing the transcription start sites at these loci (MIR159b_1 and MIR159b_2 represent two regions flanking two transcription start sites in MIR159b loci, respectively), over the no antibody ChIP that served as a negative control (Figures 6A and 6B). Pol II occupancy at these loci was significantly reduced in apc8-1 (Figure 6B). These results indicate that APC/C promotes Pol II recruitment to MIR genes.
Figure 6.
Pol II Occupancy at miRNA Loci.
(A) ChIP performed with Col-0 (light gray bars) or apc8 (dark gray bars) with no antibody as the negative control.
(B) ChIP performed with anti-RPB1 antibody. The results were reproducible in two biological replicates. The mean and sd were determined from one representative biological replicate.
Error bars in (A) and (B) show sd calculated from three technical replicates.
APC/C-Mediated miR159 Accumulation Led to DUO1 Repression
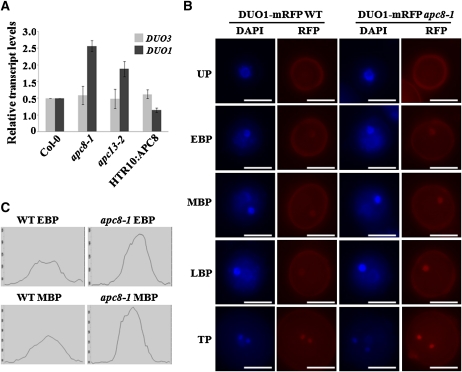
Because miR159 biogenesis was perturbed in these apc/c mutants, we next asked whether this perturbation had biological significance. Since DUO1, an miR159 target (Palatnik et al., 2007), was reported to play a role in twin sperm cell formation (Brownfield et al., 2009a), we first used qPCR to examine whether DUO1 expression was affected in the pollen of apc/c mutants. The mRNA level of DUO1 in apc8-1 was increased ~2-fold (Figure 7A), consistent with the notion that DUO1 is negatively regulated by miR159. By contrast, DUO3, which has a function similar to that of DUO1 but is not a miR159 target, had similar expression levels in apc8-1 and wild-type pollen (Figure 7A). A similar trend was observed for apc13-2 (Figure 7A). As expected, DUO1 levels were reduced in HTR10:APC8 pollen (Figure 7A). To substantiate the above result, we compared the expression level of DUO1-mRFP at different stages during pollen development in wild-type or apc8-1 plants containing the pDUO1:DUO1-mRFP construct. We consistently observed a slightly increased intensity of DUO1-mRFP signals through early bicellular stage to mature pollen in apc8-1 (Figure 7B). Therefore, our results demonstrate that APC/C plays a role in miR159 biogenesis that is important for miR159-dependent DUO1 repression.
Figure 7.
Increased DUO1 Expression in apc8-1 during Male Gametophyte Development.
(A) Expression of DUO1 and DUO3 in mature pollen from Col-0, apc8-1, apc13-2, and HTR10:APC8 as determined by qPCR. UBIQUITIN5 (UBQ5) was the loading control. All measurements represent the average of three biological replicates with error bars representing the se.
(B) Expression of pDUO1:DUO1-mRFP in the wild type and apc8-1. Plants homozygous for pDUO1:DUO1-mRFP were crossed into apc8-1 and then F3 plants homozygous for both apc8-1 and pDUO1:DUO1-mRFP were analyzed. One example for each developmental stage is shown. All images were acquired using the same exposure times. UP, unicellular pollen; EBP, early bicellular pollen; MBP, middle bicellular pollen; LBP, late bicellular pollen; TP, tricellular pollen. Bar = 10 μm.
(C) Quantitative data of RFP signal intensity at the early bicellular pollen and middle bicellular pollen stages in (B) is shown. Each x axis represents a 5-μm distance centered on the fluorescent focus. WT, wild type.
[See online article for color version of this figure.]
APC/C Regulates CYCB1;1 mRNA Levels
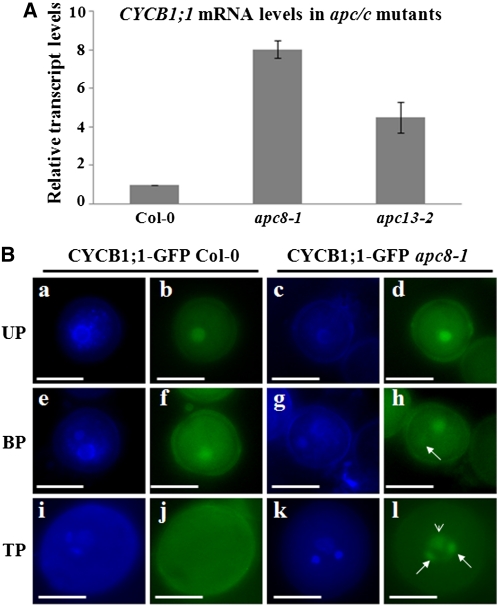
As increased DUO1 expression was observed in apc/c mutants (Figure 7) and CYCB1;1 expression was reported to be promoted by DUO1 (Brownfield et al., 2009a), we speculated that APC/C might affect not only the stability of CYCB1;1 but also expression of CYCB1;1. To test this hypothesis, we compared CYCB1;1 mRNA levels in wild-type and apc8-1 pollen by qPCR. There was significantly more CYCB1;1 mRNA in apc8-1 than in the wild type (Figure 8A). Similarly, increased CYCB1;1 mRNA was also observed in apc13-2 (Figure 8A). Taken together, these results indicate that APC/C can regulate CYCB1;1 expression.
Figure 8.
CYCB1;1 Expression Is Increased in Male Gametophytes of apc/c Mutants.
(A) Expression of CYCB1;1 in mature pollen from Col-0, apc8-1, and apc13-2. UBIQUITIN5 (UBQ5) was the loading control. All measurements represent the average of three biological replicates with error bars representing the se.
(B) Subcellular localization of CYCB1-GFP during pollen development in wild-type and apc8-1 plants harboring pCYCB1;1:CYCB1-GFP. Fifteen individual T1 plants in the wild type or the apc8-1 background were examined. DAPI staining of uninucleate microspores (UP; [a] and [c]), bicellular pollen (BP; [e] and [g]), and tricellular pollen (TP; [i] and [k]). GFP was examined in uninucleate microspores ([b] and [d]), bicellular pollen ([f] and [h]), and tricellular pollen ([j] and [l]). CYCB1;1-GFP in apc8-1 was detected weakly in generative cell nuclei (arrow) of bicellular pollen (i) and obviously accumulated in both the vegetative nucleus (arrowhead) and sperm cell nuclei (arrow) of tricellular pollen (l). Bar = 10 μm.
[See online article for color version of this figure.]
To determine if CYCB1;1 accumulates during male gametogenesis in these apc/c mutants, we introduced pCYCB1;1:CYCB1;1-GFP into wild-type, apc8-1, and apc13-2 plants. In the wild type, CYCB1;1-GFP was detected only in the nucleus of unicellular microspores and in the vegetative nucleus of bicellular pollen, but not in tricellular or mature pollen (Figure 8B), consistent with microarray results (Honys and Twell, 2004). By contrast, CYCB1;1-GFP accumulated in nuclei of both the vegetative cell and sperm cells of tricellular and mature pollen from apc8-1(Figure 8B) and apc13-2 (see Supplemental Figure 6A online). The CYCB1;1-GFP accumulation in vegetative nuclei and expanded expression in sperm cell nuclei at the tricellular stage in these apc/c mutants suggest that APC/C is necessary for removal of CYCB1;1 during male gametophyte development.
To further support the observation of accumulated CYCB1;1-GFP in apc8-1, we used immunoblots to examine the total protein levels of CYCB1;1-GFP in inflorescences from multiple individual wild-type and apc8-1 transgenic plants expressing pCYCB1;1:CYCB1;1-GFP. CYCB1;1-GFP was greatly increased in apc8-1 (see Supplemental Figure 6B online), further indicating that disruption of APC8 function caused the failure of CYCB1;1 degradation. Taken together, our data demonstrate that APC/C, in addition to its known role in regulating CYCB1;1 degradation, can regulate CYCB1;1 at the mRNA level.
DISCUSSION
We obtained evidence for a mechanism by which APC/C regulates CYCB1;1 at the transcriptional level and directly degrades CYCB1;1. To explore this dual regulation in cell cycle progression, we focused on the well-defined cell lineages in male gametophyte development and showed that disruption of APC/C resulted in increased incidence of uninucleate or binucleate pollen (Figure 4C).
Our results showed that the transcription of all three MIR159 genes was reduced in apc/c mutants (Figures 5B and 5C; see Supplemental Figures 4B and 4C online), which is consistent with the functional redundancy shown among MIR159a, MIR159b, and MIR159c (Allen et al., 2007). Interestingly, although these three genes had nearly identical expression patterns during male gametophyte development, the MIR159c expression level was higher than the expression levels of MIR159a and MIR159b, and a more obvious reduction of MIR159c transcription was seen in apc/c mutants (Figures 5B and 5C; see Supplemental Figures 4B and 4C online). Deep sequencing showed that miR159c was much less abundant in leaves, seedlings (Rajagopalan et al., 2006), and inflorescences (Fahlgren et al., 2007), consistent with the hypothesis that miR159c is pollen specific and accordingly plays a predominant role during male gametophyte development.
The temporal expression patterns of APC8 (Figure 3), MIR159 (Figure 5C), and CYCB1;1 (Brownfield et al., 2009a; Figure 8B) are similar during male gametophyte development and are reciprocal to the temporal expression pattern of DUO1. Why is it necessary that expression patterns of these genes are regulated? One of the well-known functions of the APC/C complex is to degrade cyclins to allow progression through the cell cycle. Enrichment of APC/C at the early stages of male gametophyte development correlates precisely with the period of active and continuous cell division. The most probable consequence of MIR159 expression is to restrict the expression of its target genes. DUO1, one of its target genes, functions in the formation and cell specification of male germline cells (Rotman et al., 2005). Thus, the abundance of miR159 at early stages of male gametophyte development might be mainly to limit the expression of DUO1, which is also consistent with reduced seed set of miR159ab double mutant (Allen et al., 2007). However, the expression of MIR159 and DUO1 overlaps at the bicellular stage (Brownfield et al., 2009a; Figure 5C). Other miR159 targets might play a role in male gametophyte development when their expression patterns differ from that of DUO1 (Schwab et al., 2005). Another possibility is that DUO1 expression is regulated by factors other than miR159.
DUO1 expression is initiated at late stages (Rotman et al., 2005), and DUO1 can promote CYCB1;1 expression by an unknown mechanism (Brownfield et al., 2009a), which suggests that the relationship between DUO1 and CYCB1;1 might be important and specific to the second mitosis. Moreover, overexpression of CYCB1;1 in male germlines rescued only the cell division defects of duo1/+ heterozygous plants (Brownfield et al., 2009a), indicating that CYCB1;1 is the output of a DUO1-dependent pathway during male gametophyte development. The dynamic requirements for CYCB1;1 expression in the second mitosis can be accounted for, as CYCB1;1 was expressed in the generative cell at the early bicellular stage and the late bicellular stage, but not at the middle bicellular stage or at the tricellular stage (Brownfield et al., 2009a). By contrast, we showed that CYCB1;1-GFP was detected only in the vegetative nuclei of bicellular pollen and not in tricellular or mature pollen (Figure 8B). The differences in these two studies might be due to the different constructs used: Brownfield et al. (2009a) used a possible transcriptional reporter (Glotzer et al., 1991), which included the mitotic destruction box in the first exon fused to GUS, whereas we used a fusion protein construct that encodes the entire protein. This might indicate that the remaining coding region of CYCB1;1 contains posttranscriptional information that regulates translation or protein stability. Alternatively, the possible transcriptional reporter might be more sensitive than the protein fusion construct.
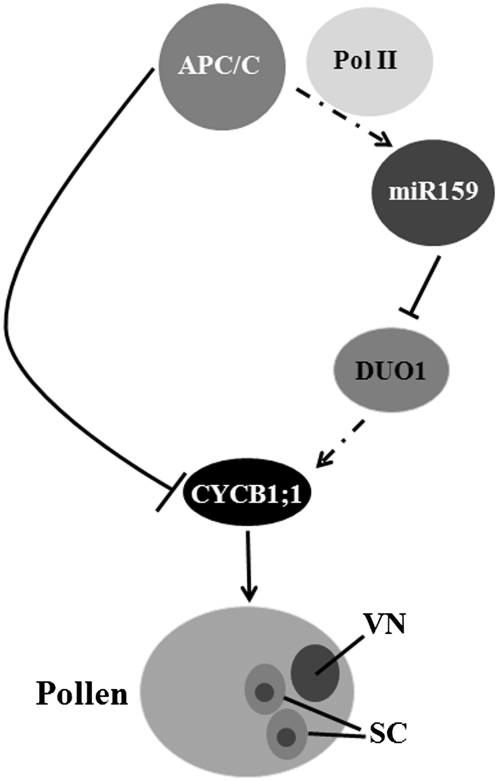
Mitotic cyclins were the first demonstrated APC/C substrates (Glotzer et al., 1991), and CYCB1;1 was shown to accumulate in several apc/c mutants (this study; Capron et al., 2003b; Kwee and Sundaresan, 2003; Pérez-Pérez et al., 2008). CYCB1;1 expression was reported to be promoted by DUO1 (Brownfield et al., 2009a). What is the relationship between APC/C-directed CYCB1;1 degradation and DUO1-mediated CYCB1;1 regulation? We propose a model in Figure 9 to explain the dual regulation of CYCB1;1 by APC/C during male gametophyte development. We showed that two pathways, integrated by APC/C, regulate CYCB1;1 expression and CYCB1;1 degradation. One outcome is that disrupted APC/C function directly causes CYCB1;1 accumulation, as CYCB1;1 is a direct substrate of APC/C (Glotzer et al., 1991). The other outcome is that reduced miR159 accumulation in apc/c mutants, accompanied with increased DUO1 expression, promotes CYCB1;1 upregulation in a direct or indirect manner because DUO1 is a transcription factor (Rotman et al., 2005), and reduced CYCB1;1 transcription was observed in duo1 heterozygous plants (Brownfield et al., 2009a). In this model, we propose that the CYCB1;1 expression promoted by DUO1 is transient, which only ensures a proper G2/M transition during pollen maturation; once that is fulfilled, CYCB1;1 must be removed immediately by APC/C. This hypothesis is consistent with the observation of dynamic expression of CYCB1;1 (Brownfield et al., 2009a). The existence of CYCB;1 transcripts at the early bicellular stage implies that CYCB1;1 from the first mitosis has not been completely degraded by APC/C. The absence of CYCB1;1 transcripts at the middle bicellular stage implies that APC/C has degraded all the CYCB1;1 that was required for the first mitosis. The reappearance of CYCB1;1 transcripts at the late bicellular stage is presumably due to the miR159-mediated DUO1 repression, which facilitates progression of the second mitosis.
Figure 9.
A Model for the Dual Roles of APC/C in Regulating Cyclin B1;1 during Male Gametophyte Development.
APC/C not only degrades CYCB1;1 directly but also directly or indirectly regulates miR159 transcription by recruiting Pol II to promoter regions. The positive regulation of miR159 by APC/C causes the correspondingly negative regulation of a miR159 target, DUO1, which ultimately leads to reduced CYCB1;1 expression. The dual role of APC/C in CYCB1;1 function ensures proper progression of the two mitoses. VN, vegetative nucleus; SC, sperm cells.
Regulation of CYCB1;1 by APC/C at the transcriptional level was unexpected. Based on the observation that APC/C is necessary for MIR159 transcription (Figure 5C), perhaps APC/C, as an ubiquitin ligase, is recruited to the transcriptional complex and acts to degrade a component specific for MIR gene transcription. It will be interesting to determine how APC/C affects the transcription of MIR genes and especially to examine whether the transcriptional machinery for MIR genes is different from that of most coding genes, although MIR genes are also transcribed by Pol II (Lee et al., 2004; Xie et al., 2005; Kim et al., 2011). Recent studies showed that a human mediator of DNA damage checkpoint protein 1 (hMDC1), which is associated with one of the core subunits of APC/C (Townsend et al., 2009), is required for normal metaphase-to-anaphase transition (Coster et al., 2007). hMDC1 is a transcription factor that belongs to the BRAC1 complex (Goldberg et al., 2003; Lou et al., 2003; Stewart et al., 2003). BRAC1 was shown to be a component of the Pol II holoenzyme (Scully et al., 1997). These relationships suggest that APC/C could be directly or indirectly involved in the transcription of MIR genes and that Arabidopsis orthologs of hMDC1 might be a bridge to recruit APC/C to transcriptional sites.
Considering that APC/C degradation of CYCB1;1 is conserved from fungi to mammals, we suspect that the dual regulation of CYCB1;1 expression by APC/C and the involvement of miRNAs in this process could also be widely conserved. A recent study showed that human immunodeficiency virus (HIV)-encoded Tat protein regulates cyclin B1 by promoting both its expression and degradation in HIV-infected lymphocytes (Zhang et al., 2010). In this case, the Tat protein appears to act as APC/C does in plants; however, the mechanism remains unknown. It will be important to understand how APC/C coordinates its proteasome activity and its role in regulating a miRNA pathway.
We showed that the Arabidopsis APC/C is a key regulatory factor in male gametophyte development, in addition to its documented role in female gametophyte development (Capron et al., 2003b; Kwee and Sundaresan, 2003). Given that APC/C is a protein complex, it is unknown why mutations in different APC/C subunits can affect one sex more profoundly than the other, and this phenomenon appears to be conserved in yeast and animals (Garbe et al., 2004; Pál et al., 2007; Jin et al., 2010). Perhaps this can be explained by target specificity of the APC/C subunits. APC/C is an E3 ubiquitin ligase, but it can only ubiquinate substrates with the help of two cofactors, the ubiquitin-activating (E1) enzyme and a ubiquitin-conjugating (E2) enzyme. The impact of a particular mutation on male or female transmission might be related to structural similarities among subunits. For example, APC6 and APC8 both have a TPR domain, while APC2 does not; mutants of APC6 and APC8 both showed defects in male transmission (Kwee and Sundaresan, 2003; Table 1), but the apc2 mutant did not (Capron et al., 2003b). These results indicate that while the overall structure of the APC/C is conserved among eukaryotes, this E3 ligase might have assumed specialized functions in different kingdoms (Lima et al., 2010). Another important layer of APC/C regulation in plants could be through subunit availability in specific tissues and/or cellular compartment, as it is known that APC subunits are differentially expressed in Arabidopsis organs (Eloy et al., 2006). Thus, in plants, the complex might be regulated by subunit availability and the different subunits could play unique regulatory roles (Eloy et al. 2006).
METHODS
Seeds, Strains, and apc8-1 Genotyping
The pDUO1:DUO1-mRFP and pHTR10:HTR10-mRFP seeds were kindly provided by David Twell and Fred Berger, respectively. The pDCL1:DCL1-YFP and pHYL1:HYL1-YFP plasmids were kindly provided by David L. Spector. T-DNA insertion mutants (SALK_027397, apc13-2; SALK_056243, dcl1-14) were obtained from the ABRC. The Schizosaccaromyces pombe temperature-sensitive apc8 strain (Cut23-174, FY9688) was obtained from the Yeast Genetic Resource Center in Japan (YGRC/NBRP). The Arabidopsis thaliana apc8-1 mutant was genotyped by ApoI digestion of the PCR fragment generated using primers APC8F2 and APC8R2; the fragment in the mutant is 25 bp smaller than that in the wild type, which is detectable when separated on a 3% agarose gel.
Plasmid Construction
APC8 was amplified from Col-0 genomic DNA with primers APC8F1 and APC8R1 (see Supplemental Table 1 online), cloned into pENTR-D/TOPO (Invitrogen), and then transferred into the plant expression destination vector pGWB40 (Nakagawa et al., 2007). The resulting plasmid was introduced into apc8-1 plants by agroinfiltration for complementation (Clough and Bent, 1998). For APC8 promoter analysis, a 2.4-kb fragment upstream of the ATG was amplified using primers APC8F1 and APC8R7 and was subcloned into pENTR-D/TOPO and then transferred into the plant expression vectors pGII-NLS3XGFP (Hellens et al., 2000) and pMDC164 (Curtis and Grossniklaus, 2003). For yeast complementation, full-length cDNAs of APC8 and APC8-1 were amplified from leaves of Col-0 and apc8-1 plants, respectively, using RT-PCR and primers APC8F3 and APC8R3, and then cloned into the pYPGE15 yeast expression vector (Brunelli and Pall, 1993). The amino acid Asp was changed to Glu by site-directed mutagenesis using primers APC8F11 and APC8R11. These plasmids were introduced into the S. pombe temperature-sensitive Cut23-174 mutant (Cullen et al., 2000). For the HTR10:APC8 overexpression construct, we first generated a modified vector pB7WG2*. The vector pB7WG2 (Karimi et al., 2002) was digested with SacI and SpeI to replace the 35S promoter with the HTR10 promoter and then digested with KpnI and ApaI to insert the LAT52-GFP cassette. A LR reaction was performed between the TOPO vector containing the full-length APC8 cDNA and pGWB7*.
For CYCB1;1 expression analysis, a genomic fragment of CYCB1;1, including a 2.5-kb fragment upstream of the ATG and downstream sequences to the stop codon, was amplified with primers CYCB1;1F1 and CYCB1;1R1 and subcloned into pENTR-D/TOPO and then transferred into the plant expression destination vector pMDC107 (Curtis and Grossniklaus, 2003). For MIR159 expression analysis, fragments of 1.9, 2.4, or 1.6 kb upstream of the first nucleotide of MIR159a, MIR159b, and MIR159c, respectively, were subcloned into pENTR-D/TOPO and then transferred into the plant expression vector pGIINLS3XGFP (Hellens et al., 2000). The primer sequences are listed in Supplemental Table 1 online.
Yeast Complementation
A single colony of cut23-174 was grown in YEA medium (5 g/L yeast extract, 30 g/L Glc, 225 mg/L adenine, 225 mg/L His, 225 mg/L Leu, 225 mg/L Lys hydrochloride) for 2 to 3 d at 28°C. One microgram of DNA of pYPGE15, pYPGE15-APC8, pYPGE15-APC8DΔE, or pYPGE15-APC8DΔN was transformed into 100 μL freshly made competent cells. After incubation at 28°C for 2 h, the cell suspension with each plasmid was divided and plated on YEA plates, one incubated at 28°C and the other at 37°C. Colonies were observed after 2 to 3 d. A single colony from each plate was diluted 1:10, 1:100, or 1:1000 and spotted onto YEA plates for 2 to 3 d of growth. Photographs were taken after 2 to 3 d. For quantitation, a single colony from each plate was cultured in YEA liquid medium for 2 to 3 d at 28°C and then inoculated into fresh YEA liquid medium to a starting OD600 of 0.2 to 0.3 and then OD600 was measured after growth for 12 or 24 h. Three colonies were analyzed from each genotype, with two biological replicates.
Microscopy Analysis and Seed Set Analysis
Pollen dissected from closed buds and open flowers from 10 to 15 individual transgenic T1 plants was stained with DAPI for 30 min at room temperature in the dark. Images were acquired with an Axiovert microscope (Zeiss) under the epifluorescence channel (the excitation/emission wavelengths are 365 nm/397 nm [DAPI], 470 nm/505 to 530 nm [GFP], 500 nm/535 nm [YFP], and 545 nm/605 nm [RFP]) using an AxioCamRM camera and AxioVision 4.8.1 software. Images were processed using Adobe Photoshop CS2 (Adobe). Seed set analysis was according to Ron et al. (2010).
Analysis of Nuclear DNA Content
DNA content was determined as described by Rotman et al. (2005) with modifications. Mature pollen from various genotypes was stained by DAPI. Relative DNA content of vegetative cell nuclei (Col-0 plants), vegetative cell-like nuclei (apc8-1), sperm cell nuclei (Col-0 plants), and generative cell-like nuclei (apc8-1 and HTR10:APC8 plants) were measured by DAPI fluorescence. Images were analyzed for fluorescence with AxioVision 4.8.1 software. A net value for each nucleus was obtained by subtracting a corresponding background reading from the cytoplasm. To standardize the relative fluorescence per C value of DNA, we first set the average net value of vegetative cell nuclei and sperm cell nuclei using wild-type mature pollen as 1C.
miRNA RNA Gel Blotting
Total RNA was isolated from inflorescences of Col-0, apc8-1, apc13-2, and APC8-YFP apc8-1. Small RNA hybridization for miRNAs was performed as described (Park et al., 2002). The 5′ end-labeled (32P) antisense oligonucleotides were used to detect U6 and miRNAs. Radioactive signals were detected with a phosphor imager.
qPCR
Total RNA was extracted using a RNeasy plant mini kit (Qiagen) and reverse transcribed with the SuperScript III first-strand synthesis system (Invitrogen) according to the manufacturer’s instructions. Real-time qPCR was performed with three technical replicates for each of three biological replicates using a MyIQ Real-Time PCR detection system (Bio-Rad) with SYBR Green PCR Master Mix (Bio-Rad). The difference between the cycle threshold (Ct) of target genes and the Ct of control primers (ΔCt = Cttarget gene – Ctcontrol) was used to obtain the normalized expression of target genes. Primers are listed in Supplemental Table 1 online.
ChIP
ChIP was performed according to Zheng et al. (2009). Pol II occupancy at several miRNA loci was determined by ChIP using anti-RPB1 antibody and 2-week-old seedlings from Col-0 and apc8-2, respectively. DNA present in the immunoprecipitates was quantified by qPCR relative to total input DNA. The results shown were consistent in two biological replicates. Commercial anti-RPB1 (Abcam) was used. The primer sets used for the PCR are listed in Supplemental Table 1 online.
GUS Staining
Tissue from 10 to 15 individual PAPC8-GUS transgenic T1 plants was fixed in 90% acetone for 2 to 3 h and then stained for 12 h in 50 mM sodium phosphate buffer, pH 7.0, containing 0.2% Triton X-100, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 1 mM X-Gluc, then washed in 70% ethanol three times. Images were taken with a Nikon SMZ800 stereoscope. Images were processed using Adobe Photoshop CS2 (Adobe).
Immunoblotting
Inflorescences were ground into powder in liquid nitrogen, agitated vigorously after adding 2× SDS-PAGE loading buffer, boiled for 5 min, and centrifuged for 5 min at 4°C. The supernatants were loaded onto 10% SDS-PAGE gels, electrophoresed for 2 to 3 h at 80 V, and transferred to nitrocellulose membrane. Mouse anti-GFP (Clontech) and anti-Hsc70 (Stressgen) antibodies were used at 1:2000 and 1:10,000 dilutions, respectively. HEN1 antibody and AGO1 antibody were made by X.C.’s lab. Detection was with an ECL plus detection kit (GE).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: APC8 (At3g48150), APC13 (At1g73177), MIR159a (At1g73687), MIR159b (At1g8075), MIR159c (At2g46255), CYCB1;1 (At4g37490), DUO1 (At3g60460), DUO3 (At1g64570), UBQ5 (At3g62250), DCL1 (At1g01040), HYL1 (At1g09700), HEN1 (At4g20910), AGO1 (At1g48410), MIR166a (At2g46685), MIR167a (At3g22886), MIR171a (At3g51375), and Actin2 (At3g18780). T-DNA insertion mutants used were dcl1-14 (SALK_056243) and apc13-2 (SALK_027397).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Isolation and Characterization of dcl1-14.
Supplemental Figure 2. Expression Analysis of APC8.
Supplemental Figure 3. Overexpression of APC8 in Male Germline Disrupts Seed Set Development and HTR10 Expression in apc8 Mature Pollen.
Supplemental Figure 4. Mature miRNA Levels and miRNA Transcription in apc/c Mutants.
Supplemental Figure 5. Expression of Known miRNA Biogenesis Genes in apc8-1.
Supplemental Figure 6. Increased CYCB1;1 Expression in apc8-1 and apc13-2.
Supplemental Table 1. Primers Used for This Study.
Supplementary Material
Acknowledgments
We thank Guang Wu for comments on the manuscript, Peng Qin and Hua Jiang for discussions and advice on phenotypic characterizations, Rebecca Haussmann for microscope assistance, and David Hantz and Julie Calfas for dedicated greenhouse maintenance. We also thank University of California-Berkeley undergraduates Daniel Park and Hugo Hua for technical assistance. We thank David Twell for pDUO1:DUO1-mRFP transgenic seeds, Fred Berger for pHTR10:HTR10-mRFP transgenic seeds, ABRC for SALK_056243 and SALK_027397 seeds, the YGRC/NBRP for providing the yeast apc8 mutant, Yuefeng Guan for modifying the pGWB7*, and Mily Ron for modifying the pGII-NLS3XGFP into a Gateway cassette vector. This work was supported by grants from the National Institutes of Health (GM61146) and the National Science Foundation (MCB-0718029) to X.C. and the USDA Current Research Information System (5335-21000-030-00D) to S.M.
References
- Allen R.S., Li J., Stahle M.I., Dubroué A., Gubler F., Millar A.A. (2007). Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc. Natl. Acad. Sci. USA 104: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley A.M., Williams B.C., Goldberg M.L., Andres A.J. (2002). Phenotypic characterization of Drosophila ida mutants: Defining the role of APC5 in cell cycle progression. J. Cell Sci. 115: 949–961 [DOI] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Borg M., Sidorova A., Mori T., Twell D. (2009a). A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 5: e1000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownfield L., Hafidh S., Durbarry A., Khatab H., Sidorova A., Doerner P., Twell D. (2009b). Arabidopsis DUO POLLEN3 is a key regulator of male germline development and embryogenesis. Plant Cell 21: 1940–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelli J.P., Pall M.L. (1993). A series of yeast/Escherichia coli lambda expression vectors designed for directional cloning of cDNAs and cre/lox-mediated plasmid excision. Yeast 9: 1309–1318 [DOI] [PubMed] [Google Scholar]
- Capron A., Okrész L., Genschik P. (2003a). First glance at the plant APC/C, a highly conserved ubiquitin-protein ligase. Trends Plant Sci. 8: 83–89 [DOI] [PubMed] [Google Scholar]
- Capron A., Serralbo O., Fülöp K., Frugier F., Parmentier Y., Dong A., Lecureuil A., Guerche P., Kondorosi E., Scheres B., Genschik P. (2003b). The Arabidopsis anaphase-promoting complex or cyclosome: Molecular and genetic characterization of the APC2 subunit. Plant Cell 15: 2370–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coster G., Hayouka Z., Argaman L., Strauss C., Friedler A., Brandeis M., Goldberg M. (2007). The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J. Biol. Chem. 282: 32053–32064 [DOI] [PubMed] [Google Scholar]
- Cullen C.F., May K.M., Hagan I.M., Glover D.M., Ohkura H. (2000). A new genetic method for isolating functionally interacting genes: High plo1(+)-dependent mutants and their suppressors define genes in mitotic and septation pathways in fission yeast. Genetics 155: 1521–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloy N.B., Coppens F., Beemster G.T.S., Hemerly A.S., Ferreira P.C. (2006). The Arabidopsis anaphase promoting complex (APC): Regulation through subunit availability in plant tissues. Cell Cycle 5: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Howell M.D., Kasschau K.D., Chapman E.J., Sullivan C.M., Cumbie J.S., Givan S.A., Law T.F., Grant S.R., Dangl J.L., Carrington J.C. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe D., Doto J.B., Sundaram M.V. (2004). Caenorhabditis elegans lin-35/Rb, efl-1/E2F and other synthetic multivulva genes negatively regulate the anaphase-promoting complex gene mat-3/APC8. Genetics 167: 663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138 [DOI] [PubMed] [Google Scholar]
- Goldberg M., Stucki M., Falck J., D’Amours D., Rahman D., Pappin D., Bartek J., Jackson S.P. (2003). MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956 [DOI] [PubMed] [Google Scholar]
- Gusti A., Baumberger N., Nowack M., Pusch S., Eisler H., Potuschak T., De Veylder L., Schnittger A., Genschik P. (2009). The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS ONE 4: e4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Honys D., Twell D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Hamamura Y., Gourgues M., Higashiyama T., Berger F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17: 1032–1037 [DOI] [PubMed] [Google Scholar]
- Iwakawa H., Shinmyo A., Sekine M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45: 819–831 [DOI] [PubMed] [Google Scholar]
- Jacobsen S.E., Running M.P., Meyerowitz E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jin F., Hamada M., Malureanu L., Jeganathan K.B., Zhou W., Morbeck D.E., van Deursen J.M. (2010). Cdc20 is critical for meiosis I and fertility of female mice. PLoS Genet. 6: e1001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 17: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim H.J., Oh S.A., Brownfield L., Hong S.H., Ryu H., Hwang I., Twell D., Nam H.G. (2008). Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature 455: 1134–1137 [DOI] [PubMed] [Google Scholar]
- Kim Y.J., Zheng B., Yu Y., Won S.Y., Mo B., Chen X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwee H.S., Sundaresan V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. Plant J. 36: 853–866 [DOI] [PubMed] [Google Scholar]
- Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. (2004). MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23: 4051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima Mde.F., Eloy N.B., Pegoraro C., Sagit R., Rojas C., Bretz T., Vargas L., Elofsson A., de Oliveira A.C., Hemerly A.S., Ferreira P.C. (2010). Genomic evolution and complexity of the Anaphase-promoting Complex (APC) in land plants. BMC Plant Biol. 10: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Z., Minter-Dykhouse K., Wu X., Chen J. (2003). MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421: 957–961 [DOI] [PubMed] [Google Scholar]
- Marrocco K., Thomann A., Parmentier Y., Genschik P., Criqui M.C. (2009). The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development 136: 1475–1485 [DOI] [PubMed] [Google Scholar]
- McCormick S. (2004). Control of male gametophyte development. Plant Cell 16 (Suppl), S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T., Kuroiwa H., Higashiyama T., Kuroiwa T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8: 64–71 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nowack M.K., Grini P.E., Jakoby M.J., Lafos M., Koncz C., Schnittger A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38: 63–67 [DOI] [PubMed] [Google Scholar]
- Okada T., Endo M., Singh M.B., Bhalla P.L. (2005). Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44: 557–568 [DOI] [PubMed] [Google Scholar]
- Pál M., Nagy O., Ménesi D., Udvardy A., Deák P. (2007). Structurally related TPR subunits contribute differently to the function of the anaphase-promoting complex in Drosophila melanogaster. J. Cell Sci. 120: 3238–3248 [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Wollmann H., Schommer C., Schwab R., Boisbouvier J., Rodriguez R., Warthmann N., Allen E., Dezulian T., Huson D., Carrington J.C., Weigel D. (2007). Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13: 115–125 [DOI] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez J.M., Serralbo O., Vanstraelen M., González C., Criqui M.C., Genschik P., Kondorosi E., Scheres B. (2008). Specialization of CDC27 function in the Arabidopsis thaliana anaphase-promoting complex (APC/C). Plant J. 53: 78–89 [DOI] [PubMed] [Google Scholar]
- Peters J.M. (2006). The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7: 644–656 [DOI] [PubMed] [Google Scholar]
- Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Ron M., Alandete Saez M., Eshed Williams L., Fletcher J.C., McCormick S. (2010). Proper regulation of a sperm-specific cis-nat-siRNA is essential for double fertilization in Arabidopsis. Genes Dev. 24: 1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman N., Durbarry A., Wardle A., Yang W.C., Chaboud A., Faure J.E., Berger F., Twell D. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15: 244–248 [DOI] [PubMed] [Google Scholar]
- Saze H., Kakutani T. (2007). Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 26: 3641–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Scully R., Anderson S.F., Chao D.M., Wei W., Ye L., Young R.A., Livingston D.M., Parvin J.D. (1997). BRCA1 is a component of the RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 94: 5605–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Wang B., Bignell C.R., Taylor A.M., Elledge S.J. (2003). MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Townsend K., Mason H., Blackford A.N., Miller E.S., Chapman J.R., Sedgwick G.G., Barone G., Turnell A.S., Stewart G.S. (2009). Mediator of DNA damage checkpoint 1 (MDC1) regulates mitotic progression. J. Biol. Chem. 284: 33939–33948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y.M., Nakaseko Y., Kumada K., Nakagawa T., Yanagida M. (1999). Fission yeast APC/cyclosome subunits, Cut20/Apc4 and Cut23/Apc8, in regulating metaphase-anaphase progression and cellular stress responses. Genes Cells 4: 445–463 [DOI] [PubMed] [Google Scholar]
- Yu H., Peters J.M., King R.W., Page A.M., Hieter P., Kirschner M.W. (1998). Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science 279: 1219–1222 [DOI] [PubMed] [Google Scholar]
- Zhang S.M., Sun Y., Fan R., Xu Q.Z., Liu X.D., Zhang X., Wang Y., Zhou P.K. (2010). HIV-1 Tat regulates cyclin B1 by promoting both expression and degradation. FASEB J. 24: 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Wang Z., Li S., Yu B., Liu J.Y., Chen X. (2009). Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 23: 2850–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.