This work provides evidence that deposition of cuticular waxes is intimately associated with plant responses to drought. The Arabidopsis MYB96 transcription factor functions as a regulator of ABA-mediated cuticular wax biosynthesis under drought conditions by binding directly to the promoters of genes encoding very-long-chain fatty acid–condensing enzymes involved in cuticular wax biosynthesis.
Abstract
Drought stress activates several defense responses in plants, such as stomatal closure, maintenance of root water uptake, and synthesis of osmoprotectants. Accumulating evidence suggests that deposition of cuticular waxes is also associated with plant responses to cellular dehydration. Yet, how cuticular wax biosynthesis is regulated in response to drought is unknown. We have recently reported that an Arabidopsis thaliana abscisic acid (ABA)–responsive R2R3-type MYB transcription factor, MYB96, promotes drought resistance. Here, we show that transcriptional activation of cuticular wax biosynthesis by MYB96 contributes to drought resistance. Microarray assays showed that a group of wax biosynthetic genes is upregulated in the activation-tagged myb96-1D mutant but downregulated in the MYB96-deficient myb96-1 mutant. Cuticular wax accumulation was altered accordingly in the mutants. In addition, activation of cuticular wax biosynthesis by drought and ABA requires MYB96. By contrast, biosynthesis of cutin monomers was only marginally affected in the mutants. Notably, the MYB96 protein acts as a transcriptional activator of genes encoding very-long-chain fatty acid–condensing enzymes involved in cuticular wax biosynthesis by directly binding to conserved sequence motifs present in the gene promoters. These results demonstrate that ABA-mediated MYB96 activation of cuticular wax biosynthesis serves as a drought resistance mechanism.
INTRODUCTION
Drought or water deficit conditions profoundly affect plant growth and productivity and reduce crop yield worldwide. Therefore, plants have evolved versatile mechanisms to cope with drought stress. Molecular and cellular responses to drought stress and underlying regulatory mechanisms have been extensively studied via molecular genetic and physiological approaches and gene expression studies.
Gene transcriptional regulation is the most important step during plant adaptation to environmental fluctuations (Zhu, 2002; Shinozaki et al., 2003; Chinnusamy et al., 2004; Yamaguchi-Shinozaki and Shinozaki, 2006). Particularly, roles of various transcription factors, such as basic leucine zipper, MYC, MYB, and NAC (NAM, ATAF1/2, CUC2) members, have been widely documented in drought stress response and signaling cascades (Kang et al., 2002; Abe et al., 2003; Fujita et al., 2004; Tran et al., 2004; Seo et al., 2009). The transcription factors interact with specific cis-acting elements existing in target gene promoters and regulate elaborate networks of drought-responsive genes and associated physiological responses (Yamaguchi-Shinozaki and Shinozaki, 2006).
Plant resistance responses to drought conditions are exerted via several distinct mechanisms, such as synthesis of osmoprotectants and antioxidants, maintenance of root growth and water uptake, and adjustments of aerial development (Schroeder et al., 2001; Seki et al., 2007; Mouillon et al., 2008). Previous studies have shown that stomatal regulation of transpiration is a primary defense mechanism that prevents water loss under drought conditions (Schroeder et al., 2001; Sirichandra et al., 2009). Abscisic acid (ABA) is a key component of signaling networks that induce stomatal closure, minimizing water transpiration from the leaves (Christmann et al., 2006; Sirichandra et al., 2009). In addition, a number of transcription factors and their target genes are involved in mediating ABA perception and signaling and have been shown to regulate drought stress responses by modulating stomatal movement (Kang et al., 2002; Abe et al., 2003).
Notably, recent studies strongly support the idea that cuticular wax accumulation is also closely associated with drought resistance responses (Aharoni et al., 2004; Zhang et al., 2005, 2007; Kosma et al., 2009). The cuticle is an outermost thin hydrophobic layer that covers the surface of aerial plant parts, such as leaves and stems. It protects plants from nonstomatal water loss (Riederer and Schreiber, 2001), pathogen infection (Barthlott and Neinhuis, 1997), insect attack (Eigenbrode and Espelie, 1995), and UV radiation (Reicosky and Hanover, 1978; Solovchenko and Merzlyak, 2003). The cuticle framework is provided by cutin, a plant-specific polyester that consists of C16 and C18 hydroxy and epoxy-hydroxy fatty acids and glycerol monomers (Kolattukudy, 2001; Nawrath, 2006; Pollard et al., 2008).
Within the cuticle framework, the cutin matrix is surrounded by intracuticular waxes and overlaid with epicuticular waxes, which consist mainly of very-long-chain fatty acids (VLCFAs; C20 to C34) and their derivatives, such as alcohols, aldehydes, alkanes, ketones, and wax esters (Post-Beittenmiller, 1996; Kunst and Samuels, 2003; Samuels et al., 2008). The C16 and C18 fatty acids synthesized in the plastids are exported to the cytoplasm, where they are further elongated to VLCFAs through sequential addition of two-carbon units in a reaction catalyzed by fatty acid elongase complexes at the endoplasmic reticulum (Millar et al., 1999; Hooker et al., 2002; Joubès et al., 2008; Samuels et al., 2008). Fatty acid elongation involves a series of four reactions: condensation of the two-carbon units to acyl-CoA by 3-ketoacyl-CoA synthetases (KCSs), reduction of 3-ketoacyl-CoA by 3-ketoacyl-CoA reductases (KCRs), dehydration of 3-hydroxyacyl-CoA by 3-hydroxyacyl-CoA dehydratases, and reduction of trans-2,3-enoyl-CoA by trans-2-enoyl-CoA reductases (Todd et al., 1999; Fiebig et al., 2000; Kunst and Samuels, 2003, 2009; Beaudoin et al., 2009). Cuticular wax biosynthesis is regulated by diverse environmental signals, and few transcription factors have been implicated in cuticular wax biosynthesis and accumulation (Aharoni et al., 2004; Zhang et al., 2005, 2007; Cominelli et al., 2008). However, little is known about how cuticular wax biosynthesis is regulated, particularly under drought stress conditions.
We recently reported that the MYB96 transcription factor plays a role in drought stress responses (Seo et al., 2009). MYB96 mediates ABA signaling in the modulation of lateral root development. It regulates lateral root meristem activation under drought conditions via an ABA-auxin signaling crosstalk. In this signaling scheme, the MYB96-mediated ABA signals are incorporated into an auxin signaling pathway that involves a subset of GH3 genes encoding auxin-conjugating enzymes (Seo et al., 2009). MYB96 also regulates the RD22 gene, possibly in modulating stomatal movement. Whereas an activation-tagged myb96-1D mutant exhibits enhanced drought resistance, the MYB96-deficient mutant (myb96-1) is susceptible to drought stress (Seo et al., 2009). It has been proposed that the MYB96 transcription factor contributes to optimization of plant growth and development under drought conditions, although the underlying molecular mechanisms have been unexplored.
In this study, we demonstrate that the MYB96 transcription factor confers drought resistance by modulating cuticular wax biosynthesis. A microarray assay revealed that whereas expression of a group of genes encoding cuticular wax biosynthetic enzymes was greatly upregulated in the myb96-1D mutant, they were downregulated in the myb96-1 mutant. Accordingly, epicuticular waxes accumulated to a high level in myb96-1D leaves and stems, but epicuticular wax deposition was significantly reduced in myb96-1 leaves and stems. Notably, the MYB96 transcription factor binds directly to the promoters of genes encoding enzymes involved in VLCFA biosynthesis, demonstrating that MYB96-mediated cuticular wax biosynthesis is intimately associated with drought resistance responses.
RESULTS
Wax Biosynthetic Genes Are Upregulated in myb96-1D
The MYB96 transcription factor promotes drought resistance: whereas the myb96-1D mutant is resistant to drought, the myb96-1 mutant is susceptible to drought (Seo et al., 2009). Accordingly, stomatal aperture is slightly altered in the mutants. However, we expected that additional traits would also contribute to drought resistance. To obtain clues as to how MYB96 promotes drought resistance, we performed microarray assays using the Affymetrix GeneChip representing ~24,000 genes, and differentially expressed genes were identified after statistical analysis (>2-fold change, P < 0.05). The P values were corrected for multiple testing using false discovery rate (FDR) methodology (see Methods).
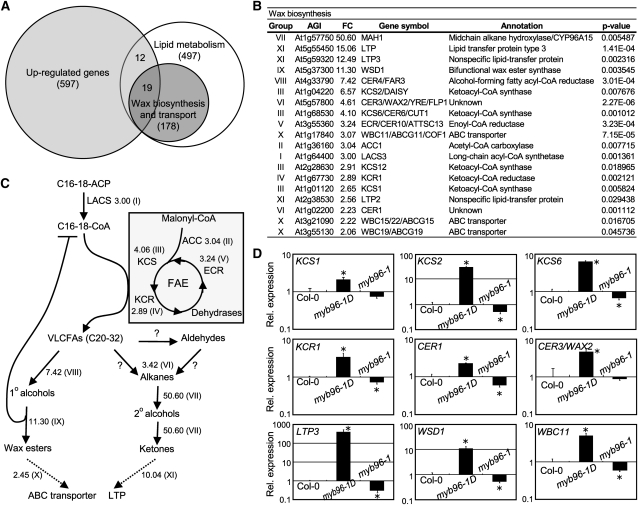
Approximately 600 genes were upregulated in the myb96-1D mutant (Figure 1A; see Supplemental Data Set 1A online). A major functional category of the upregulated genes included those encoding a subset of wax biosynthetic enzymes (Figure 1B; see Supplemental Data Set 1B online), such as KCS1, KCS2, KCS6, KCR1, ECERIFERUM1 (CER1), and CER3 (Aarts et al., 1995; Todd et al., 1999; Fiebig et al., 2000; Rowland et al., 2007; Beaudoin et al., 2009; Lee et al., 2009b). Genes encoding putative wax transporters were also upregulated in the mutant (Figures 1B and 1C), suggesting that cuticular wax biosynthesis and transport are broadly influenced in the myb96-1D mutant. The microarray data were verified by quantitative real-time RT-PCR (qRT-PCR). Whereas the wax biosynthetic genes were upregulated in the myb96-1D mutant, they were downregulated in the myb96-1 mutant (Figure 1D). Furthermore, MYB96 was expressed to a high level in stem epidermal cells (see Supplemental Figure 1 online), where cuticular waxes are synthesized (Suh et al., 2005), supporting the idea that the MYB96 transcription factor is related to cuticular wax biosynthesis.
Figure 1.
Upregulation of Cuticular Wax Biosynthetic Genes in myb96–1D.
(A) Venn diagrams showing the distribution of overlapping and nonoverlapping genes encoding lipid metabolic and wax biosynthetic enzymes with genes upregulated in myb96-1D. Three independent experiments were performed. The microarray data set was deposited into the ArrayExpress database with accession number E-MEXP-2965 at http://www.ebi.ac.uk/at-miamexpress.
(B) List of wax biosynthetic genes upregulated in myb96-1D. Genes upregulated >2-fold in the myb96-1D mutant compared with the wild type are listed in Supplemental Data Set 1A online. The P values were corrected for multiple testing using FDR methodology. The group of genes was classified based on their biochemical functions. AGI, Arabidopsis Genome Initiative number; FC, fold changes.
(C) Simplified cuticular wax biosynthetic pathway. Adapted from Kunst and Samuels (2009). Numbers indicate mean fold change of the genes belonging to individual gene groups (I to XI), as marked in (B). The question marks and dotted lines indicate unidentified enzymes and processes, respectively. FAE, fatty acid elongation.
(D) qRT-PCR of wax biosynthetic gene expression. Total RNAs were extracted from 2-week-old whole plants grown on MS-agar. Transcript levels were examined via qRT-PCR. Biological triplicates were averaged and statistically analyzed using a student t test (*P < 0.01). Bars indicate se of the mean. The vertical axis is displayed on a logarithmic scale to obtain better comparison of transcript levels.
Epicuticular Wax Deposition Is Altered in myb96-1D and myb96-1 Mutants
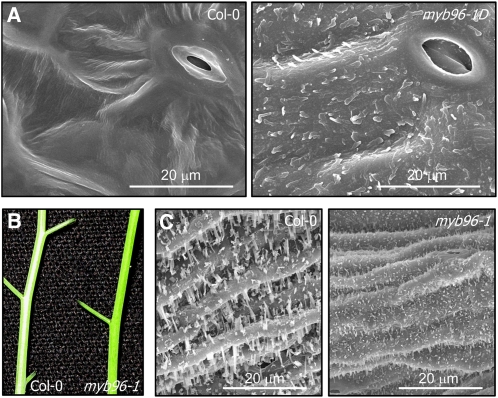
We found that expression of cuticular wax biosynthetic genes was altered in myb96-1D and myb96-1 mutants. We therefore analyzed deposition of epicuticular wax crystals on the leaf surface by scanning electron microcopy. Strikingly, a large amount of epicuticular wax crystals was observed on myb96-1D leaves, unlike wild-type leaves (Figure 2A). By contrast, they were considerably reduced on myb96-1 stems. The whitish appearance observed in the wild type largely disappeared on myb96-1 stems (Figure 2B). Scanning electron microscopy and Nile red staining revealed that epicuticular wax crystals were reduced accordingly on mutant stems (Figure 2C; see Supplemental Figure 2 online).
Figure 2.
Scanning Electron Microscopy Analysis of Epicuticular Wax Deposition on the Surfaces of Plant Organs in myb96-1D and myb96-1 Mutants.
(A) Scanning electron microscopy images of epicuticular wax crystals on the leaves. Rosette leaves of 4-week-old wild-type (Col-0) and myb96-1D plants grown in soil were used for scanning electron microscopy analysis. Bars = 20 μm.
(B) Glossy appearance indicating wax deficiency of the myb96-1 stem. Inflorescence stems of 5-week-old wild-type (Col-0) and myb96-1 plants grown in soil were photographed.
(C) Scanning electron microscopy images of epicuticular wax crystals on the stems. Inflorescence stems of 5-week-old wild-type (Col-0) and myb96-1 plants grown in soil were subject to scanning electron microscopy. Bars = 20 μm.
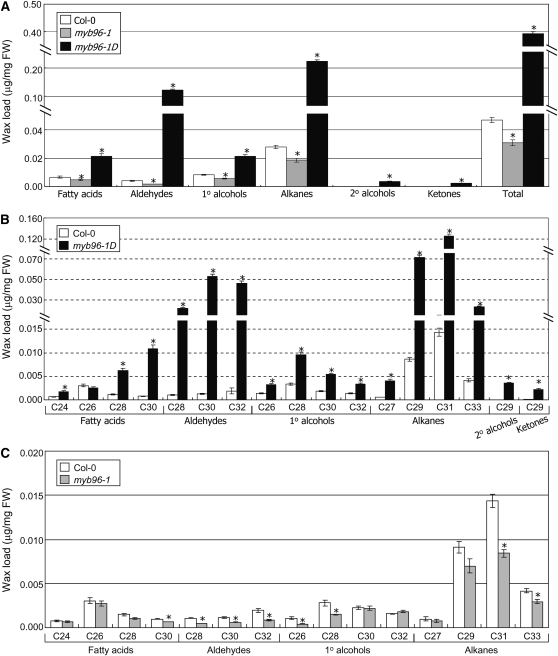
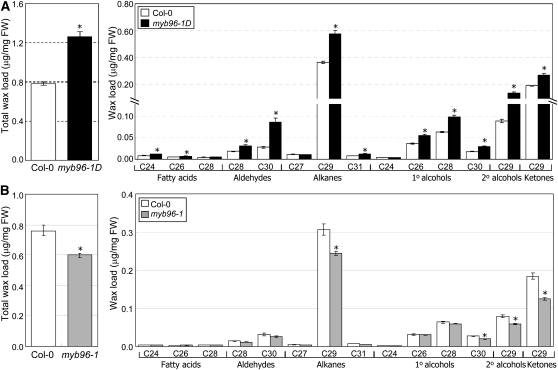
Measurements of cuticular wax content and composition by gas chromatography with flame ionization detection and gas chromatography with mass spectrometry, respectively, showed that the total wax load was elevated 8.6-fold in myb96-1D leaves (Figure 3A). Cuticular wax composition was also altered in mutant leaves (Figure 3B). Alterations in the contents of aldehydes and alkanes were the most prominent changes (Figures 3A and 3B). By contrast, the total wax load was decreased by ~34% in myb96-1 leaves (Figure 3C; see Supplemental Figure 3 online). Similar to the alterations in cuticular wax contents and compositions in the leaves, the total wax load was increased ~1.6-fold in myb96-1D stems (Figure 4A), whereas it was decreased ~25% in myb96-1 stems (Figure 4B; see Supplemental Figure 4 online).
Figure 3.
Cuticular Wax Amount and Composition in myb96-1D and myb96-1 Leaves.
Rosette leaves of 4-week-old wild-type (Col-0) and mutant plants grown in soil were used for analysis of cuticular wax composition and loads. Biological triplicates, each with technical duplicates, were averaged and statistically analyzed using a student t test (*P < 0.01). Bars indicate se of the mean. FW, fresh weight.
(A) Cuticular wax amount in myb96-1D and myb96-1 leaves compared with the wild type.
(B) and (C) Cuticular wax composition and loads in the leaves of the myb96-1D (B) and myb96-1 mutants (C) compared with the wild type. The loads of cuticular waxes of the myb96-1 mutant were also expressed as μg/cm2 in Supplemental Figure 3 online.
Figure 4.
Cuticular Wax Composition in myb96-1D and myb96-1 Stems.
Inflorescence stems of 5- to 10-week-old wild-type and mutant plants grown in soil were used for analysis of cuticular wax composition and loads. Biological triplicates, each with technical duplicates, were averaged and statistically analyzed using a Student’s t test (*P < 0.01). Bars indicate se of the mean. Cuticular wax composition and loads in the stems of myb96-1D (A) and myb96-1 mutants (B). The loads of cuticular waxes of the myb96-1 mutant were also expressed as μg/cm2 in Supplemental Figure 4 online. FW, fresh weight.
However, deposition and composition of cutin monomers, which are the major components of the cuticle (Pollard et al., 2008), and expression of cutin biosynthetic genes (Wellesen et al., 2001; Schnurr et al., 2004; Xiao et al., 2004) were not discernibly altered in myb96-1D leaves (see Supplemental Figures 5 and 6 online), indicating that MYB96 specifically regulates biosynthesis and accumulation of cuticular waxes.
Chlorophyll leaching assays, which are frequently used to examine cuticular defects in leaves (Chen et al., 2003), showed that chlorophyll leaching occurred more slowly in myb96-1D leaves but more rapidly in myb96-1 leaves in comparison to wild-type leaves (see Supplemental Figure 7 online), which corresponds with the differential accumulation of epicuticular wax crystals. Collectively, these observations support the role of MYB96 in biosynthesis and accumulation of cuticular waxes.
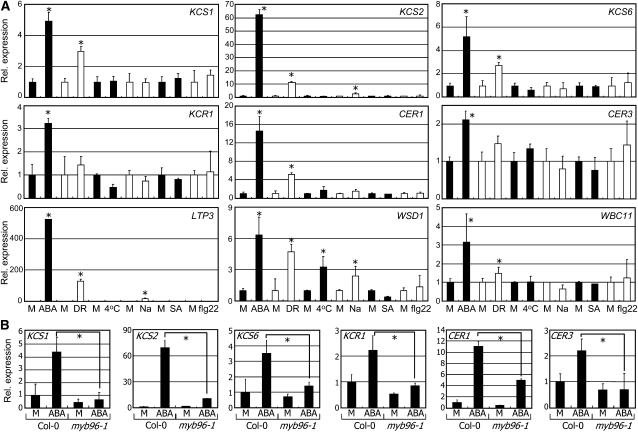
Drought Induction of Cuticular Wax Biosynthetic Genes Requires MYB96
The MYB96 transcription factor induces drought resistance via an ABA signaling pathway (Seo et al., 2009). Therefore, we asked whether the selected wax biosynthetic genes are influenced by drought and ABA. The KCS1, KCS2, KCS6, CER1, LIPID TRANSFER PROTEIN3 (LTP3), WAX ESTER SYNTHASE/ACYL-COA: DIACYLGLYCEROL ACYLTRANSFERASE1 (WSD1), and WHITE-BROWN COMPLEX HOMOLOG PROTEIN11 (WBC11) genes were significantly induced by exogenous application of ABA and exposure to drought (Figure 5A). KCR1 and CER3 were also induced by ABA but not by drought. Furthermore, the effects of ABA on wax biosynthetic gene expression were greatly reduced in the myb96-1 mutant (Figure 5B; see Supplemental Figure 8A online), showing that ABA induction of the genes is at least partially dependent on MYB96. However, PHOSPHOLIPASE C1 and HYDROXYSTEROID DEHYDROGENASE1 encoding lipid biosynthetic enzymes were induced normally by ABA in the myb96-1 mutant (see Supplemental Figure 8B online; Sanchez and Chua, 2001; Li et al., 2007), indicating that the ABA induction of these genes is independent of MYB96.
Figure 5.
Drought Induction of Cuticular Wax Biosynthetic Genes.
Two-week-old plants grown on MS-agar plates were treated with growth hormones, such as 20 μM ABA (6 h) and 100 μM SA (6 h), and stress conditions, including cold (4°C, 24 h), 150 mM NaCl (Na, 6 h), and 5 μM flagellin22 (flg22, 24 h), before whole-plant materials were harvested for extraction of total RNAs. Drought stress (DR) was induced in 20-d-old plants grown in soil by halting watering. Transcript levels were examined as described in Figure 1D. Bars indicate se of the mean (t test, *P < 0.01). M, mock treatment.
(A) Effects of growth hormones and stresses on gene expression. Statistical significance of the measurements was determined using a Student’s t test by comparing with mock treatment.
(B) Effects of ABA on gene expression in myb96-1. Statistical significance of the measurements was determined using a Student’s t test by comparing Col-0 and mutant plants treated with ABA.
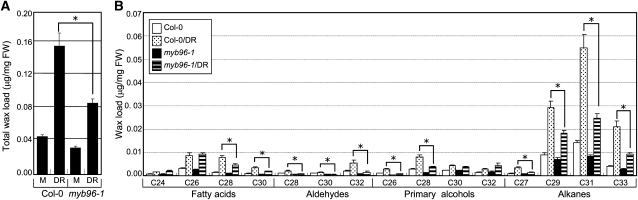
We next examined whether drought induces cuticular wax biosynthesis. Consistent with the ABA induction of wax biosynthetic genes, the total cuticular wax load was elevated ~4-fold in wild-type leaves under drought conditions (Figure 6A). However, the positive effects of drought on wax accumulation were significantly reduced in the leaves of the drought-susceptible myb96-1 mutant, which is in agreement with the notion that cuticular wax accumulation confers drought resistance (Aharoni et al., 2004; Zhang et al., 2005; Seo et al., 2009). These observations indicate that MYB96 regulates, at least in part, drought stress induction of cuticular wax accumulation. The composition of cuticular waxes was also significantly altered in the mutant leaves. Under drought conditions, whereas the levels of VLCFAs, aldehydes, primary alcohols, and alkanes were elevated in wild-type leaves, accumulation of aldehydes and alkanes was considerably reduced in myb96-1 leaves (Figure 6B).
Figure 6.
Cuticular Wax Accumulation in the Leaves under Drought Conditions.
(A) Accumulation of cuticular waxes under drought. Biological triplicates, each with technical duplicates, were averaged. Statistical significance of the measurements was determined using a Student’s t test (t test, *P < 0.01) by comparing Col-0 and mutant plants treated with drought (DR). Bars indicate se of the mean. FW, fresh weight; M, mock.
(B) Cuticular wax composition in the leaves under drought. Three-week-old leaves were used. Biological triplicates, each with technical duplicates, were averaged. Statistical significance of the measurements was determined using a Student’s t test (t test, *P < 0.01) by comparing Col-0 and mutant plants treated with drought (DR). Bars indicate se of the mean.
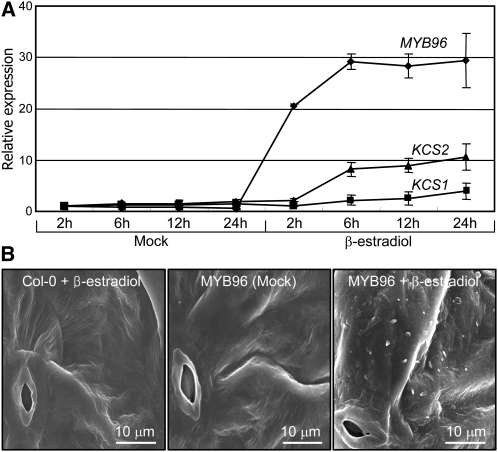
To examine more directly the role of MYB96 in cuticular wax biosynthesis, we produced transgenic Arabidopsis thaliana plants expressing the MYB96 gene under the control of a β-estradiol–inducible promoter and examined deposition of epicuticular waxes on the leaves. Genes involved in VLCFA biosynthesis, such as KCS1, KCS2, KCS6, and KCR1, were induced within 6 h after the inducer was added (Figure 7A; see Supplemental Figure 9 online), confirming that MYB96 regulates the transcription of these genes. However, the CER1, WBC11, WSD1, and LTP3 genes were induced after 6 h, suggesting that the inductive effects of MYB96 may be secondary responses (see Supplemental Figure 9 online). Accordingly, deposition of epicuticular wax crystals was increased on the transgenic leaves only after induction of the MYB96 gene (Figure 7B). These observations demonstrate that the induction of cuticular wax biosynthesis and accumulation by drought is mediated by MYB96.
Figure 7.
Effects of MYB96 Induction on Epicuticular Wax Deposition.
(A) Induction of wax biosynthetic genes. Two-week-old transgenic plants expressing the pER8-MYB96 gene fusion under the control of a β-estradiol–inducible promoter were incubated in MS liquid cultures supplemented with 10 μM β-estradiol. Whole plants were harvested at the indicated time points (h) after β-estradiol application for total RNA extraction. Transcript levels were examined by qRT-PCR. Biological triplicates were averaged. Bars indicate se of the mean.
(B) Elevation of epicuticular wax crystals after β-estradiol induction of MYB96. One-week-old plants grown in soil were sprayed every 3 d with 10 μM β-estradiol solution. Epicuticular wax crystals were examined by scanning electron microscopy 3 weeks after induction. Bars = 10 μm.
Cuticular Wax Accumulation Contributes to Drought Resistance
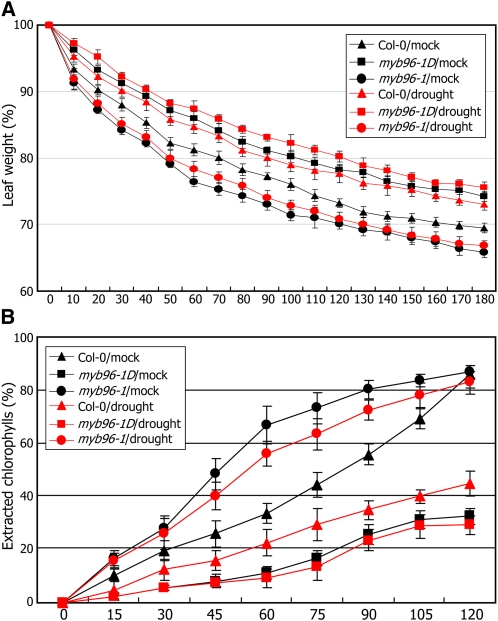
Our data support the idea that MYB96 promotes drought resistance by inducing cuticular wax biosynthesis and accumulation. We examined more directly whether cuticular wax accumulation is linked with drought resistance response by measuring cuticular transpiration and chlorophyll leaching. Four-week-old plants grown under either normal or drought conditions were dark acclimated for 6 h to assure stomatal closure and soaked in water for 1 h in the dark. The leaves were subject to water loss measurements and chlorophyll leaching assays (Kosma et al., 2009). The results showed that cuticular transpiration occurred more slowly in myb96-1D leaves but more rapidly in myb96-1 leaves compared with the wild type (Figure 8A), demonstrating that cuticular wax accumulation contributes to drought resistance.
Figure 8.
Cuticular Transpiration in myb96-1D and myb96-1 Leaves.
Four-week-old plants grown under either normal (mock) or drought conditions were acclimated in the dark for 6 h and used for subsequent treatments.
(A) Water loss assays. Whole rosettes of the dark-acclimated plants were excised and soaked in water for 60 min in the dark. They were dried and weighed at the indicated time points. Three measurements were averaged at each time point. Bars indicate se of the mean.
(B) Chlorophyll leaching assays. Whole rosettes of the dark-acclimated plants were soaked in 80% ethanol for the indicated time periods (min). Extracted chlorophyll contents at individual time points were expressed as percentages of that at 24 h after initial immersion. Three measurements were averaged. Bars indicate se of the mean.
Chlorophyll leaching assays further supported the linkage between cuticular wax accumulation and drought resistance. In wild-type leaves, chlorophylls were extracted more slowly from drought-treated leaves (Figure 8B), certainly due to cuticular wax accumulation. It was also found that whereas chlorophyll extraction occurred more slowly from myb96-1D leaves, it occurred more rapidly from myb96-1 leaves, regardless of drought treatments. These observations indicate that MYB96-mediated cu-ticular wax accumulation is linked with the drought resistance response.
MYB96 Binds to Consensus Motifs in the Promoters of Wax Biosynthetic Genes
Transcriptional control of cuticular lipid biosynthesis has been documented (Zhang et al., 2005, 2007; Kannangara et al., 2007; Cominelli et al., 2008; Raffaele et al., 2008). However, no specific transcription factors have been unequivocally proven to bind directly to the biosynthetic gene promoters. We therefore asked whether the MYB96 transcription factor regulates directly the cuticular wax biosynthetic genes identified from the microarray assays.
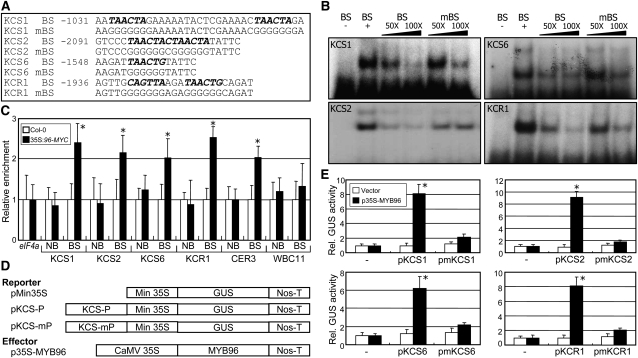
Sequence analysis revealed that promoters of the genes that are related to wax biosynthesis, such as KCS1, KCS2, KCS6, KCR1, CER1, CER3, LTP3, WSD1, and WBC11, contain conserved sequence motifs (Figure 9A; see Supplemental Figure 10A online), which are analogous to the MYB binding consensus sequence (BS) (Urao et al., 1993; Abe et al., 2003). Electrophoretic mobility shift assays using a recombinant maltose binding protein (MBP)-MYB96 fusion protein produced in Escherichia coli cells showed that the MYB96 protein bound specifically to the BS sequence motifs of KCS1, KCS2, KCS6, KCR1, CER3, and WSD1 gene promoters (see Supplemental Figure 10B online). The MBP protein alone did not bind to the consensus sequences (see Supplemental Figure 11 online). Whereas the MYB96 binding of these genes was considerably reduced in the presence of excess amounts of unlabeled BS fragments, it was reduced to a lesser degree by mutated DNA fragments (mBS), supporting the specific binding of MYB96 to the BS sequences (Figure 9B).
Figure 9.
Binding of MYB96 to Consensus Motifs in the Promoters of Wax Biosynthetic Genes
(A) MYB binding consensus sequences (BSs). Core binding sequences, marked in bold, were mutated, resulting in mBSs, to verify specific binding.
(B) In vitro binding of MYB96 to the consensus sequences. The (−) lanes are controls without recombinant MBP-MYB96 proteins. Excess amounts (50× or 100×) of unlabeled BS or mBS DNA fragments were added as competitors.
(C) ChIP assays. Total protein extracts from 35S:96-MYC transgenic plants grown on MS-agar plates for 2 weeks were immunoprecipitated with an anti-MYC antibody. Fragmented genomic DNA was eluted from the protein-DNA complexes and subjected to quantitative PCR analysis. Three measurements were averaged for individual assays. Statistical significance of the measurements was determined using a Student’s t test (t test, *P < 0.01) by comparison with the value of Col-0. Bars indicate se of the mean. In each measurement, the measurement values in Col-0 were set to 1 after normalization against eIF4a for quantitative PCR analysis. NB, nonbinding sequence.
(D) Expression constructs used. The promoter sequence elements of the KCS genes listed in (A) were fused to minimal 35S promoter (Min 35S) and GUS reporter gene. In the effector construct, the MYB96 gene was fused to the CaMV 35S promoter. Nos-T, Nos terminator.
(E) Transcriptional activation activity assays in Arabidopsis protoplasts. The expression constructs in (D) were expressed transiently in Arabidopsis protoplasts, and GUS activities were determined fluorimetrically. Luciferase gene expression was used to normalize the GUS activities. Three measurements were averaged (t test, *P < 0.01). Bars indicate se of the mean.
Chromatin immunoprecipitation (ChIP) assays were employed to confirm the binding of MYB96 to the gene promoters in 35S:96-MYC transgenic plants. Total proteins extracted from 35S:96-MYC transgenic plants were immunoprecipitated with an anti-MYC antibody. Quantitative real-time ChIP-PCR assays showed that the MYB96 protein binds to the gene promoters in planta in identical patterns to those observed in the electrophoretic mobility shift assays (Figure 9C; see Supplemental Figure 12 online).
We next performed transient β-glucuronidase (GUS) expression assays in Arabidopsis protoplasts to investigate further the MYB96 regulation of wax biosynthetic genes. KCS1-BS and -mBS DNA fragments were transcriptionally fused to a cauliflower mosaic virus (CaMV) 35S minimal promoter in the pCAMBIA1305.1 plasmid containing the GUS reporter gene, resulting in pKCS-P or pKCS-mP (Figure 9D). The reporter plasmids and an effector plasmid, p35S-MYB96, were cotransformed into Arabidopsis protoplasts. A vector construct containing the luciferase gene was included to monitor transformation efficiencies (Miura et al., 2007). The cotransformation of p35S-MYB96 with pKCS-P was found to elevate the reporter gene expression >6-fold (Figure 9E). By contrast, cotransformation with pKCS-mP did not elevate the reporter gene expression, confirming that MYB96 acts as a transcriptional activator of the KCS genes.
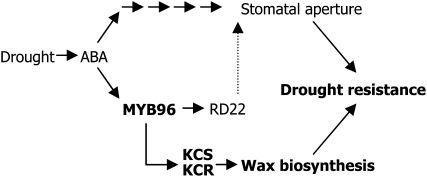
Altogether, our data demonstrate that the MYB96 transcription factor regulates directly the genes involved in cuticular wax biosynthesis under drought (Millar and Kunst, 1997; Kunst and Samuels, 2009), thus providing resistance to drought (Figure 10). This signaling pathway is distinct from the ABA signaling pathways governing stomatal regulation (Zhang et al., 2005; Cominelli et al., 2008; Kosma et al., 2009), although it is still possible that the MYB96 transcription factor contributes at least in part to stomatal regulation via the RD22 gene (Seo et al., 2009).
Figure 10.
Schematic Working Model of MYB96 Function in Cuticular Wax Biosynthesis under Drought Conditions.
MYB96 transcription is upregulated in response to ABA-mediated drought signals. The MYB96 protein activates the cuticular wax biosynthetic genes by binding directly to the gene promoters, resulting in accumulation of cuticular waxes. MYB96 may also contribute to drought resistance via the RD22 pathway by regulating stomatal aperture (Seo et al., 2009).
DISCUSSION
It has been documented that biosynthesis and accumulation of cuticular waxes are closely linked with drought resistance responses in many plant species. Water deficit and osmotic stresses induce significant increases in the amount of leaf cuticular lipids (Kosma et al., 2009). Cuticular wax accumulation is elevated by ~75% under osmotic stress conditions, primarily by accumulation of wax alkanes, in Arabidopsis (Kosma et al., 2009). Accumulation of cuticular waxes under osmotic stress or water deficit conditions have also been observed in other plant species, such as tobacco (Nicotiana tabacum), sesame (Sesamum indicum), and soybean (Glycine max) (Cameron et al., 2006; Kim et al., 2007a, 2007b), suggesting that cuticular wax accumulation is an important stress adaptation strategy that minimizes cellular and organismal dehydration under drought stress conditions in plants (Kosma et al., 2009).
To date, ~190 genes have been implicated in cuticular wax biosynthesis and processing in Arabidopsis (Beisson et al., 2003; Li-Beisson et al., 2010), and some of them have been biochemically characterized in several plant species (Aarts et al., 1995; Todd et al., 1999; Fiebig et al., 2000; Rowland et al., 2007; Samuels et al., 2008; Beaudoin et al., 2009; Lee et al., 2009b). In addition, several transcription factors have been implicated in the functional relationship between cuticle formation and leaf surface permeability. It has been observed that Arabidopsis transgenic plants overexpressing a plant-specific AP2/EREBP transcription factor gene SHINE1/WIN1 accumulate cuticular waxes and show enhanced drought resistance (Aharoni et al., 2004). Furthermore, leaf ultrastructures, such as epidermal cell structure, trichome number, and stomatal index that contribute to drought resistance, have also been altered in the transgenic plants. The soybean AP2/EREBP transcription factors WXP1 and WXP2 also affect cuticular wax accumulation. Cuticular waxes accumulate to a high level in Arabidopsis transgenic plants overexpressing either WXP1 or WXP2 that exhibit a drought-resistant phenotype (Zhang et al., 2005, 2007). A MYB transcription factor has been also implicated in cuticle-mediated drought resistance. Overexpression of MYB41 influences leaf surface permeability by altering cuticle biosynthesis (Cominelli et al., 2008). However, the gene regulatory networks and underlying molecular mechanisms governing cuticular wax biosynthesis have been unexplored at the molecular level in most cases.
Here, we found that the MYB96 transcription factor, which is a part of an ABA signaling cascade in the drought resistance response (Seo et al., 2009), modulates cuticular wax biosynthesis in response to drought by regulating cuticular wax biosynthetic genes. The MYB96 transcription factor binds directly to conserved sequences existing in the promoters of the cuticular wax biosynthetic genes and transcriptionally activates the target genes. These observations are consistent with altered cuticular wax loads in the MYB96-overexpressing and myb96-1 mutant plants. It has been observed that ABA induces accumulation of cuticular waxes, although it is possible that cuticular wax biosynthesis is also induced by abiotic stresses in an ABA-independent manner (Kosma et al., 2009). We found that the cuticular wax biosynthetic genes regulated by MYB96 are also induced by ABA and drought stress. Furthermore, our results demonstrate that the inductive effects of ABA and drought stress are, at least in part, dependent on MYB96, confirming that MYB96 regulates cuticular wax biosynthesis via ABA-mediated drought stress signaling.
It has been shown that changes in cuticular wax accumulation and composition influence epidermal properties (Gray et al., 2000; Aharoni et al., 2004). In addition, the effects of cuticular transpiration on drought resistance are relatively smaller than those of stomatal transpiration (Kerstiens, 1996). Therefore, another possibility is that the drought-resistant and -susceptible phenotypes of the myb96-1D and myb96 mutants, respectively, would be caused by alterations in epidermal properties rather than by changes in cuticular transpiration. We previously observed that stomatal density and index and trichome morphology and distribution are not altered in the myb96-1D and myb96-1 mutants (Seo et al., 2009). We found that cuticular transpiration is detectably reduced in the drought-resistant myb96-1D leaves but increased in the drought-susceptible myb96-1 leaves. The previous and our own data support that MYB96 does not affect leaf epidermal properties. We therefore believe that cuticular wax accumulation is associated intimately with drought resistance, in which MYB96 plays a role by modulating cuticular wax biosynthesis. However, it is still possible that certain aspects of stomatal properties, such as microstructures and assembly, would be altered due to changes in cuticular wax accumulation and composition in the mutants. This uncertainty could be clarified by measuring stomatal conductance in the leaves of Columbia-0 (Col-0) and mutant plants grown under normal and drought conditions.
It is interesting that MYB96 also plays a role in pathogen resistance (Seo and Park, 2010). Cuticular waxes modulate plant–microbe interactions by providing mechanical strength and viscoelastic properties (Baker et al., 1982; Lee et al., 2009a). Therefore, it is likely that MYB96 promotes pathogen resistance by inducing cuticular wax accumulation. Notably, it has been observed that the salicylic acid (SA) content is elevated and transcription of the SA biosynthetic SA INDUCTION DEFICIENT2 gene is upregulated in the myb96-1D mutant (Seo and Park, 2010). It is yet unknown whether SA biosynthesis and cuticular wax accumulation are interrelated or not. SA biosynthesis may be induced indirectly because the disease resistance response is triggered due to cuticular wax accumulation in the mutant. Alternatively, the two processes may be independently regulated by MYB96.
Cutin biosynthesis and deposition are also influenced by drought stress. Endogenous contents of cutin monomers are elevated in Arabidopsis leaves that are exposed to drought conditions (Kosma et al., 2009). However, contents of cutin monomers were not discernibly changed in the MYB96-overexpressing and deficient mutant plants, and cutin monomer biosynthetic genes were not regulated by MYB96. It is notable that the levels of cutin monomers are largely unaffected by ABA treatments (Kosma et al., 2009). Previous and our own data support that MYB96 is not directly involved in cutin biosynthesis.
We found that MYB96 regulates the KCS genes by directly binding to the gene promoters. Transcription of the KCS genes was upregulated in the myb96-1D mutant but downregulated in the myb96-1 mutant. Roles of several KCS enzymes have been reported in fatty acid chain elongation. Among the 22 KCS isoforms identified in Arabidopsis (Beisson et al., 2003; Li-Beisson et al., 2010), KCS1, KCS2/DAISY, KCS6/CER6/CUT1, KCS5/CER60, and KCS20 have been shown to be involved in cuticular wax biosynthesis (Millar and Kunst, 1997; Millar et al., 1999; Todd et al., 1999; Fiebig et al., 2000; Clemens and Kunst, 2001; Hooker et al., 2002; Lee et al., 2009b), although their biochemical activities are not well understood. Of particular interest is KCS2. It is induced by osmotic stress and ABA (Lee et al., 2009b). In addition, the KCS2 enzyme is involved in the VLCFA biosynthesis required for suberin accumulation in the root endodermis and seed chalaza-micropyle (Franke et al., 2009; Lee et al., 2009b). It is supposed that the KCS2-mediated regulation of cuticular wax biosynthesis via MYB96 may be related with the role of MYB96 in lateral root development and drought resistance response (Seo et al., 2009).
Genes encoding four major components of the fatty acid elongation complex, including KCS, KCR, ECR, and 3-hydroxyacyl-CoA dehydratase (PAS2), are transcriptionally upregulated in the myb96-1D mutant. Those enzymes involved in the subsequent reactions, such as CER3 and MAH1 in the decarbonylation pathway and CER4 and WSD1 in the acyl reduction pathway (Rowland et al., 2006, 2007; Greer et al., 2007; Li et al., 2008), are also induced in the mutant. In addition, WBC11, which encodes a plasma membrane–localized ABC transporter functioning as the exporter of cutin monomers and waxes (Bird et al., 2007; Luo et al., 2007; Panikashvili et al., 2007; Ukitsu et al., 2007), is also regulated in a similar manner by MYB96. Furthermore, additional genes encoding LTP and ABC transporters are also upregulated in the myb96-1D mutant. We therefore propose that the MYB96 transcription factor may act as a master transcriptional regulator for cuticular wax biosynthesis and accumulation in response to drought.
Alkane biosynthesis is one of the key responses of plants under osmotic stress conditions (Kosma et al., 2009). Alkanes, primary alcohols, and aldehydes are known to confer resistance to water movement through artificial membranes more efficiently than do fatty acids. It has been shown that alkanes constitute the most impermeable crystalline barrier against water movement (Grncarevic and Radler, 1967; Kosma et al., 2009). Consistent with the role of alkanes serving as a barrier against water movement, alkane contents were dramatically elevated in the drought-resistant myb96-1D mutant but reduced in the drought-susceptible myb96-1 mutant. It is particularly noteworthy that endogenous contents of alkanes, a major constituent of gasoline and diesel, were increased >8-fold in the myb96-1D mutant, suggesting that our findings would be applicable to engineering of alkane mass production in terrestrial plants (Jetter and Kunst, 2008; Schirmer et al., 2010).
METHODS
Plant Materials and Growth Conditions
All Arabidopsis thaliana lines used were in the Col-0 background. Plants were grown in a controlled culture room or on half-strength Murashige and Skoog (MS)-agar plates (hereafter referred to as MS-agar plates) at 22°C under long-day conditions (16 h light and 8 h dark). White light illumination (120 μmol photons m−2 s−1) was provided by fluorescent FLR40D/A tubes (Osram). The activation-tagged myb96-1D and T-DNA insertional myb96-1 mutants have been described previously (Seo et al., 2009).
The primers used for subcloning of the MYB96 gene under the control of a β-estradiol–inducible promoter were MYB96 (pER8)-F and MYB96 (pER8)-R (see Supplemental Table 1 online). The PCR product was subcloned into the XhoI and PacI sites of the pER8 vector (Zuo et al., 2000). Two-week-old transformants grown on MS-agar plates were used for induction of the MYB96 gene by 10 μM β-estradiol.
To examine the effects of growth hormones and stress conditions on gene expression, 2-week-old plants grown on MS-agar plates were transferred to MS liquid cultures supplemented with 20 μM ABA or 100 μM SA and incubated for 6 h. To examine the effects of drought, plants grown in soil for 20 d were subject to drought treatment by halting watering. To examine the effects of high salinity, plants were soaked in MS liquid cultures containing 150 mM NaCl and incubated for 6 h. For cold treatments, plants were exposed to 4°C for 24 h. For flagellin22 (flg22) treatments, plants were soaked in MS liquid cultures containing 5 μM flg22 and incubated for 24 h.
Microarray Assays
Two-week-old whole plants grown on MS-agar plates at 22°C under long-day conditions (16 h light and 8 h dark) were used for extracting total RNAs using the RNeasy plant mini kit (Qiagen). The total RNA samples were pretreated with RNase-free DNase I and cleaned up using the Plant Total RNA isolation kit (Qiagen). Three independent RNA samples were assayed and statistically treated. Probe preparation, hybridization to the GeneChip Arabidopsis ATH1 Genome Arrays (Affymetrix), and subsequent processing steps were performed according to the manufacturer’s procedure. Global normalization and expression estimates were analyzed by GC-robust multiarray implemented in the bioconductor (http://www.bioconductor.org) and R software (Wu et al., 2004). Wild-type (Col-0) plant values were used as the baseline to calculate the intensity ratio/fold changes of the mutant plants. FDRs were calculated by significance analysis of microarrays algorithm (Tusher et al., 2001; Gentleman et al., 2004), and genes with fold change of at least 2 and a FDR-corrected P value lower than 0.05 were identified. The microarray data set is deposited into ArrayExpress with accession number E-MEXP-2965 at http://www.ebi.ac.uk/at-miamexpress.
Analysis of Transcript Levels
qRT-PCR reactions were performed in 96-well blocks with an Applied Biosystems 7500 Real-Time PCR system using the SYBR Green I master mix in a volume of 25 μL. The reactions were performed in biological triplicates using RNA samples extracted from three independent plant materials and gene-specific primers listed in Supplemental Table 1 online. Data processing and determination of the reaction specificities were performed as described previously (Seo et al., 2009).
Scanning Electron Microscopy and Nile Red Staining
For epicuticular wax crystal observation, the fourth rosette leaves or inflorescence stem segments from tip to 2 cm of 4- to 5-week-old plants grown in soil were examined by scanning electron microscopy as described previously (Lee et al., 2009b).
For Nile red staining, plant materials were incubated for 20 min in a 5 μg mL−1 Nile red solution. Stained samples were washed with deionized water and analyzed using a TCS SP5 AOBS/Tandem confocal laser scanning microscope (Leica). Nile red was excited with a 488-nm laser and collected with a 560- to 615-nm filter as described previously (Pighin et al., 2004).
Analysis of Cuticular Wax Composition and Loads
Cuticular waxes were extracted from the leaves (200 to 1000 mg) and stems (200 mg) of 4-week-old plants in chloroform for 30 s at room temperature. n-Octacosane, docosanoic acid, and 1-tricosanol were added to the extracted chloroform solvent as internal standards. The solvent was subsequently evaporated under a gentle stream of nitrogen and redissolved in a mixture of 100 μL of pyridine and 100 μL of bis-N,N-(trimethylsilyl)trifluoroacetamide. The wax mixtures were heated at 90°C for 30 min to convert waxes into trimethylsilyl derivatives. Qualitative and quantitative composition analyses were conducted as described previously (Lee et al., 2009a, 2009b). The P values from each comparison were corrected for multiple tests using FDR control (Benjamini and Hochberg, 1995).
Analysis of Cutin Polyester Monomers
Rosette leaves of 4-week-old plants grown in soil were used to quantify cutin polyester monomers. Methyl heptadecanoate and ω-pentadecalactone were added as internal standards into the delipidated and dried leaves and then depolymerized by hydrogenolysis with LiAlH4 or by methanolysis with NaOCH3. Cutin polyesters were analyzed by gas chromatography–mass spectrometry (GCMS-QP2010; Shimazu) with a HP-5 column (60 m, 0.32 mm inner diameter, film thickness 0.1 mm; Agilent). The analysis system was maintained at 110°C. The temperature was increased to 300°C at a rate of 2.5°C min−1 and maintained at 300°C for 3 min.
Electrophoretic Mobility Shift Assays
The MYB96 gene was subcloned into the pMAL-c2X Escherichia coli expression vector (NEB), which has an MBP coding sequence, using MYB96 (MBP)-F and MYB96 (MBP)-R primers (see Supplemental Table 1 online). The MBP-MYB96 fusion protein was purified according to the manufacturer’s procedure using the pMAL Protein Fusion and Purification System (#E8000S). DNA fragments were end-labeled with [γ-32P]dATP using T4 polynucleotide kinase. Labeled probes were then incubated with ~0.5 μg of the purified MBP- MYB96 protein for 30 min at 25°C in a binding buffer (10 mM Tris-HCl, pH 7.6, 50 mM NaCl, 1 mM EDTA, 5 mM DTT, and 5% glycerol) with or without competitor DNA fragments. The reaction mixtures were electrophoresed on 6% native PAGE gels. The gels were dried on Whatman 3MM paper and exposed to x-ray films.
ChIP Assays
A MYC-coding sequence was fused in frame to the 3′ end of the MYB96 gene, and the gene fusion was subcloned under the CaMV 35S promoter (Kim et al., 2006). The expression construct was transformed into Col-0 plants. Two-week-old 35S:96-MYC transgenic plants grown on MS-agar plates were used for extraction of total proteins. The processing of plant materials and qRT-PCR were performed as described previously (Lawrence et al., 2004). The qRT-PCR primers used are listed in Supplemental Table 2 online.
Transcriptional Activation Activity Assays
The MYB96 gene sequence was fused in frame to the 3′ end of the GAL4 DNA binding domain-coding sequence in the effector plasmid. The activity of the MYB96 transcription factor was examined by a GAL4 transient expression system using Arabidopsis protoplasts as described previously (Miura et al., 2007).
Chlorophyll Leaching Assays
Rosette leaves of 4-week-old plants grown in soil were used. The leaves were weighed, and ~2 g of each leaf sample was incubated on ice for 30 min and immersed in 30 mL of 80% ethanol in 50 mL conical tubes at room temperature. Aliquots of 100 μL were removed from the solution at every 10 min after initial immersion. The amount of extracted chlorophylls was quantified by measuring absorbance at 647 and 664 nm using a diode array spectrophotometer (WPA Biowave) as described previously (Lolle et al., 1998).
Accession Numbers
Sequence data from this article can be obtained from the Arabidopsis Genome Initiative database under the following accession numbers: MYB96 (At5g62470), KCS1 (At1g01120), KCS2 (At1g04220), KCS6 (At1g68530), KCR1 (At1g67730), CER1 (At1g02205), and CER3 (At5g57800). The microarray data set is deposited into ArrayExpress with accession number E-MEXP-2965 at http://www.ebi.ac.uk/at-miamexpress.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of MYB96 in Epidermal Cells of Stems.
Supplemental Figure 2. Epicuticular Wax Deposition in myb96-1 Stems.
Supplemental Figure 3. Cuticular Wax Composition in myb96-1 Leaves.
Supplemental Figure 4. Cuticular Wax Composition in myb96-1 Stems.
Supplemental Figure 5. Cutin Biosynthesis in myb96-1D Leaves.
Supplemental Figure 6. Expression of Cutin Biosynthetic Genes in myb96-1D and myb96-1.
Supplemental Figure 7. Rate of Chlorophyll Leaching in myb96-1D and myb96-1 Leaves.
Supplemental Figure 8. MYB96 Regulation of Wax Biosynthetic Genes in Response to ABA.
Supplemental Figure 9. Effects of MYB96 Induction on Wax Biosynthetic Gene Expression.
Supplemental Figure 10. Binding of MYB96 to the Consensus Sequences in the Promoters of Wax Biosynthetic Genes in Vitro.
Supplemental Figure 11. In Vitro Binding of MBP Alone to the Consensus Sequences.
Supplemental Figure 12. Direct Binding of MYB96 to the Promoters of Wax Biosynthetic Genes in Vivo.
Supplemental Table 1. Primers Used in qRT-PCR and Subcloning.
Supplemental Table 2. Primers Used in ChIP Assays.
Supplemental Data Set 1A. Upregulated Genes in the myb96-1D Mutant.
Supplemental Data Set 1B. Ontologies of Upregulated Genes in the myb96-1D Mutant.
Supplementary Material
Acknowledgments
We thank Fred Beisson and Yonghua Li-Beisson for scientific discussion. This work was supported by the Leaping Research Program (20100014373) provided by the National Research Foundation of Korea and by grants from the Plant Signaling Network Research Center (20100001457), the National Research Foundation of Korea (20100028147), and from the Agricultural R&D Promotion Center (309017-05-2-HD130), Korea Ministry for Food, Agriculture, Forestry, and Fisheries. It was also supported in part by the World Class University (R31-2009-000-20025-0 to M.C.S.) and Basic Research Programs (20090064298 to M.C.S.) from the National Research Foundation of Korea and a grant (PJ0067152010 to M.C.S.) from the Korean Rural Development Administration. P.J.S. was supported by the Seoul Science Fellowship. We thank Emily Wheeler for editorial assistance.
References
- Aarts M.G., Keijzer C.J., Stiekema W.J., Pereira A. (1995). Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7: 2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni A., Dixit S., Jetter R., Thoenes E., van Arkel G., Pereira A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.J., McCormick S.L., Bateman D.F. (1982). Effects of purified cutin esterase upon the permeability and mechanical strength of cutin membranes. Phytopathology 72: 420–423 [Google Scholar]
- Barthlott W., Neinhuis C. (1997). Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta 202: 1–8 [Google Scholar]
- Beaudoin F., Wu X., Li F., Haslam R.P., Markham J.E., Zheng H., Napier J.A., Kunst L. (2009). Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F., et al. (2003). Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Bird D., Beisson F., Brigham A., Shin J., Greer S., Jetter R., Kunst L., Wu X., Yephremov A., Samuels L. (2007). Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Cameron K.D., Teece M.A., Smart L.B. (2006). Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 140: 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Goodwin S.M., Boroff V.L., Liu X., Jenks M.A. (2003). Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot. 55: 225–236 [DOI] [PubMed] [Google Scholar]
- Christmann A., Moes D., Himmelbach A., Yang Y., Tang Y., Grill E. (2006). Integration of abscisic acid signalling into plant responses. Plant Biol. (Stuttg.) 8: 314–325 [DOI] [PubMed] [Google Scholar]
- Clemens S., Kunst L., inventors (February 1, 2001). A plant long chain fatty acid biosynthetic enzyme. International Patent Application No. WO 01/07586 [Google Scholar]
- Cominelli E., Sala T., Calvi D., Gusmaroli G., Tonelli C. (2008). Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53: 53–64 [DOI] [PubMed] [Google Scholar]
- Eigenbrode S.D., Espelie K.E. (1995). Effects of plant epicuticular lipids on insect herbivores. Annu. Rev. Entomol. 40: 171–194 [Google Scholar]
- Fiebig A., Mayfield J.A., Miley N.L., Chau S., Fischer R.L., Preuss D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R., Höfer R., Briesen I., Emsermann M., Efremova N., Yephremov A., Schreiber L. (2009). The DAISY gene from Arabidopsis encodes a fatty acid elongase condensing enzyme involved in the biosynthesis of aliphatic suberin in roots and the chalaza-micropyle region of seeds. Plant J. 57: 80–95 [DOI] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J.E., Holroyd G.H., van der Lee F.M., Bahrami A.R., Sijmons P.C., Woodward F.I., Schuch W., Hetherington A.M. (2000). The HIC signalling pathway links CO2 perception to stomatal development. Nature 408: 713–716 [DOI] [PubMed] [Google Scholar]
- Greer S., Wen M., Bird D., Wu X.M., Samuels L., Kunst L., Jetter R. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145: 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grncarevic M., Radler F. (1967). The effect of wax components on cuticular transpiration-model experiments. Planta 75: 23–27 [DOI] [PubMed] [Google Scholar]
- Hooker T.S., Millar A.A., Kunst L. (2002). Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetter R., Kunst L. (2008). Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. Plant J. 54: 670–683 [DOI] [PubMed] [Google Scholar]
- Joubès J., Raffaele S., Bourdenx B., Garcia C., Laroche-Traineau J., Moreau P., Domergue F., Lessire R. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67: 547–566 [DOI] [PubMed] [Google Scholar]
- Kang J.Y., Choi H.I., Im M.Y., Kim S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannangara R., Branigan C., Liu Y., Penfield T., Rao V., Mouille G., Höfte H., Pauly M., Riechmann J.L., Broun P. (2007). The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstiens G. (1996). Cuticular water permeability and its physiological significance. J. Exp. Bot. 47: 1813–1832 [Google Scholar]
- Kim K., Park S., Jenks M. (2007a). Influence of water deficit on leaf cuticular waxes of soybean. Int. J. Plant Sci. 168: 307–316 [Google Scholar]
- Kim K.S., Park S.H., Jenks M.A. (2007b). Changes in leaf cuticular waxes of sesame (Sesamum indicum L.) plants exposed to water deficit. J. Plant Physiol. 164: 1134–1143 [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Kim S.G., Park J.E., Park H.Y., Lim M.H., Chua N.H., Park C.M. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18: 3132–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy P.E. (2001). Polyesters in higher plants. Advances in Biochemical Engineering Biotechnology: Biopolyesters, Babel W., Steinbuechel A., (Berlin: Springer-Verlag: ), pp. 1–49 [DOI] [PubMed] [Google Scholar]
- Kosma D.K., Bourdenx B., Bernard A., Parsons E.P., Lü S., Joubès J., Jenks M.A. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Samuels A.L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Kunst L., Samuels L. (2009). Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12: 721–727 [DOI] [PubMed] [Google Scholar]
- Lawrence R.J., Earley K., Pontes O., Silva M., Chen Z.J., Neves N., Viegas W., Pikaard C.S. (2004). A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol. Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Lee S.B., Go Y.S., Bae H.J., Park J.H., Cho S.H., Cho H.J., Lee D.S., Park O.K., Hwang I., Suh M.C. (2009a). Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol. 150: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.B., Jung S.J., Go Y.S., Kim H.U., Kim J.K., Cho H.J., Park O.K., Suh M.C. (2009b). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60: 462–475 [DOI] [PubMed] [Google Scholar]
- Li F., Asami T., Wu X., Tsang E.W., Cutler A.J. (2007). A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 145: 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wu X., Lam P., Bird D., Zheng H., Samuels L., Jetter R., Kunst L. (2008). Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 148: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., et al. (2010). Acyl-lipid metabolism. The Arabidopsis Book 8: e0133, doi/10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle S.J., Hsu W., Pruitt R.E. (1998). Genetic analysis of organ fusion in Arabidopsis thaliana. Genetics 149: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B., Xue X.Y., Hu W.L., Wang L.J., Chen X.Y. (2007). An ABC transporter gene of Arabidopsis thaliana, AtWBC11, is involved in cuticle development and prevention of organ fusion. Plant Cell Physiol. 48: 1790–1802 [DOI] [PubMed] [Google Scholar]
- Millar A.A., Clemens S., Zachgo S., Giblin E.M., Taylor D.C., Kunst L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11: 825–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.A., Kunst L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12: 121–131 [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.J., Hasegawa P.M. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillon J.M., Eriksson S.K., Harryson P. (2008). Mimicking the plant cell interior under water stress by macromolecular crowding: Disordered dehydrin proteins are highly resistant to structural collapse. Plant Physiol. 148: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. (2006). Unraveling the complex network of cuticular structure and function. Curr. Opin. Plant Biol. 9: 281–287 [DOI] [PubMed] [Google Scholar]
- Panikashvili D., Savaldi-Goldstein S., Mandel T., Yifhar T., Franke R.B., Höfer R., Schreiber L., Chory J., Aharoni A. (2007). The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiol. 145: 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin J.A., Zheng H., Balakshin L.J., Goodman I.P., Western T.L., Jetter R., Kunst L., Samuels A.L. (2004). Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Pollard M., Beisson F., Li Y., Ohlrogge J.B. (2008). Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Post-Beittenmiller D. (1996). Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 405–430 [DOI] [PubMed] [Google Scholar]
- Raffaele S., Vailleau F., Léger A., Joubès J., Miersch O., Huard C., Blée E., Mongrand S., Domergue F., Roby D. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reicosky D.A., Hanover J.W. (1978). Physiological effects of surface waxes: I. Light reflectance for glaucous and nonglaucous Picea pungens. Plant Physiol. 62: 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M., Schreiber L. (2001). Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 52: 2023–2032 [DOI] [PubMed] [Google Scholar]
- Rowland O., Lee R., Franke R., Schreiber L., Kunst L. (2007). The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581: 3538–3544 [DOI] [PubMed] [Google Scholar]
- Rowland O., Zheng H.Q., Hepworth S.R., Lam P., Jetter R., Kunst L. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels L., Kunst L., Jetter R. (2008). Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59: 683–707 [DOI] [PubMed] [Google Scholar]
- Sanchez J.P., Chua N.H. (2001). Arabidopsis PLC1 is required for secondary responses to abscisic acid signals. Plant Cell 13: 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer A., Rude M.A., Li X., Popova E., del Cardayre S.B. (2010). Microbial biosynthesis of alkanes. Science 329: 559–562 [DOI] [PubMed] [Google Scholar]
- Schnurr J., Shockey J., Browse J. (2004). The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J.I., Kwak J.M., Allen G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410: 327–330 [DOI] [PubMed] [Google Scholar]
- Seki M., Umezawa T., Urano K., Shinozaki K. (2007). Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 10: 296–302 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Park C.M. (2010). MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186: 471–483 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Xiang F., Qiao M., Park J.Y., Lee Y.N., Kim S.G., Lee Y.H., Park W.J., Park C.M. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K., Seki M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Sirichandra C., Wasilewska A., Vlad F., Valon C., Leung J. (2009). The guard cell as a single-cell model towards understanding drought tolerance and abscisic acid action. J. Exp. Bot. 60: 1439–1463 [DOI] [PubMed] [Google Scholar]
- Solovchenko A., Merzlyak M. (2003). Optical properties and contribution of cuticle to UV protection in plants: Experiments with apple fruit. Photochem. Photobiol. Sci. 2: 861–866 [DOI] [PubMed] [Google Scholar]
- Suh M.C., Samuels A.L., Jetter R., Kunst L., Pollard M., Ohlrogge J., Beisson F. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139: 1649–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J., Post-Beittenmiller D., Jaworski J.G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17: 119–130 [DOI] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis–element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukitsu H., et al. (2007). Cytological and biochemical analysis of COF1, an Arabidopsis mutant of an ABC transporter gene. Plant Cell Physiol. 48: 1524–1533 [DOI] [PubMed] [Google Scholar]
- Urao T., Yamaguchi-Shinozaki K., Urao S., Shinozaki K. (1993). An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K., Durst F., Pinot F., Benveniste I., Nettesheim K., Wisman E., Steiner-Lange S., Saedler H., Yephremov A. (2001). Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega -hydroxylation in development. Proc. Natl. Acad. Sci. USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99: 909–917 [Google Scholar]
- Xiao F., Goodwin S.M., Xiao Y., Sun Z., Baker D., Tang X., Jenks M.A., Zhou J.M. (2004). Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 23: 2903–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., Broeckling C.D., Blancaflor E.B., Sledge M.K., Sumner L.W., Wang Z.Y. (2005). Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42: 689–707 [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., Broeckling C.D., Sumner L.W., Wang Z.Y. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64: 265–278 [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.