This work presents a high-resolution time-course analysis of gene expression during development of a leaf from expansion through senescence. Enrichment in ontologies, sequence motifs, and transcription factor families within genes showing altered expression over time identified both metabolic pathways and potential regulators active at different stages of leaf development and senescence.
Abstract
Leaf senescence is an essential developmental process that impacts dramatically on crop yields and involves altered regulation of thousands of genes and many metabolic and signaling pathways, resulting in major changes in the leaf. The regulation of senescence is complex, and although senescence regulatory genes have been characterized, there is little information on how these function in the global control of the process. We used microarray analysis to obtain a high-resolution time-course profile of gene expression during development of a single leaf over a 3-week period to senescence. A complex experimental design approach and a combination of methods were used to extract high-quality replicated data and to identify differentially expressed genes. The multiple time points enable the use of highly informative clustering to reveal distinct time points at which signaling and metabolic pathways change. Analysis of motif enrichment, as well as comparison of transcription factor (TF) families showing altered expression over the time course, identify clear groups of TFs active at different stages of leaf development and senescence. These data enable connection of metabolic processes, signaling pathways, and specific TF activity, which will underpin the development of network models to elucidate the process of senescence.
INTRODUCTION
During leaf senescence, the plant recovers and recycles valuable nutrient components that have been incorporated during growth that would otherwise be lost when the leaf dies or is shed. Efficient senescence is essential to maximize viability in the next season or generation, but premature senescence, a protective mechanism employed when plants are stressed, results in reduced yield and quality of crop plants. During the senescence process, viability of cells within the leaf is actively maintained until maximum remobilization has occurred (Hörtensteiner and Feller, 2002). This requires meticulous control of each step of the process, regulated by internal and external signals via a series of interlinking signaling pathways involving gene expression changes and influenced by the balance of hormones and metabolites. Thus, senescence is a very complex process involving the expression of thousands of genes and many signaling pathways (Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006). Elucidation of the relative influences of each pathway and the crosstalk between them is crucial in identifying the key regulatory genes that control senescence.
To date, genes with a role in leaf senescence have been identified either by forward genetic screening to find mutants with altered senescence rates followed by cloning of the genes involved or by using reverse genetics for functional analysis of genes that show differential expression during senescence (reviewed by Lim et al., 2007). Many of these altered senescence phenotypes occur as a result of altered hormone signaling, such as reduced ethylene signaling (Grbic and Bleecker, 1995) or increased cytokinin signaling (Kim et al., 2006), both of which result in delayed senescence. However, traditional molecular biology approaches in which one gene or mutant at a time is identified and analyzed have resulted in interesting information but have generally failed to reveal a global picture of senescence regulatory networks, including likely feed-forward, feedback, and crosstalk mechanisms. To understand a system as complex as senescence, where the influence of many external and internal signals is balanced to allow controlled disassociation and dispersal of cellular components, it is essential to study the system in its entirety rather than focus on small parts. The first step in this global analysis is to identify the dynamic changes that are occurring in transcript levels as senescence progresses. Obviously, transcripts are only one part of the regulatory process; factors such as RNA stability, translation rates, protein processing and stability, metabolite concentrations, and many others will have essential roles in the fine-scale moderation of cellular activity. However, transcription plays a key role in regulating both senescence and hormone signaling; therefore, identification of regulatory networks based on transcript levels is an ideal starting point in identifying key switch points in senescence.
Here, we use high-resolution time series microarray data, collected over many time points during the development of the leaf, to identify and characterize the gene expression changes during the different steps that make up the senescence process. The resulting detailed measurement of transcript levels for 22 time points during the developmental process is highly valuable for the investigation of numerous complex processes, such as the discovery of metabolic pathway switches, the identification of key regulatory genes that are active at different time points, and the inference of gene regulatory networks. Analysis of the expression patterns has enabled us to propose a detailed chronology of transcriptional and functional changes during leaf senescence. Promoter motif and transcription factor (TF) analysis has revealed a progression of regulatory genes that influence gene expression at different times during development. Finally, a preliminary model, generated with selected genes from the array data, is presented to illustrate the value of this data set for future network inference.
RESULTS
Growth and Biochemical Changes during Senescence
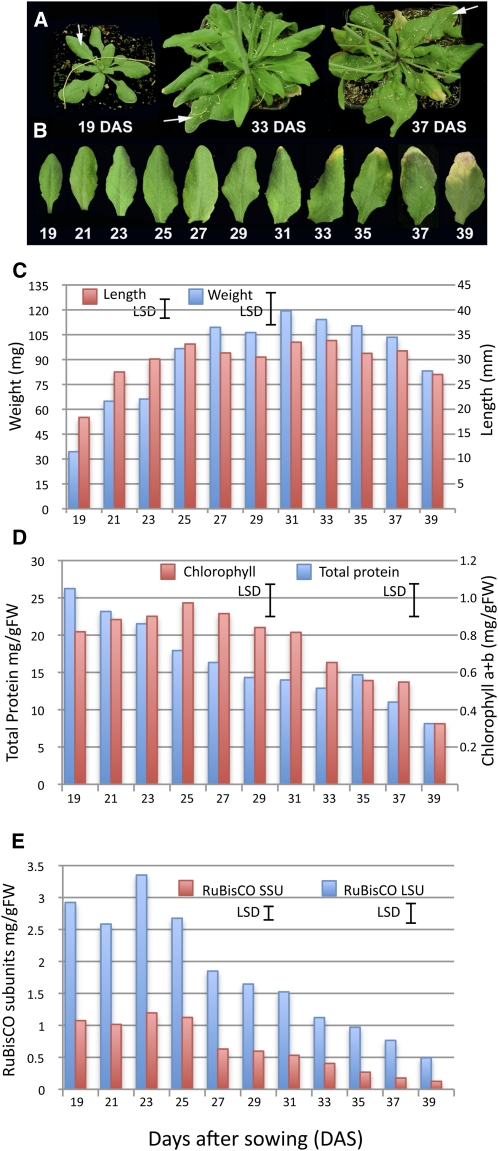
All measurements in this study were made on samples collected from leaf 7, chosen because senescence and mobilization of constituents from this leaf occur concurrently with flower development and silique filling in our growth conditions and are thus likely to be controlled by developmental signals. Each sample was harvested from an independent Arabidopsis thaliana plant at each time point (Figure 1A), and samples were not pooled for any of the analyses. Arabidopsis Col-0 plants were grown in controlled conditions until leaf 7 was ~50% of its final size (19 d after sowing [DAS]). This leaf was harvested at defined time points until 39 DAS when it was visibly senescent (~50% of leaf area being yellow, Figure 1B). Samples were taken in the morning (7 h into the light period) and afternoon (14 h into the light period) every other day, resulting in 22 time points in total. Sampling was carried out at these two time points each day to allow us to distinguish genes that are altered in a diurnal rhythm, as well as being differentially expressed over time; the times were selected based on likely maximum changes in expression. Plants started flowering from around 21 DAS. Leaf 7 started to show yellowing at the tip at around 31 DAS and was 25 to 50% yellow by 37 DAS. By the final sample time (39 DAS), the plants were fully flowering, and siliques were filling. Physiological parameters were measured in the morning samples only (i.e., 11 time points). Sampled leaves reached full expansion by 23 DAS (Figure 1C). However, leaf weight increased significantly between time points up to 25 DAS (P < 0.01) and continued to increase, reaching a maximum at 31 DAS when the first signs of yellowing were visible and then declined rapidly after 37 DAS (P < 0.05). Once the leaf is fully expanded, weight may continue to increase due to synthesis of macromolecules, expansion of organelles, and water uptake. Similarly, loss of fresh weight is primarily due to the decline in macromolecules and the loss of water as the leaf begins to dry.
Figure 1.
Plant Growth Parameters and Protein and Chlorophyll Measurements.
(A) Appearance of the Arabidopsis plants at three different stages of development, 19, 33, and 37 DAS. White arrows indicate leaf 7, the sampled leaf from each plant.
(B) An example of leaf 7 harvested from plants at 19 to 39 DAS (picture shows the morning sample only).
(C) Length (mm, red bars) and weight (mg, blue bars) of the sampled leaves over the time course. Least significant differences (LSD; 5%, 71 [length] and 99 [weight] df) calculated based on the minimum sample size of 6 (length) and consistent sample size of 10 (weight) for comparing pairs of means is shown for each variable, calculated from the ANOVA.
(D) Total protein (blue bars) and chlorophyll a+b (red bars) levels were measured in leaf samples at each stage of development. LSD (5%, 42 [protein] and 43 [chlorophyll] df) is shown for both variables. FW, fresh weight.
(E) Levels of the large (LSU) and small (SSU) subunits of Rubisco were estimated from stained polyacrylamide gels. LSD (5%, 42 df) is shown for both variables. Values shown in (D) and (E) represent the means of five independent biological replicates per time point.
Protein and chlorophyll levels are often used as markers for the progression of senescence since both are degraded during the senescence process. Levels of total chlorophyll and protein were measured (Figure 1D). Chlorophyll levels did not change significantly until after 31 DAS, when levels started to fall (P < 0.001 from maximum). However, relative protein levels started to drop considerably earlier at 23 DAS (P < 0.05 from maximal), which is before the time at which maximum leaf weight is reached. This implies that the leaf weight increase seen up to 31 DAS is not due to new protein synthesis but is probably due to increased water content and possibly continuing increases in cell wall density and membrane and other structural developments. Levels of the small and large subunit (SSU and LSU, respectively) of the photosynthetic carbon-fixation enzyme ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) increased to maximum at 23 DAS (LSU) and 25 DAS (SSU) and then fell steadily during senescence (P < 0.001 from maximum; Figure 1E). Rubisco is abundant in a mature green leaf and has been suggested to have some role as a storage protein (Staswick, 1994). Early degradation of this protein may provide building blocks for synthesis of additional proteins required for senescence without affecting the rate of photosynthesis.
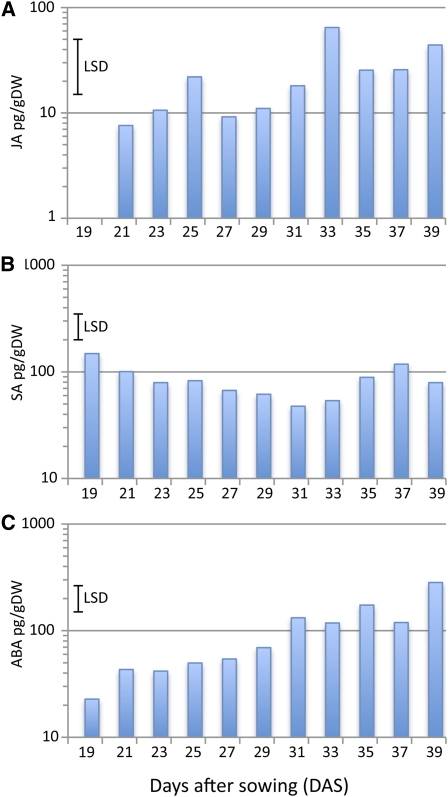
Senescence results in activation of signaling pathways involving the stress-related plant hormones salicylic acid (SA), jasmonic acid (JA), and ABA (Weaver et al., 1998; Morris et al., 2000; He et al., 2002). Levels of these three hormones were measured in the leaf 7 developmental time series and showed phased increases during senescence (Figure 2). SA levels were high in immature leaves, gradually decreased to minimal levels at 31 DAS (P < 0.001 from initial maximum) and then rose significantly (P < 0.05) from a relatively late stage (35 DAS). ABA levels significantly increased earlier at around 31 DAS (P < 0.05), with a subsequent increase to maximum at 39 DAS; JA levels showed a complex pattern with peaks at 25, 33, and 39 DAS.
Figure 2.
Hormone Levels during Leaf Development.
Levels of JA (A), SA (B), and ABA (C) were measured in leaf 7 harvested at different times during plant development. LSD (5%, 41 df) for comparisons between pairs of means are shown for each hormone, calculated from the ANOVA of log10-transformed data. Values represent the means of five independent biological replicates per time point. DW, dry weight.
[See online article for color version of this figure.]
Microarray Analysis over Multiple Time Points Identified Thousands of Differentially Expressed Genes
Four biological replicates for each time point were used for RNA isolation (88 samples in total), each hybridized as four technical replicates to the two channel microarrays. The resulting gene expression profiles were analyzed, and time point means were extracted using a local adaptation of the MAANOVA (MicroArray ANalysis Of VAriance) package, which quality checks and normalizes the data and produces data files containing predicted means for each gene; in essence, a single normalized value for each gene for each biological replicate measured at each time point (Wu et al., 2003; Churchill, 2004). Two different data sets were obtained in this way following the completed analysis: the first contained predicted mean values for each of the four biological replicates at each of the 22 distinct time points, therefore including time-of-day variation, whereas the second contained predicted mean values for eight biological replicates for each of 11 d, the values calculated by omitting the time-of-day and day/time-of-day interaction effects from the fitted model. These data sets were both used in the further analysis described below.
F tests, constructed from the variance estimates obtained from the MAANOVA model-fitting process, were used to assess each gene for significant changes in gene expression between time points. The model fitting allowed separate assessments of the variation due to differences between days (averaged across time-of-day samples), differences between time of day (averaged across days), and the interaction between these terms. Significance levels for all tests were adjusted across genes for multiple testing using a step-down false discovery rate (FDR) controlling procedure (Westfall et al., 1998; Benjamini and Liu, 1999), resulting in 8878 genes showing significant (P < 0.05) variation due to day of sampling (19–39 DAS). Additional genes were identified as showing significant (P < 0.05) variation due to the time of day or the interaction between day of sampling and the time of day, and the numbers of genes having significant test results for combinations of these terms are summarized in a Venn diagram, together with sample expression profiles for each combination (see Supplemental Figure 1 online). The selection process that was used to identify the list of differentially expressed genes used in all further analyses is described in the Methods section and combined information about the adjusted significance levels of the statistical tests with visual examination of expression patterns. The final list of genes used for the analysis described below contains probes for 6323 genes (see Supplemental Data Set 1 online).
We have generated a web tool that illustrates the expression levels of each individual probe on the Complete Arabidopsis thaliana MicroArray (CATMA; Allemeersch et al., 2005) array using the two alternative summaries of the senescence data. Expression patterns for each gene in the 22– and 11–time point data can be viewed in the Data section at http://go.warwick.ac.uk/presta.
Clustering Genes by Expression Pattern Illustrates the Extensive Metabolic Changes Occurring during Leaf Senescence
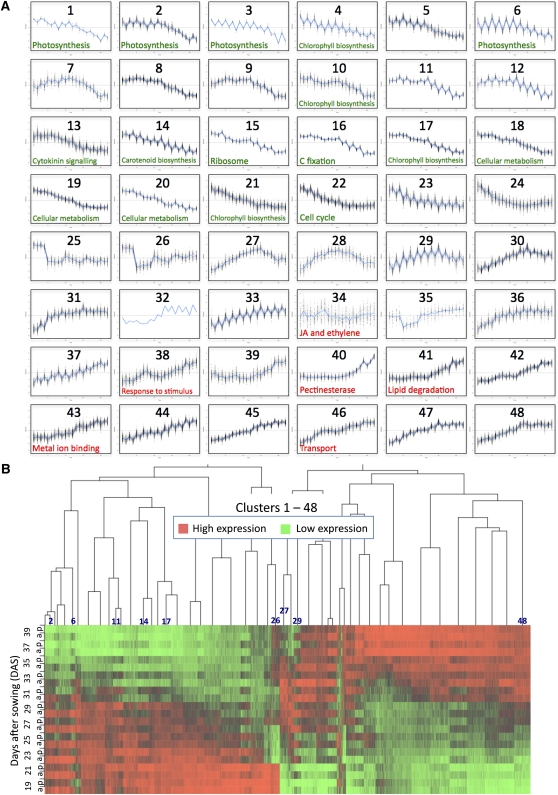
The 6323 differentially expressed genes were clustered using the time series-clustering software SplineCluster (Heard et al., 2006). Clustering analysis of both the 22– and 11–time point data was performed, and 48 and 74 clusters were obtained respectively. Supplemental Data Set 1 online shows the cluster number for each differentially expressed gene in both the 11– and 22–time point clusters. For reasons of space, only the 22–time point clusters are analyzed in this article (Figure 3A). The heat map (Figure 3B) indicates that there are changes in gene expression at each time point but that there are several time points at which an obvious step change in the transcriptome occurs. Overall, the major switch in gene expression in leaf 7, both in genes upregulated and downregulated, occurs between 29 and 33 DAS, and the genes identified as differentially expressed can be divided into two major groups, genes in clusters 1 through 24, which are downregulated during this period, and genes in clusters 27 through 48, which are upregulated. Some of the clusters in the center of the heat map show a more complex pattern; for example, cluster 26 genes are downregulated initially and then increase in expression, and genes in clusters 27, 28, and 29 show an initial increase followed by a decrease in expression (Figures 3A and 3B).
Figure 3.
SplineCluster Analysis of Differentially Expressed Genes.
SplineCluster analysis was performed on the 22–time point data using normalized data for the 6323 differentially expressed gene probes (average of the four biological replicates).
(A) Forty-eight clusters were obtained. The blue line on each plot represents the mean expression profile for the cluster. The genes present in each cluster may be viewed in Supplemental Data Set 1 online. Selected enriched GO terms (data shown in Supplemental Table 1 online) are indicated, in green for downregulated and red for upregulated genes.
(B) The heat map illustrates the expression profiles for genes in each cluster, with red representing high expression and green representing low expression. Morning (a) and afternoon (p) data are shown for 19 to 39 DAS. Darker shades show intermediate levels of expression. A few cluster positions are identified to compare with the cluster profiles shown in (A).
A clear diurnal variation in expression is seen with many of the differentially expressed genes, which show higher expression in either the morning or the afternoon samples from the same sample day. Other genes show a distinct morning to night rhythm that did not alter significantly over the 22 d of the experiment. These genes were not selected as being differentially expressed over time but were identified in the MAANOVA analysis as being significantly affected by the time of day term only (1086 genes; see Supplemental Figure 1 online). These genes show clearly that the clock does not decline as late senescence is reached. Morning genes include well-characterized clock genes such as LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK ASSOCIATED1 (CCA1), and PSEUDO-RESPONSE REGULATOR 7 (PRR7; Harmer, 2009) as well as genes involved in light signaling such as PHYTOCHROME A (PHYA), CRYPTOCHROME 1 (CRY1), and PIF4, a phytochrome-interacting factor (see Supplemental Figure 2A online). Afternoon genes include EARLY FLOWERING 4 (ELF4) and PHYTOCLOCK1 (PCL1), both of which are negatively regulated by CCA1 and LHY (Hazen et al., 2005; Kikis et al., 2005; see Supplemental Figure 2B online).
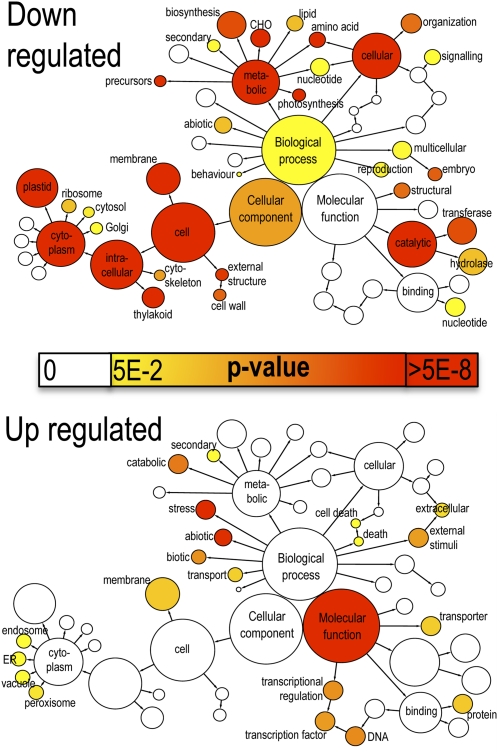
The 48 clusters of genes identified from SplineCluster analysis of the 22–time point data were analyzed using the gene ontology (GO) enrichment tool BiNGO (Maere et al., 2005). Initially, the two groups of genes showing either decreasing (clusters 1–24) or increasing (clusters 27–48) expression during leaf development were analyzed for overrepresented functions using the GoSlim Plants annotation. The BiNGO-derived graph (Figure 4) illustrates the most highly significant enrichment of specific functions. Downregulated genes are significantly enriched for genes linked to the plastid and thylakoid, and with functions in metabolic processes, particularly photosynthesis and carbohydrate and amino acid metabolism. A more detailed investigation using all GO terms (see Supplemental Table 1 online) shows overrepresentation for genes involved in chloroplast activity such as photosystem (PS) I and II, carbon fixation, chlorophyll (tetrapyrrole) biosynthesis, and amino acid metabolism. All these functions are essential for a growing and active leaf but are downregulated during senescence, when cellular structures are dismantled.
Figure 4.
Enriched GO Terms in Genes Upregulated and Downregulated during Senescence.
The network graphs show BiNGO visualization of the overrepresented GO terms for the combined clusters of genes either downregulated (clusters 1–24, 2849 genes) or upregulated (clusters 27–48, 3292 genes) during senescence. Categories in GoSlimPlants (Maere et al., 2005) were used to simplify this analysis and the same nodes are shown on both graphs. Uncolored nodes are not overrepresented, but they may be the parents of overrepresented terms. Colored nodes represent GO terms that are significantly overrepresented (Benjamini and Hochberg corrected P value < 0.05), with the shade indicating significance as shown in the color bar. A more detailed analysis of the GO categories is shown in Supplemental Table 1 online. ER, endoplasmic reticulum.
Upregulated genes show a very different picture, illustrating clearly the protective steps the plant takes to respond to the stress generated by the degradative and mobilization functions that occur during senescence (Figure 4). Only two cellular component terms are overrepresented, peroxisome and vacuole. Within the molecular function annotations, only transporter, protein binding, and transcription are overrepresented, whereas there is significant enrichment for stress response and catabolic processes in the biological process terms. In more detail (see Supplemental Table 1 online), enrichment is seen for genes involved in response to stimulus, particularly ABA and ethylene, and many stress responses such as osmotic, salt, and water stress. Enrichment of genes involved in metal ion binding is interesting. Many of these genes (64/222) encode zinc finger (C3H4-type RING) proteins, which may be involved in targeting specific proteins for ubiquitination and degradation. Other zinc binding proteins present have DNA binding activity and may act as TFs. Also, copper chaperones, metallothioneins, calcium binding proteins, and metal ion transporters are represented, which may illustrate the importance of the remobilization of valuable metal ions. Autophagy genes are a significant group; 15 Arabidopsis genes involved in autophagy are upregulated during senescence, showing the key role of autophagy in the controlled degradation of cellular components.
This global analysis is highly informative, as it shows broad classes of genes altered in expression during senescence and indicates the processes that are changing. Analysis of enriched GO terms in individual clusters should help to elucidate the chronology of gene expression and associated metabolic activities (Figure 3, see Supplemental Data Set 2 online). Not surprisingly, there was a very strong representation of photosynthesis-related genes in many of the clusters of downregulated genes. Clusters 2, 3, and 6, all of which show a strong diurnal variation with higher morning expression (Figure 3A), are highly enriched with photosynthesis genes, particularly those for the light reaction. Cluster 16, which shows less diurnal change, contains genes encoding enzymes such as Rubisco that are involved in carbon fixation. Clusters 4, 10, 17, and 21 are enriched for chlorophyll biosynthesis genes, and clusters 18 and 19 contain genes involved in cellular biosynthesis such as those for amino acid, polysaccharide, and lipid metabolism. Cluster 15 contains many genes encoding ribosomal proteins. Downregulation of these groups of genes reflects the shutdown of cellular biosynthetic activity as senescence occurs, and the coregulation is an indication of the organized control of this process. Cluster 13 is enriched for cytokinin signaling genes; a reduced level of cytokinin is a key signal that initiates the senescence process (Noodén et al., 1990).
GO terms enriched in the clusters of genes showing increased expression during senescence are less informative than those for the downregulated gene clusters. Certain clusters are enriched for stress-related genes, e.g., genes involved in JA and ethylene signaling are overrepresented in cluster 34. Other clusters are enriched for genes involved with macromolecule degradation, such as clusters 40 and 41 containing genes involved in carbohydrate and lipid degradation, respectively. Metal ion binding, particularly calcium binding, is overrepresented in cluster 43 and transporter genes in cluster 46.
Distinct Pathways Become Active at Different Times during Senescence
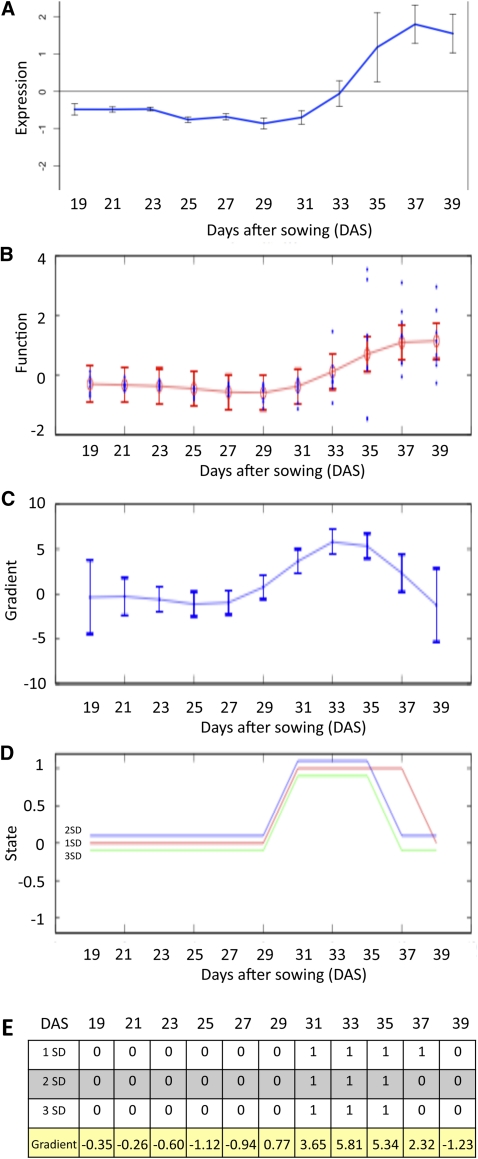
Although SplineCluster is useful in identifying groups of genes that are coexpressed and hence may be coregulated across the entire time series, it is not easy to divide these clusters according to their time of differential expression because the overall pattern of expression is the driving factor for cluster membership. To identify an ordering of events, the rate of change of gene expression (gradient) was inferred using Gaussian process (GP) regression applied to the 11–time point data set (described in detail in Supplemental Methods 1 online). Where data are sufficiently time resolved, this method can be used to identify the time points at which the gradient of a gene’s expression profile is significantly positive (increased), significantly negative (decreased), or not statistically different from zero (steady), whereas for less resolved data, it will identify times of significant change to the derivative of the global trend. The results are illustrated using the well-characterized, senescence-enhanced gene SAG12 (Figure 5). Expression profiles (Figure 5A) are used to train a GP model of gene expression (Figure 5B), after which a GP model of the gradient is obtained (Figure 5C) and used to identify whether the gradient at any time is sufficiently far from zero at three different significance thresholds (Figure 5D). A numeric representation of Figure 5C is shown in Figure 5E and suggests that SAG12 expression first becomes significantly enhanced around 31 DAS. This method can also be used to show when the gene expression gradient is maximal, i.e., the time of most rapid change. For example, the maximum change of the expression for SAG12 occurs between 33 and 35 DAS (Figure 5E).
Figure 5.
Gradient Analysis of SAG12 to Identify the Time of First Significant Change in Expression.
(A) Expression levels from microarray data (output from Gene Viewer). The blue line shows the mean of the eight replicates (n = 8; error bars = standard deviation, sd).
(B) Expression levels used by the GP regression, showing the eight biological replicates and the 95% confidence interval.
(C) GP model of the gradient showing 95% confidence interval.
(D) Change in gradient measured at each time point, shown at three different significance values: 3 sd, 2 sd, and 1 sd. Positive value of 1 shows an increased expression, 0 shows no significant change in expression, and −1 shows a significant decrease in expression.
(E) Data output for each significance value, i.e., whether gradient is significantly positive or steady at each time point, with the actual gradient value shown below.
[See online article for color version of this figure.]
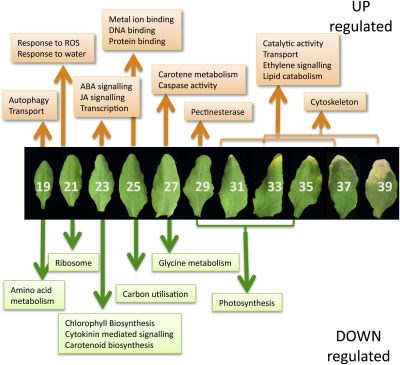
After examining the results for a number of genes, a significance stringency of two standard deviations was taken to represent a sufficient distance from zero and was used to generate discrete representations of the state of a gene (Figure 5D) for the 6323 differentially expressed genes. The resulting data were then sorted according to the time of first differential expression to identify 19 clusters (see Supplemental Data Set 3 online). GO term enrichment within the 19 clusters or subsets provided more clarity on the cellular and metabolic activities, showing step changes at each time point during the experiment than was gained from the cluster analysis described above (Figure 6, see Supplemental Data Set 4 online).
Figure 6.
Metabolic Processes Initiated or Repressed at Different Time Points during Development.
Enriched GO terms were identified using BiNGO (Maere et al., 2005) in groups of genes that show first significant upregulation or downregulation at each time point during leaf development and senescence. ROS, reactive oxygen species; white numbers indicate DAS.
Downregulated Gene Clusters Show Step Changes in Cellular Dismantling
It is clear that there are progressive changes in genes being downregulated as senescence progresses, and these are highly informative in indicating changes in metabolic pathways. Genes downregulated from the first time point (19 DAS, cluster D1; see Supplemental Data Sets 3 and 4 online), before the leaf is fully expanded, are enriched for genes involved in amino acid metabolism, including those for biosynthesis of Arg, Trp, Lys, and Gln. Genes involved in tRNA aminoacylation and over 30 ribosomal protein genes are downregulated at 21 DAS (cluster D2), indicating that expansion of the ribosomal content of the cells has slowed down. This suggests that large-scale de novo protein synthesis has ceased and that leaf cells are fully developed and equipped for activity.
Many tetrapyrrole or chlorophyll biosynthesis genes are first downregulated at 23 DAS (cluster D3), including the two genes encoding HEMA (glutamyl- tRNA reductase), which catalyzes the rate-limiting and first committed step in tetrapyrrole biosynthesis, and two genes encoding the D subunit of Mg-chelatase, part of the enzyme that diverts the tetrapyrrole pathway toward chlorophyll biosynthesis (Tanaka and Tanaka, 2007). Thus, the requirement for de novo chlorophyll biosynthesis appears to cease at 23 DAS, indicating that all chloroplasts are fully developed. Three genes involved in a branch of the carotenoid biosynthesis pathway (LUT1, 2, and 5) show a correlated drop in expression at this stage. These genes encode enzymes in the pathway leading from trans-lycopene via α-carotene to lutein, the major carotenoid component in the leaf with an important role in light-harvesting complex-II structure and function and in photoprotection (Kim and DellaPenna, 2006). In addition, expression of three cytokinin-inducible transcription repressors (response regulators ARR4, 6, and 7) that mediate a negative feedback loop in cytokinin signaling (Hwang and Sheen, 2001) also drops at this time point.
At the next stage (25 DAS, cluster D4), there is significant overrepresentation of genes involved in fixation of carbon dioxide or the Calvin cycle, including two Rubisco small subunit genes and sedoheptulose bisphosphatase, a key enzyme involved in the regeneration of the CO2 acceptor molecule, ribulose-1,5-bisphosphate. The reduction in expression of the two Rubisco small subunit genes correlates with the reduction in protein levels shown in Figure 1E and indicates that photosynthetic activity probably starts to drop at this stage. At 27 DAS (cluster D5), expression of genes involved in Gly metabolism declines, including Gly decarboxylase and Ser trans hydroxymethyl transferase 1, both involved in photorespiration, which presumably is also less important as photosynthesis becomes less active. Interestingly, five genes designated as HIGH CHLOROPHYLL FLUORESCENCE PHENOTYPE (HCF101, 109, 152, 173, and 208) are downregulated together at 27 DAS. Several such genes have been shown to have a role in maintaining the stability of chloroplast-encoded transcripts (Meurer et al., 1996; Meierhoff et al., 2003), and it may be that reduced expression of these genes enables enhanced degradation of photosynthetically related transcripts in the chloroplast. Finally, gene clusters that show expression that declines at 29, 31, and 33 DAS (clusters D6, D7, and D8) are significantly overrepresented for photosynthesis-related genes. Cluster D7 includes the gene-encoding TF GOLDEN2-LIKE (GLK2) that, together with its functional homolog GLK1, has been shown to coordinate expression of the photosynthesis apparatus genes in Arabidopsis (Waters et al., 2009). Inducible expression of GLK2 resulted in significantly increased expression of many photosynthesis-related genes (Waters et al., 2009), including those for the PSII chlorophyll binding proteins LHCB 2.2, 4.2, and 6 that are found in the same cluster of downregulated genes as GLK2, together with many others encoding subunits of the PSI and PSII complexes. The observation that expression of many photosynthesis-related genes is maintained until this late stage of development implies that there must be a continued requirement of these transcripts to retain sufficient energy production for the senescence process to occur.
Upregulated Gene Clusters Illustrate the Complexity of the Senescence Process and Reveal Novel Groups of Coregulated Genes
Genes that show increased expression at different time points during senescence were divided into clusters based on the time of first significant increase, but these clusters were also subdivided further depending on the subsequent expression patterns (see Supplemental Data Sets 3 and 4 online). This separation revealed additional enriched GO terms, as shown in Supplemental Data Set 4 online and Figure 6.
Many autophagy-related (ATG) genes are enhanced in expression from the start of the experiment (cluster U1), indicating that there may be a role for these proteins even before the leaf is fully expanded. Autophagy has a key role in the senescence process, and accelerated senescence has been observed in a number of autophagy-defective mutants (Doelling et al., 2002; Hanaoka et al., 2002; Yoshimoto et al., 2004). Nine of the 15 upregulated autophagy genes show increased expression from the first time point, with five others upregulated at 21 or 23 DAS and one, ATG7, being upregulated at 29 DAS. Investigation of the overall expression patterns of the autophagy genes shows four genes, ATG7, ATG8H, ATG8A, and ATG8B that show correlated and rapidly increased expression between 29 and 31 DAS (see Supplemental Figure 3A online). In yeast, ATG7p has been shown to be required for activation of ATG8p to allow conjugation with phosphatidylethanolamine (Ichimura et al., 2000), and the resulting ATG8p-phosphatidylethanolamine conjugates assist in the formation of the autophagosome. Thus, the timing of expression of ATG7 (around 29 DAS) may be the key control point for autophagy activation in senescing leaf cells.
Genes induced at 21 DAS (cluster U2) are enriched for response to oxidative stress. These include TFs such as DREB2A, a key regulator in drought and heat stress responses (Sakuma et al., 2006), and LSD1, a zinc finger that monitors superoxide levels and regulates cell death (Epple et al., 2003). Increased expression of the mitogen-activated protein kinase MPK7, which is also induced by hydrogen peroxide treatment and enhances plant defense responses (Dóczi et al., 2007), and heat shock proteins such as HSP70, a stress-enhanced heat shock chaperone with a protective role, also indicates that the plant is protecting itself from the detrimental effects of oxidative stress caused by the early stages of senescence. Genes involved in response to water deprivation are also overrepresented; DREB2A described above, Arabidopsis HISTIDINE KINASE3 (AHK3), a stress-responsive gene that has been shown to influence cytokinin control of leaf longevity (Kim et al., 2006), dehydration-responsive genes such as ERD1 and ERD14, and RAB18, ABF2, and other ABA-responsive genes.
Genes involved in both JA and ABA responses are clearly overrepresented in those whose expression increases at 23 DAS (cluster U3). Many of these show a peak of activity at this time point followed by a drop, and this subset is highly enriched for JA biosynthetic genes (cluster U3_1). JA-related genes upregulated at this time point include genes required for JA biosynthesis such as two lipoxygenases, two allene oxide cyclase genes, AOC1 and AOC4, and 12-oxophytodienoate reductase. This increase correlates with a peak in levels of JA detected at 25 DAS (Figure 2). Also, genes implicated in controlling JA responses are upregulated, including the TF MYC2, and jasmonate ZIM-Domain genes, JAZ1, JAZ6,and JAZ8. JAZ proteins are repressors of JA signaling, binding to MYC2 and preventing its action (reviewed in Staswick, 2008). MYC2 is also involved in expression of ABA response genes, and this may be the cause of the increased expression of ABA-related genes at this time point. ABA levels only show a large increase later in senescence (Figure 2), but several ABA-signaling genes (e.g., ABI1 and AFP1) and dehydration response genes whose expression is induced by ABA (e.g., RD20 and RD26; Fujita et al., 2004; Choudhury and Lahiri, 2011) are upregulated at 25 DAS. This suggests a potential coordination of JA and ABA responses at this early stage of senescence.
Many genes encoding TFs are first upregulated at 23 DAS, including four WRKY factors, eight NAC domain (for Petunia hybrida NAM and for Arabidopsis ATAF1, ATAF2, and CUC2) proteins, 10 zinc finger proteins, and the Nuclear Factor Y subunit NF-YA4, which has been implicated in regulating endoplasmic reticulum stress (Liu and Howell, 2010). Many of these genes show an increased expression followed by a fall in expression later in senescence (cluster U3_3), whereas others show a continued increase from this time until later in senescence (see Supplemental Data Set 3 online). These are likely candidates for the control of later senescence-related processes.
At 25 DAS, the cluster of genes upregulated (cluster U4) is enriched for metal ion binding proteins, including many genes encoding DNA binding proteins, TFs, calcium-signaling genes, etc. The subgroup of this cluster that shows a pattern of increased followed by decreased expression (cluster U4_1) is enriched for genes with a protein binding function. There are five C3HC4-type RING finger protein binding genes in this group, which presumably have a role in regulating specific protein levels via the ubiquitination pathway.
At 27 DAS, there is an interesting overrepresentation of genes involved in carotene metabolism (cluster U5). The three genes involved are a β-carotene hydroxylase and two carotenoid cleavage dioxygenase genes, CCD7 and CCD8. Carotenoids are precursors of signaling molecules that regulate shoot branching in Arabidopsis, and CCD7 and CCD8 mutants, max3 and max4, respectively, show increased lateral branching (Ongaro and Leyser, 2008). These genes are involved in the production of a strigolactone-related signaling molecule (Gomez-Roldan et al., 2008). Interestingly, another gene that has a shoot-branching role, MAX2, was originally identified as ORE9, encoding an F box leucine-rich repeat protein required for normal leaf senescence (Woo et al., 2001). Mutants in MAX2 show increased branching, indicating that this protein is a regulator of the strigolactone signal. The ORE9/MAX2 gene also shows senescence-enhanced expression and is upregulated at 21 DAS. Enhanced expression of all three genes that regulate shoot branching in a senescing leaf and the fact that a mutant in ORE9/MAX2 shows delayed senescence indicate that there may be a role for the novel strigolactone-like hormone in regulating an aspect of leaf senescence.
Caspase activity is also an enriched GO term in genes upregulated at 27 DAS due to increased expression of two of the nine Arabidopsis genes encoding potential caspase counterparts, metacaspases MC6 and MC9. The other seven Arabidopsis metacaspase genes do not show differential expression during senescence. In other organisms, caspases play an essential role in controlling and executing programmed cell death (PCD), and two metacaspase genes, MC1 and MC2, have been shown recently to control pathogen-induced PCD in Arabidopsis (Coll et al., 2010). Links between senescence and plant PCD are tenuous, but the coregulated expression of these two caspase-like genes at this time point may indicate that they have a role in the degradative processes and/or cell death that occur in leaf senescence. The autophagy gene ATG7 also shows initial enhanced expression at this time point as described above, and this may be a significant link showing that, under our growth conditions, the first degradative processes of senescence are initiated at 27 DAS.
At the next time point (29 DAS, cluster U6), senescence-related degradation processes are shown by enrichment in genes for cell wall degradation. Six upregulated genes encode pectinesterase, involved in the degradation of plant cell wall pectin components. These enzymes and others such as xylosidase, glucosyl hydrolase, β-glucosidase, pectate lyase, and pectin methylesterase inhibitor, may have a role in controlling the degradation of cell wall components and releasing sugars for respiration (Lee et al., 2007). Similarly, genes upregulated at 31 DAS (cluster U7) are highly enriched for catalytic activity, reflecting the considerable degradation that is underway. These include additional carbohydrate-degrading enzymes such as pectinesterases, glycosyl and glucosyl transferases, and polygalacturonase, and several proteases including the well-characterized senescence-enhanced Cys protease SAG12, which may have a role in chloroplast degradation (Martínez et al., 2008). Two upregulated genes, LACS6, encoding a long-chain acyl-CoA synthetase (Shockey et al., 2002), and ACX1 (acyl-CoA oxidase), encoding the enzyme that catalyzes the first step in fatty acid β-oxidation in the peroxisome (Fulda et al., 2002), are involved in mobilizing membrane lipids via β-oxidation, likely to provide an energy source to fuel the senescence process. This could act as an initiator of precursors for jasmonate biosynthesis because levels of this hormone increase after this time point (Figure 2).
By the later time points in this experiment, after 31 DAS, the senescing leaf becomes more and more heterogeneous, with some cells within the leaf being at a more advanced stage of senescence than others and more variability between biological replicates. This means that there is less clarity in the functions of different groupings of genes that are differentially expressed at each time point after 31 DAS. GO term enrichment analysis of the 31, 33, and 35 DAS groups combined (clusters U7, U8, and U9) illustrates the degradation and mobilization of nutrients, with 44% of these genes involved in catalytic activity, with lipid catabolism highly represented, and 10% involved in transport. Response to chemical stimulus is also high, with two of the three Arabidopsis genes annotated detection of ethylene stimulus, i.e., ethylene ETR1 and ACC OXIDASE2 (ACO2) being upregulated late in senescence, indicating that the ethylene regulation of senescence may have a significant role at this time.
A surprising group of genes identified by this analysis is downregulated for most of the time course followed by a significant increase in expression at 35 or 37 DAS (clusters U8_1 and U9). This group is highly enriched for genes involved in the cytoskeleton (see Supplemental Figure 3B online), with members of the α-tubulin family (TUA2, 4, and 5), actin genes, ACT3 and ACT11, and two aurora genes (AURORA1 and AURORA2) encoding kinase proteins that have a role in histone phosphorylation and have been reported to be associated with microtubule spindles and abundantly transcribed only in dividing cells (Demidov et al., 2005, 2009). Downregulation of this group of genes after completion of the cell division and expansion stages of leaf development is to be expected, but the increase in expression at the end of senescence is unanticipated. AURORA1 has been shown to phosphorylate histone H3 at Ser10 (Demidov et al., 2009), and, in mammalian cells, this modification has been suggested to have a crucial role in transcription and apoptosis as well as in cell division (Prigent and Dimitrov, 2003). These proteins may alter chromatin structure in late senescence to allow DNA fragmentation and eventual degradation. Histone modification and chromatin restructuring is a key regulator in Arabidopsis stress responses (Kim et al., 2010), and H3 phosphorylation increased in response to salinity, osmotic stress, and ABA treatment of cultured cells (Sokol et al., 2007).
The increased expression of actin and tubulin genes late in senescence could reflect an autophagy role (Monastyrska et al., 2009). Evidence from yeast and mammalian systems indicates that efficient autophagy requires microtubule action to facilitate autophagosome movement, and actin microfilaments have a role in selective types of autophagy in yeast. The Arabidopsis autophagy ATG8 gene family shows significant homology to mammalian microtubule binding proteins and bind to microtubules in vitro (Ketelaar et al., 2004). Therefore, plant autophagy may involve the action of microtubules and microfilaments, explaining the increased expression of these genes late in senescence when the autophagic degradation of cellular compounds is active.
Chlorophyll degradation is a key step in the senescence process, and several of the genes involved are under transcriptional control (Hörtensteiner, 2009). The STAYGREEN gene, SGR1 regulates the first step in the dismantling of chlorophyll from the chlorophyll binding proteins. Key genes involved in chlorophyll degradation, SGR1, SGR2, NYC1 (chlorophyll b reductase), and PaO (pheophorbide a oxygenase), all show enhanced expression during senescence. All of these genes increase in expression during early time points, leveling out between 25 and 29 DAS, followed by a sudden increase in expression after 29 DAS (see Supplemental Figure 3C online). It is likely that it is the expression of SGR after 29 DAS that initiates the dismantling of the protein chlorophyll complexes, releasing chlorophyll for detoxification.
Additional information can be gained from the gradient analysis described above if the time of maximum gradient is also considered. For example, statistical analysis of gene clusters based on time of first differential expression indicated that photosynthetic genes were overrepresented in clusters showing downregulation at 29 to 33 DAS. However, if all the downregulated genes annotated as photosynthesis are examined, many of these show initial significant downregulation earlier in the time series (see Supplemental Figure 4A online), but the size of the clusters at these time points means this annotation does not show up as being significantly enriched. When the maximum absolute gradient for each of the photosynthesis genes was calculated (see Supplemental Figure 4B online), the vast majority of photosynthesis genes showed the most rapid drop in expression between 31 and 35 DAS, confirming the observation that photosynthesis-related gene activity is maintained until late in the leaf’s development (Figure 6).
The maximum absolute gradient analysis was also applied to investigate genes responding to JA and ABA stimulus. In both cases, the majority of genes were first significantly upregulated at 21 and 23 DAS, early in senescence (see Supplemental Figures 4C and 4E online). However, although several of the genes had a maximum gradient early, at 23 DAS, there were also many showing maximum gradient much later in the time series, up to 35 and 37 DAS for the JA response genes and 33 and 35 DAS for the ABA response genes (see Supplemental Figures 4D and 4F online). This correlates with the data on the levels of JA and ABA shown in Figure 2 where a maximum level of both hormones is measured late in the process (increasing at 33 and 31 DAS, respectively). The timing of expression of specific hormone biosynthesis genes (see Supplemental Figure 5 online) clearly illustrates the rapid increase in JA biosynthesis genes between 23 and 25 DAS, whereas ABA and SA biosynthesis genes show a later increase in expression with a maximum at the final stage of senescence. Although ethylene levels were not measured during the time course, the ethylene biosynthesis genes ACS2 and ACS7 also show increased expression from around 29 to 31 DAS, with a steady increase as senescence progresses. Thus, ABA, ethylene, and probably SA synthesis appear to be coordinately regulated in senescence, whereas JA synthesis shows a different pattern. Interestingly, some JA biosynthesis and signaling genes are only expressed at the early time point (e.g., OPR3), whereas others (e.g., LOX3) are also upregulated late, presumably enabling the accumulation of JA later during senescence.
Our detailed expression profiles and novel tools have enabled us to distinguish biological processes initiated at different stages of senescence and hence tease apart some of the components of this complex phenomenon. We now have a timeline that can be built upon to link these different processes and to identify the overarching regulatory mechanisms as well as candidate genes for specific senescence processes.
TF Binding Motifs Show Specific Enrichment in Differentially Expressed Gene Clusters
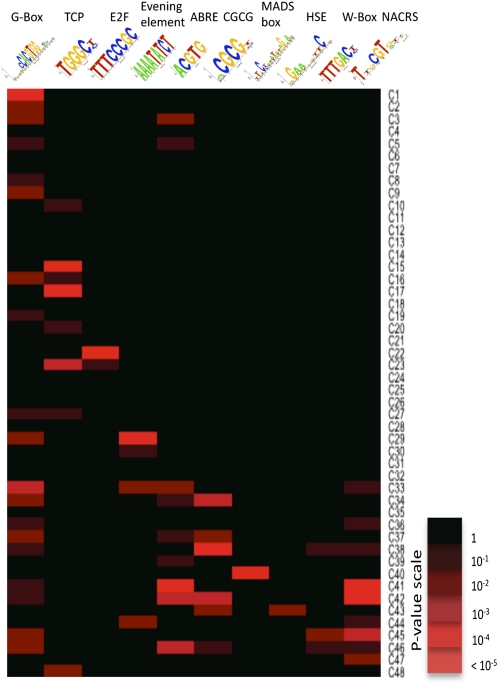
The SplineCluster analysis of differentially expressed genes (Figure 3) identified groups of genes that exhibit similar expression profiles and thus may be coregulated. Analysis of such coregulated gene sets should help pinpoint potential TF binding motifs important for gene expression during leaf development. To gain an initial understanding of the regulatory mechanisms of genes differentially expressed during senescence, promoters corresponding to 500 bp upstream of the predicted transcription start site of genes in each cluster were screened for overrepresentation of known TF binding motifs.
This analysis shows clearly that certain sequence motifs are selectively enriched in clusters that exhibit similar expression patterns (Figure 7, data shown in Supplemental Data Set 5 online), and there is an obvious difference in the range of motifs distributed over the different clusters. Consistent with the GO term analysis results, several of the downregulated clusters (clusters 1–24) are significantly enriched for sequence motifs associated with the regulation of photosynthesis and cell growth. For example, the G box variant motif is linked with the regulation of photosynthetic genes and responses to light (Martínez-García et al., 2000) and the TCP motif binds members of the TCP family of TFs, which have been implicated in the regulation of growth and cell division (Li et al., 2005). Binding sites for E2F TFs, key regulators of cell proliferation (Ramirez-Parra et al., 2003) are enriched in cluster 22, which is consistent with this cluster being enriched with genes annotated with the GO term cell cycle. Genes in this cluster are downregulated from the start of the measured time course, and this would be expected since cell division has ceased before the leaf is fully expanded.
Figure 7.
Over-Representation of Known TF Binding Motifs in Promoters of Coexpressed Genes.
Logo representations of known TF binding motifs are on the horizontal axis, and expression profile for each cluster (see Figure 3) is on the vertical axis. Colored boxes represent pairs of motif and expression cluster with a significant statistical link. Shown are a limited number of representative motifs and clusters (see Supplemental Data Set 5 online for full results).
Sequence regions upstream of genes in upregulated clusters (clusters 27–48) contain a number of sequence motifs that can bind TF families that are themselves upregulated during senescence. For example, NAC domain and WRKY TFs constitute a large proportion of the senescence-regulated TFs and are known to play significant roles in regulating leaf senescence in Arabidopsis (Miao et al., 2004; Guo and Gan, 2006; Kim et al., 2009). Binding sites for NAC and WRKY TF families are overrepresented in several upregulated clusters sharing similar expression profiles. Sequence motifs associated with stress responses are also enriched. The heat shock element is overrepresented in a single cluster, and several heat shock factors are upregulated during senescence. The CGCG motif, which has been implicated as a calcium-signaling element in a range of stresses, is enriched in several upregulated clusters. This motif has been shown to bind CAMTA TFs (Yang and Poovaiah, 2002) involved in signaling responses to wounding, cold, and other stresses (Walley et al., 2007; Doherty et al., 2009). The ABA-responsive element (ABRE) is overrepresented in multiple upregulated clusters, and enrichment correlates with the observed increase in levels of ABA during senescence. ABRE-binding factors are known to activate target genes in an ABA-dependent manner (Nakashima et al., 2006). The ABRE contains an ACGT-core and, therefore, is a subset of the G box sequence (CACGTG). However, the pattern of overrepresentation of these two similar motifs across the senescence clusters is different, suggesting that divergent functional roles can be identified. G box–like motifs can bind many members of the bZIP and bHLH TF superfamilies (Toledo-Ortiz et al., 2003; Jakoby et al., 2002), and TFs from both these families are upregulated during senescence.
TF Families Are Active at Different Times during Senescence
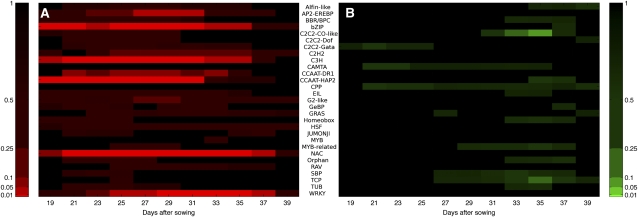
To complement the analysis of TF binding motifs above, we investigated whether specific families of TFs were differentially expressed at particular times during senescence. The groups of genes identified as having the same initial timing of differential expression by the GP gradient tool analysis were further analyzed to identify time periods when families were overrepresented for genes with a positive or negative gradient (i.e., expression significantly increasing or decreasing). A heatmap, mapped to the significance of each family’s activity, is shown in Figure 8, with the numerical data shown in Supplemental Data Set 6 online.
Figure 8.
TF Families Significantly Over-Represented with Positive or Negative Gene Expression Gradients Highlighting Distinct Periods of Activity.
(A) A number of TF families significantly upregulated early, including the NAC and bZip families (19–21 DAS), remaining strongly upregulated throughout the experiment, and a small number of families only upregulated toward the middle and end of the experiment (27+ DAS), such as WRKY, AP2, and G2-like.
(B) A small number of weakly significant TF families downregulated early, and several families (C2C2-CO-like and TCP) significantly downregulated toward the end of the experiment (33+ DAS).
Color bars indicate P value (after FDR correction), with a range of significance thresholds (0.01, 0.05, 0.1, 0.25, and 0.5). Numerical data used to derive this figure are shown in Supplemental Data Set 6 online.
A number of TF families were significantly overrepresented for upregulated genes (adjusted P < 0.01), indicating a large amount of similar transcriptional activity and potential coregulation within these families. Specific members of the bZIP family have been shown to participate in defense against pathogens, development, stress treatments such as cold and drought, ABA signaling, and phenylpropanoid biosynthesis (Weisshaar and Jenkins, 1998; Jakoby et al., 2002). Another significantly overrepresented upregulated family is the large C3H superfamily, of which little is known of the function of many of its members. Several subfamilies related to the CCAAT box binding factor family were also significantly upregulated; factors in this family form the heterotrimeric NF-Y binding complex (consisting of NF-YA, NF-YB, and NF-YC subunits) that has been shown to influence flowering time and stress responses in plants (Wenkel et al., 2006; Liu and Howell, 2010). Interestingly, it is the NF-YA subunits specifically that are enriched in the senescence-enhanced gene lists, with nine of the 10 genes in the genome showing increased expression during senescence. In comparison, few of the NF-YB and NF-YC genes are altered in expression, with only three NF-YB genes and one NF-YC gene being upregulated. This implies that it may be the regulated expression of the NF-YA subunit that controls the activity of the NF-Y complex during senescence.
The large NAC family also had a significant early overrepresentation, with over 30 of the members of this family being altered in expression at various times during senescence. Members of this family are known to have a large number of regulatory interactions with a diverse range of biological processes, including senescence, defense, and abiotic stress (Olsen et al., 2005, and references quoted above). Several other families show upregulated transcription later in the time course. The WRKY family shows an overrepresentation, with many members of this family being upregulated from around 25 DAS. WRKY TFs have been shown to be important for senescence (Robatzek and Somssich, 2001; Miao et al., 2004); others are induced by infection by viruses or bacteria (Eulgem et al., 2000) and are downstream of defense-signaling mitogen-activated protein kinase pathways and involved in the regulation of SA- and JA-dependent defense signaling pathways (Ülker and Somssich, 2004; Eulgem and Somssich, 2007). The large AP2-EREBP family becomes significantly overrepresented around 27 DAS; members of this family are induced in several cases by hormones such as JA, SA, and ethylene, along with other signals related to pathogens, wounding, and abiotic stresses, and have influence on other stress and disease resistance pathways (Kizis et al., 2001; Gutterson and Reuber, 2004). Therefore, cascades of cellular information flow during the progress of leaf senescence can be predicted by this analysis, such as upregulation of NAC or WRKY genes influencing various hormone responses, followed by upregulation of AP2-EREBP TFs. This knowledge is key for future modeling of senescence transcriptional networks.
ANAC092 Target Genes Are Highly Enriched in Clusters Overrepresented for NAC Binding Motifs
The motif and TF analyses described above pinpoint NAC domain genes as being of key importance in regulation of senescence and we follow this observation up in more detail as an example of the increased understanding that this data set provides. A recent publication (Balazadeh et al., 2010) describes an elegant experiment using inducible expression of ANAC092 to identify likely target genes. Of the 170 genes identified in that study as being upregulated after induced expression of ANAC092, 102 of these are senescence enhanced in our time course experiment; of these, 75%, including ANAC092 itself, are to be found in the clusters enriched for NAC domain motifs (clusters 41, 42, 44, and 45; see Supplemental Data Set 7 online). This provides clear evidence that the detection of enriched motifs within clusters is providing biologically relevant information and also indicates that ANAC092, probably together with other NAC domain proteins, has an influential role in regulating the expression of many genes at this stage of senescence. This information might be used in preliminary modeling experiments to predict interactions that regulate gene expression in these clusters.
DISCUSSION
In this article, we describe a high-resolution, highly replicated time-course analysis of gene expression during Arabidopsis leaf development from before complete expansion to full senescence. Over this time, the leaf develops from a sink that is importing nutrients for growth into an active source organ, performing maximum photosynthesis and exporting fixed carbon for further growth of the plant. This is followed, relatively rapidly in this short-lived plant, by the initiation of senescence whereby the leaf is converted from a source of photosynthetic carbon to a source of valuable macromolecules such as nitrogen, phosphorus. and minerals, as cellular components become degraded and mobilized from the leaf. Thus, in this short time period, the leaf undergoes enormous changes in metabolism and transport of metabolites.
To obtain insight into the timing and potential coregulation of the changes in genes and pathways in the complex process of senescence, it is essential to sample highly controlled replicate leaves and to measure at many time points. This is also essential if these data are to be used for network inference analysis. In the experiment reported here, we harvested the same leaf (leaf 7) from individual plants at different times over 3 weeks. Previous transcript profiling studies on developmental leaf senescence in Arabidopsis have analyzed a more limited number of time points (e.g., Buchanan-Wollaston et al., 2005; van der Graaff et al., 2006). Analysis of gene expression changes in field-grown Populus leaves has been performed over several time points during late summer and autumn (Andersson et al., 2004). In these examples, pooled leaves were used as biological samples; and replicates were limited; although these experiments gave a picture of the overall changes in gene expression that occur during senescence, they provide little information on the timing of the changes during the process. Senescing leaves are, by their nature, quite variable, particularly in mid or late senescence; therefore, to accurately determine coregulation of genes through senescence, it is essential to control and allow estimation of other sources of variability, such as may be caused by plant-to-plant differences and environmental factors by using a carefully designed sampling strategy.
Methods Developed to Enable Large-Scale, Two-Color Microarray Analysis and Identification of Differentially Expressed Genes
This large-scale microarray experiment with both biological and technical replication across multiple time points required the development of a novel and complex design approach to take full advantage of the two-color microarray system, providing efficient estimation of the differences in response between adjacent time points while still allowing effective comparison of all samples. The highly replicated experiment allowed the application of stringent statistical analysis to identify and characterize genes differentially expressed at different time points during senescence, which has not been possible with data from previous studies. The quantity of data generated necessitated the adaptation of existing analysis methods/algorithms, as well as the development of some new analysis tools.
The Bioconductor package MAANOVA was adapted to meet the specifications of the CATMA arrays, providing monitoring of slide and data quality and using information from the four technical replicates of each sample to remove the influence of the occasional outliers. The mixed model-fitting algorithm then enabled the estimation and testing of the differences caused by the treatment factors (day, time of day, the interaction [combined effect] of these factors, and the biological replicates), allowing for the complex design structure and the sources of variability (between slides and between dyes) imposed by using the two-color microarray system. There are considerable advantages to the experimental and analysis approaches used. The use of two-color arrays allows direct comparisons of samples between key time points and, through careful design of the pairs of samples compared on each array, across all time points by indirect association. By contrast, many applications of two-color arrays compare experimental samples by calculating the ratio of expression responses of each with a control sample hybridized on every slide, thus halving the amount of useful data obtained per slide (or doubling the cost of obtaining the same data).
Identification of genes showing interesting differential expression patterns was achieved by first assessing the significance of the variation due to different model terms (day, time of day, and the interaction between them) relative to the between-biological replicate variation using the MAANOVA analysis, and then conducting a visual inspection of gene expression responses over time for genes giving a less significant test result (0.0001 < P < 0.05) for the effect of day. Of course, high levels of biological (between-plant) variability can lead to large changes in gene expression not being identified as statistically significant and, hence, genes not identified as being differentially expressed. Further exploration of approaches to control this biological variation, through both the statistical design of future experiments and the development of novel analysis methods, is important for future successful research.
The mixed model-fitting algorithm implemented in the MAANOVA package allows separation of the variation due to different sources within the treatment combinations (i.e., day, time of day, the interaction between these factors, and biological replicates), and hence the identification of genes showing different generic patterns of differential expression (see Supplemental Figure 1 online). An advantage of this approach, over a simple comparison of the responses across all 22 time points, is that genes showing only diurnal (time of day) variation can be easily identified and ignored in subsequent modeling of potential senescence-related gene networks, with those showing combined effects of day and time of day also easily identified and included. A disadvantage is that the analysis does not formally include any allowance for the ordering of the samples through time (e.g., the effect of day is essentially just an assessment of the average variability between the 11 mean values, and reordering the days would not change the test statistic and hence level of significance). A further development of this approach, allowing both separation of effects within a factorial treatment structure and estimation of the underlying shape of response over time, possibly following the approach proposed by Eastwood et al. (2008), should lead to a more reliable identification of genes showing important patterns of differential expression, although issues with high levels of biological variability would still result in some false-negative test results. Better estimation of the shape of expression profiles could also contribute to improved clustering of genes with similar shapes of expression profiles. In the absence of such a modeling approach, the approach used here, combining the highly significant results of the formal analysis with a visual inspection, is likely to result in the identification of most of the important genes showing differential expression related to senescence, while minimizing the number of false positives.
Analysis of Differentially Expressed Genes Revealed a Chronology of Processes and Signals
Analysis of individual clusters identified in the SplineCluster analysis, particularly those for downregulated genes, identified groups of genes involved in a common process such as photosynthesis, chlorophyll metabolism, etc. It seems likely that genes involved in the same process, with similar expression profiles, are coregulated rather than simply coexpressed during senescence, and this prediction is strengthened by the promoter motif analysis.
The GP gradient analysis, developed to enable more effective dissection of gene expression changes over time, identified groups of genes that showed their first significant change in expression between the same pair of adjacent time points. The resulting clusters present a highly informative picture of the timeline of senescence, showing when individual pathways are upregulated or downregulated (Figure 6). Knowledge of such timing will prove a powerful tool for separation of pathways into groups to allow identification of upstream genes that control them.
Comparing the two approaches used to cluster the differentially expressed genes, it is clear that they will generate different sets of clusters. SplineCluster groups genes with overall profile shapes that are similar based on the fitted regression coefficients, which should therefore mean that genes in the same cluster will have similar changes in expression between every pair of adjacent time points. However, the approach does not take any account of the biological (between-plant) variability, so that only the initial filter will determine the significance associated with the overall differential expression, and so a gene with highly significant variation in expression could be clustered with one just breaking the significance threshold. By contrast, the GP gradient analysis groups genes that have the first significant changes in expression in the same direction at the same time. However, unless the gradient information is also taken into account, these may not always be showing the most dramatic change in expression at the same time point. Therefore, both approaches have value in identifying coregulated genes, but both have the potential to inappropriately group genes.
The clustering results have been discussed in detail above and have identified groupings of genes that had not been observed previously with more limited time series data. For example, it is clear that the extensive overall reduction in expression of genes involved in chloroplast activities occurs via a timed process. Chlorophyll biosynthesis genes are downregulated before carbon fixation genes, and these are downregulated well before the majority of key genes encoding proteins involved in photosynthesis, including chlorophyll binding proteins and components of PSI and II. Autophagy genes are enhanced from the start, but the level of the key gene ATG7 starts to rise at 29 DAS. This is also the time at which chlorophyll degradation genes show a rapid induction of expression. Other metabolic pathways such as strigolactone synthesis, hormone biosynthesis, cell wall degradation, cytoskeleton, and microtubule activity, to name just a few, are implicated at different times during the senescence process.
Microarray Data Analysis Tools Used to Develop and Test Hypotheses for Transcriptional Control during Senescence
Analysis of the core promoters of coexpressed genes revealed potential regulatory sequence motifs that are likely to contribute to the coregulation of genes involved in the senescence process. Known sequence motifs are enriched in the promoters of genes that share similar expression profiles and correlate with the biological processes associated with such genes. The importance of the NAC and, to a lesser extent, the WRKY TF families in promoting senescence in Arabidopsis is illustrated through the specific and highly significant enrichment for potential binding sites for these regulators in the promoters of certain clusters of upregulated genes. In addition, most of the genes implicated as under the control of the senescence-enhanced ANAC092 TF (Balazadeh et al., 2010) occur in these same clusters, showing the importance of this TF and other NAC family members in regulating gene expression during senescence. The identity of specific TFs that target these known motifs is unknown, and further bioinformatic analysis and modeling as well as laboratory experiments is required to characterize them fully. Regulation via known cis-regulatory elements is not sufficient to explain the expression patterns of all genes, and unknown sequence motifs likely contribute toward regulating specific groups of genes within the senescence process.
In comparison with previous senescence gene expression studies, the study reported here collected highly replicated gene expression responses at a high temporal resolution across the period during which the senescence response develops. Thus, these data are more suitable than previously collected data sets for applying statistical analyses aimed at predicting the gene regulatory networks operating during senescence. This is of immense value for the next step in the development of a model for the regulation of leaf senescence. However, even this substantial microarray experiment still imposes data limitations that make the application of network inference nontrivial; gene expression measurements of just 88 biological samples are insufficient to accurately model the regulation of thousands of genes. As the number of possible networks grows superexponentially with the number of genes involved, the correlations in the patterns of response across these 88 samples are certainly insufficient to accurately identify a unique network model describing the regulation of the 6326 genes identified as differentially expressed. This is known as the “curse of dimensionality” (Bellman, 1961). Therefore, the design of future studies needs to consider the balance between experimental cost and informative data for network inference, especially as a greater number of samples brings diminishing returns with respect to the number of additional genes it allows a researcher to model. It is also important to balance the need for good information about the biological (between-plant) variability, technical replicate variability, and temporal changes in gene expression during the senescence process.
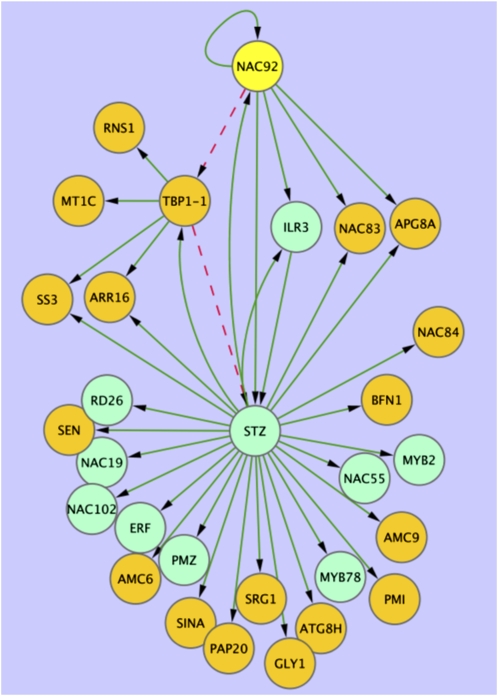
The data presented here can be used to produce constrained network models for many small sets of genes. An example of the type of network model that can be inferred from these data is shown (Figure 9). A variational Bayesian state space modeling method (Beal et al., 2005) was applied to a selection of genes using the 11–time point data series (see Supplemental Table 2 online). Genes selected were present in clusters enriched for the NAC motif (i.e., clusters 41–45) and also present in the ANAC092 overexpression data described above (Balazadeh et al., 2010; see Supplemental Data Set 7 online) or were annotated as TFs. The resulting model correctly predicts a positive influence of ANAC092, either direct or indirect, on multiple known downstream target genes (Figure 9, orange nodes) The model also makes several new hypotheses for experimental testing. For example, it predicts the influence of a zinc finger protein (STZ) in the expression of ANAC092 and its downstream genes. Although no experimental evidence exists for this regulatory link, both STZ and ANAC092 are induced in expression during salt stress, and knock out mutants in both genes have enhanced tolerance to stress in Arabidopsis (Mittler, et al., 2006; Balazadeh et al., 2010). Regulation by ANAC092 of several other TFs known to be stress related is also predicted in this network model. ANAC019, ANAC055, and RD26 (ANAC072) have all been shown to have a role in drought stress (Tran et al., 2004); MYB2 has a role in ABA signaling and salt stress (Abe et al., 2003; Yoo et al., 2005), and PMZ, a zinc finger protein, has a role in stress-induced senescence (Breeze et al., 2008). The gene regulatory network model also predicts feedback and feed-forward connections between ANAC092, STZ, and TBP1-1, which encodes a telomere binding protein. These types of interactions are crucial for the robustness of gene regulatory networks and would be almost impossible to predict from biological data alone. Thus, this model, generated with a small subset of the array data, correctly predicts known gene-gene interactions and generates complex novel predictions for experimental testing.
Figure 9.
Network Model Inferred from Microarray Data.
Variational Bayesian state space modeling was used to generate a network model using senescence-enhanced genes selected from SplineClusters 41, 42, 44, or 45 in the microarray data. Genes showing induced expression in an ANAC092 (yellow node) inducible overexpression experiment (see Supplemental Data Set 6 online; Balazadeh et al., 2010; orange nodes) were combined with selected TFs from the same clusters (green nodes). Green edges represent positive interactions, while red dashed edges predict negative effects. Genes are identified in Supplemental Data Set 7 online.
Many different models can be obtained with different collections of genes, and these transcriptional network models can be expanded using information on coregulated pathways and promoter motif analysis to identify likely downstream targets of key TFs. This model development and experimental testing is underway to generate validated gene regulatory network models underlying senescence.
METHODS
Plant Growth
Arabidopsis thaliana plants were grown as described in Breeze et al. (2008). Leaf 7 was tagged with thread 18 DAS. Sampling of leaf 7 started at 19 DAS and continued every other day until full senescence was reached (39 DAS). Leaves were harvested twice on each sampling day, 7 and 14 h into the light period. This resulted in samples being obtained at 22 distinct time points. At each time point, leaf 7 was sampled from 20 plants among the 720 being grown in the controlled-environment growth room, the plants being randomly selected to avoid any potential effects of position within the growth room. Leaves were rapidly weighed and photographed with a size scale before being frozen in individual tubes in liquid nitrogen. Leaf length was estimated against this scale from the photographs.
Protein and Chlorophyll Measurements
Total protein was extracted from five individual leaf samples by grinding the sample in liquid nitrogen before the addition of 500 μL of extraction buffer (50 mM lithium phosphate [pH 7.2], 1 mM monoiodoacetic acid, 120 mM mercaptoethanol, 5% [v/v] glycerol, 1 mM PMSF, and 0.2% lithium dodecyl sulfate). At this stage, a 100-μL aliquot of the extract was taken for chlorophyll analysis. The protein extract was boiled for 45 s and centrifuged for 20 min at 12,800g. Total protein was measured using the RC DC protein assay (Bio-Rad) according to the manufacturer’s instructions. In addition, levels of the small and large subunits of Rubisco were assessed by diluting the protein extracts to normalize for leaf weight and then running an equal volume of each extract (equivalent to 0.5 mg of fresh tissue) on polyacrylamide gels (Invitrogen Novex 4-12% Bis-Tris gel), staining with Coomassie blue, and scanning the relevant protein band. Protein levels were assessed densitometrically using image analysis software (GeneTools; Syngene) against a calibration curve of bovine serum albumin (LSU) and lysozyme (SSU).
Chlorophyll was measured from five individual leaves using the total protein extracts. The chlorophyll was extracted using 80% acetone, vortexed, and then stored at −20°C for 1 h in the dark. The samples were then centrifuged for 3 min at 12,800g, and the absorbance of 1 mL was measured at 663 and 646 nm. Chlorophyll concentrations were calculated using the equations: total chlorophyll (mg/L) = 20.2A646 + 8.02A663, chlorophyll a (mg/L) = 13.19A663 − 2.57A646, and chlorophyll b (mg/L) = 22.1A646 − 5.26A663.
Hormone Measurements
The hormones ABA, SA, and JA were measured in five individual leaves. Each leaf was freeze dried and 10 mg of freeze dried tissue was used for hormone extraction as described in Forcat et al. (2008). Analysis was performed using an HPLC-ESI/MS-MS.
Statistical Treatment of Leaf Morphological and Biochemical Measurements
Leaf morphology (weight and length) and biochemical assay (total protein, chlorophyll a+b, Rubisco LSU and SSU, and hormone) data were subjected to ANOVA to assess for differences in response over the time course using GenStat (VSN International). Data from hormone assays were subjected to a log10 transformation (including the addition of a small constant to cope with zero observations) prior to analysis to satisfy the assumption of homogeneity of variance. LSDs were calculated at a 5% significance level to allow easy comparison of differences between adjacent time points. Significant effects noted in the results relate to either F tests for the overall variability over time or t tests for comparisons between adjacent time points.
Microarray Analysis
RNA Preparation and Labeling
Total RNA was isolated from four individual leaves from each sampled time point (arbitrarily labeled as biological replicates A, B, C, and D) using TRIzol reagent (Invitrogen), purified with RNeasy columns (Qiagen), and amplified using the MessageAmp II aRNA Amplification kit (Ambion) in accordance with the kit protocol with a single round of amplification. Cy3- and Cy5-labeled cDNA probes were prepared by reverse transcribing 5 μg of aRNA with Cy3- or Cy5-dCTP (GE Healthcare) and a modified dNTP mix (10 mM each dATP, dGTP, and dTTP; 2 mM dCTP) using random primers (Invitrogen) and SuperScript II reverse transcriptase (Invitrogen), with the inclusion of RNase inhibitor (RNaseOUT; Invitrogen) and DTT. Labeled probes were purified using QiaQuick PCR Purification columns (Qiagen), freeze-dried, and resuspended in 50 μL of hybridization buffer (25% formamide, 5× SSC, 0.1% SDS, and 0.5μg/μL yeast tRNA; Invitrogen).
Microarray Experiments
The microarray experiments were performed using the CATMA (version 3) microarray (Allemeersch et al., 2005; http://www.catma.org). CATMA probe annotations were updated using the TAIR9 release: oligo sequences of CATMA array probes were mapped to individual mRNA sequences of transcripts from the TAIR9 genome assembly using BLASTn (Altschul et al., 1997), with e-value cutoff of 0.01. Additionally, results were filtered to exclude alignments shorter than 30 bp or with less than 80% sequence identity. The best matching gene model (by e-value of hit to transcript) was identified for each probe. In addition, probe sequences were mapped to TAIR9 genomic DNA to clarify cases where a probe had been designed to a region of an earlier genome assembly now unannotated in TAIR9.
A novel experimental design strategy (A. Mead, unpublished data), based on the principle of the “loop design” (Kerr and Churchill, 2001), was developed to enable efficient extraction of information about key sample comparisons using a two-color hybridization experimental system. With 88 distinct samples (four biological replicates at each of 22 time points) to be compared, the experimental design included 176 two-color microarray slides, allowing four technical replicates of each sample to be observed. The detailed structure of the design, indicating how pairs of treatments were allocated to arrays, is described in Supplemental Methods 1 online, with an illustrative diagram shown in Supplemental Figure 6 online. According to a randomization of this experimental design, pairs of labeled samples were hybridized to slides overnight at 42°C. Following hybridization, slides were washed and scanned using an Affymetrix 428 array scanner at 532 nm (Cy3) and 635 nm (Cy5). Scanned data were quantified using Imagene 7.5.0 software (BioDiscovery, Inc.).
MAANOVA Analysis
A local adaptation of the MAANOVA package (Wu et al., 2003) was used to analyze the quantified microarray data, providing data quality assurance, slide normalization through LOWESS data transformation, mixed model fitting, and identification of genes showing significant differential expression via F tests of fixed (treatment) terms included within the model. MAANOVA was selected to analyze the data because it is able to provide an accurate analysis of the effects on gene expression of multiple sources of variation (both fixed, treatment, terms, and random sources of background variation) in the experimental design, harnessing the power of direct comparisons between pairs of samples obtained using two-channel microarrays (Churchill, 2004). Full details of the data quality checking procedures, of the mixed model fitting approach to describe the observed gene expression data, and of the construction of F tests for fixed treatment terms are given in Supplemental Methods 1 online. Having fitted the mixed model to each gene, predicted means were calculated for each of the 88 samples, assuming the full treatment model (effects of day, time of day, the interaction between them, and the nested biological replicates) to produce a four-replicate 22–time point data set for each gene, or assuming a reduced treatment model (effects of day and the nested biological replicates) to produce an eight-replicate 11–time point data set for each gene. These data sets were then used in subsequent analyses.
Selection of Differentially Expressed Genes
The most significant differentially expressed genes were identified initially and this was followed by visual analysis of genes close to the borderline of significance. First, CATMA probes with no corresponding gene model in the TAIR9 annotation were ignored; also, replicate CATMA probes were removed (the most gene-specific probe being identified in each case). In total, 4989 genes had an adjusted day main effect F test P-value < 0.0001 (after multiple testing correction via a step-down FDR-controlling procedure (Westfall et al., 1998; Benjamini and Liu, 1999), equivalent to responses showing a significant test result at an FDR of P < 0.0001), and these were included in the initial list of differentially expressed genes. The patterns of expression of all genes with an adjusted day main effect F test P-value between 0.0001 and 0.05 (further responses showing a significant test result at an FDR of P < 0.05) were then screened visually to remove any showing either a small or a very variable change in expression over time. The final list of 6323 differentially expressed genes, with adjusted F test statistics, is shown in Supplemental Table 1 online.
Gene Expression Profile Clustering
Clustering of coregulated genes was performed by the application of SplineCluster (Heard et al., 2006), a Bayesian model-based hierarchical clustering algorithm for time series data, using the mean of the biological replicates for each gene. Recent functionality added to SplineCluster, (Heard, 2011) improves the gene allocation to clusters. Where a gene has become an outlier for its allocated cluster, it is reallocated to alternative clusters to maximize the log marginal likelihood once more. This option was used on all SplineCluster analyses presented in this article. The 22–time point data (averaged across the four biological replicates) was clustered using a prior precision of 5×10−4, while the other data set composed of 11 time points was averaged across all eight morning and afternoon biological replicates before being clustered using a prior precision of 1×10−4. These prior precisions were selected as they produce ~50 clusters for each of the two data sets.
GP Gradient Analysis
To identify an ordering of events, the rate of change of gene expression (gradient) was inferred using a GP regression approach (see Supplemental Methods 1 online), which has the notable advantage of incorporating all biological replicates. Furthermore, since the marginal distribution of a GP is itself a Gaussian distribution, the probability that the gradient (at any particular time) lies sufficiently far from zero may be calculated analytically. When data are sufficiently time resolved, the GP model may therefore be used to identify times when the gradient of a gene expression profile is significantly positive (increased), negative (decreased), or not statistically different from zero (steady), whereas for less time-resolved data, it may identify times of significant change to the global trend.
GO Analysis
GO annotation analysis on gene clusters was performed using the BiNGO 2.3 plugin tool in Cytoscape version 2.6 with GO_full and GO_slim categories, as described by Maere et al. (2005). Over-represented GO_Full categories were identified using a hypergeometric test with a significance threshold of 0.05 after a Benjamini and Hochberg FDR correction (Benjamini and Hochberg, 1995).
Promoter Analysis
Plant position-specific scoring matrices (PSSMs) were collected from the TRANSFAC database, version 2010.3, (Matys et al., 2006) and the PLACE database (Higo et al., 1999). This set was supplemented with PSSMs for a heat shock element (TRANSFAC matrix record M00146) and two NAC TF binding sites (Olsen et al., 2005) since these important motifs were absent from the databases. PSSMs were clustered, and a representative of each cluster was chosen for screening. Promoter regions corresponding to 500 bp upstream of the transcription start site were retrieved from the Ensembl Plants sequence database (release 50).
For any given PSSM and promoter, we scanned the sequence and computed a matrix similarity score (Kel et al., 2003) at each position on both strands. P values for each score were computed from a score distribution obtained by applying the PSSM to a random sequence of 100 million bases in length generated by a 3rd order Markov model learned from the whole Arabidopsis genome. We took the top k nonoverlapping hits and performed the binomial test for the occurrence of k sites with observed n values within a sequence of length 500 bp. The parameter k is optimized within the range 1 to 5 for minimum binomial P-value. This allows detection of binding sites without a fixed threshold per binding site. Using a threshold (P < 0.05), the presence or absence of a PSSM was scored for each promoter based on the binomial probability.
For each PSSM, its frequency in promoters of each cluster was compared with its occurrence in all promoters in the entire genome. Motif enrichment was calculated using the hypergeometric distribution (phyper function in the R stats package). Hypergeometric P-values were corrected for the number of clusters tested using Bonferroni correction. Corrected P-values ≤ 0.05 were considered significant. Sequence logos were generated using code modified from Lenhard and Wasserman (2002). Sequence analysis was performed within the APPLES software framework (S. Ott, unpublished data).
TF Family Analysis
Gene expression activity was analyzed for 1733 TFs, grouped into 50 families defined in the Arabidopsis thaliana Transcription Factor Database, AtTFDB (Palaniswamy et al., 2006; 1843 TF, 50 families as of June 2010). Of these, 1733 were probes on the CATMA array using the GP gradient model. Families overrepresented for genes with significantly positive or negative gradients at each time point, using all genes within the experiment as a reference, were identified using the hypergeometric distribution (computed using the hypgeomdist function in MS Excel 12.2.3) with Benjamini and Hochberg FDR correction. A heatmap of adjusted P-values, using five levels of significance (0.01, 0.05, 0.1, 0.25, and 0.5) was then generated, using only those values that correspond to overrepresented counts (e.g., the proportion of positive/negative gradient TFs for a given family is larger than the proportion of positive/negative gradient genes in the entire data set for each time point).
Variational Bayesian State Space Modeling
Data for eight biological replicates from the 11–time point data series were used to generate a network model using the method published in Beal et al., 2005. ANAC092 was used as the gene on which to base the model and two groups of genes were selected to accompany it. First, several genes were selected that showed rapidly increased expression following induced expression of ANAC092 in green leaves (from Balazadeh et al., 2010), many of which were also in clusters enriched for NAC domains. Therefore, these are likely to be direct or indirect targets of ANAC092 activation. Second, a group of TFs that show coexpression with ANAC092 selected from the clusters 41 through 45 was included. Ten models were run from different random seeds and connections occurring in more than 50% of models at a confidence level of >95% were included in the network shown in Figure 9.
Data Repository
The microarray data used in this article have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and have been given a GEO Series accession number, GSE22982.
Accession Numbers
Arabidopsis gene names and identifiers referred to in this article are: ANAC092 (At5g39610), WRKY53 (At4g23810), ANAC029 (At1g69490), SAG12 (At5g45890), LHY (At1g01060), CCA1 (At2g46830), PRR7 (At5g02810), PHYA (At1g09570), CRY1 (At4g08920), PIF4 (At2g43010), ELF4 (At2g40080), PCL1 (At3g46640), LUT1 (At3g53130), LUT2 (At5g57030), LUT5 (At1g31800), ARR4 (At1g10470), ARR6 (At5g62920), ARR7 (At1g19050), sedoheptulose bisphosphatase (At3g55800), HCF101 (At3g24430), HCF109 (At5g36170), HCF152 (At3g09650), HCF173 (At1g16720), HCF208 (At5g52110), GLK2 (At5g44190), GLK1 (At2g20570), LHCB2.2 (At2g05070), LHCB4.2 (At3g08940), LHCB6 (At1g15820), ATG7 (At5g45900), ATG8H (At3g06420), ATG8A (At4g21980), ATG8B (At4g04620), DREB2A (At5g05410), LSD1 (At4g20380), AtMPK7 (At2g18170), HSP70 (At3g12580), (AHK3 (At1g27320), ERD1 (At5g51070), ERD14 (At1g76180), RAB18 (At5g66400), ABF2 (At1g45249), AOC1 (At3g25760), AOC4 (At1g13280), 12-oxophytodienoate reductase (At2g06050), MYC2 (At1g32640), JAZ1 (At1g19180), JAZ6 (At1g72450), JAZ8 (At1g30135), COI1 (At2g39940), LOX3 (At1g17420), ABI1 (At4g26080), AFP1 (At1g69260), RD20 (At2g33380), RD26 (At4g27410), NF-YA4 (At2g34720), CCD7 (At2g44990), CCD8 (At4g32810), ORE9 (At2g42620), AtMC6 (At1g79320), AtMC9 (At5g04200), LACS6 (At3g05970), ACX1 (At4g16760), ETR1 (At1g66340), ACO2 (At1g62380), TUA2 (At1g50010), TUA4 (At1g04820), TUA5 (At5g19780), ACT3 (At3g53750), ACT11 (At3g12110), AtAURORA1 (At4g32830), AtAURORA2 (At2g25880), SGR1/NYE1 (At4g22920), SGR2 (At4g11910), NYC1 (At4g13250), and PaO (At3g44880).
Author Contributions
All authors had a role in discussion of results. V.B.-W., J.B., K.D., B.T., S.J., D.R., S.O., and D.L.W. designed the research; E.B., E.H., C.Z., K.M., A.T., and C.J. did the experimental work; A.M. designed the array experiment; S.M., L.H., and A.M. developed microarray data extraction and primary analysis methods; S.M. designed Gene Viewer; S.M., D.L.W., and S.K. developed and applied clustering methods; Y.-s.K., C.H., V.B.-W., E.B., R.H., C.A.P., and D.J. performed data analysis and biological interpretation; C.A.P. and D.L.W. developed the gradient analysis tool; R.H. and S.O. did the promoter motif analysis; D.J. developed the TF family analysis method; J.D.M. and R.L. organized data handling and web page; and V.B.-W. wrote the majority of the paper with input from E.B., E.H., S.M., A.M., C.H., Y.-s.K., R.H., S.O., D.J., C.A.P., S.K., K.D., J.D.M., and D.L.W.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Venn Diagram Summarizing Numbers of Genes Showing Significantly Different Expression for Different Combinations of Treatment Terms in the MAANOVA Fixed Model.
Supplemental Figure 2. Expression Patterns of Selected Genes Showing Time-of-Day Changes Only.
Supplemental Figure 3. Expression Patterns of Selected Genes during Leaf Senescence.
Supplemental Figure 4. Gradient Analysis on Selected Groups of Genes.
Supplemental Figure 5. Expression Patterns of Selected Hormone Biosynthesis Genes.
Supplemental Figure 6. Representation of the Experimental Design for the Microarray Experiment.
Supplemental Table 1. Enriched GO Terms in Genes Downregulated or Upregulated during Senescence.
Supplemental Table 2. Genes Used in the Variational Bayesian State Space Modeling Method Model Shown in Figure 9.
Supplemental Data Set 1. Genes Differentially Expressed during Senescence.
Supplemental Data Set 2. Enriched GO Terms in Each Cluster Shown in Figure 3.
Supplemental Data Set 3. Gradient Data for Differentially Expressed Genes.
Supplemental Data Set 4. Cluster Patterns Identified from the Gradient Analysis Showing Enriched GO terms.
Supplemental Data Set 5. Known DNA Sequence Motif Enrichment.
Supplemental Data Set 6. TF Family Analysis Data.
Supplemental Data Set 7. Senescence-Enhanced Genes Differentially Expressed in Inducible ANAC092 Line.
Supplemental Methods 1. Microarray Experiments and GP Regression and Gradient.
Supplementary Material
Acknowledgments
We thank Mary Coates (University of Warwick) for helping with manuscript preparation and Miriam Gifford (University of Warwick) for critical reading. We thank Mark Bennett (Imperial College) and Prof. Murray Grant (Exeter University) for performing the hormone measurements. E.B., E.H., C.Z., C.J., and K.M. were funded for this work by a Biotechnology and Biological Sciences Research Council (BBSRC) core strategic grant to Warwick HRI; S.M., S.K., and R.H. are funded by the Engineering and Physical Sciences Research Council/BBSRC-funded Warwick Systems Biology Doctoral Training Centre; L.H. was funded by a BBSRC studentship; J.B., K.D., V.B.-W., D.R., D.L.W., S.O., C.H., Y.-s.K., C.A.P., D.J., J.D.M., R.L., and A.T. are part of the BBRSC-funded grant Plant Response to Environmental Stress Arabidopsis (BB/F005806/1).
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemeersch J., et al. (2005). Benchmarking the CATMA microarray. A novel tool for Arabidopsis transcriptome analysis. Plant Physiol. 137: 588–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A., et al. (2004). A transcriptional timetable of autumn senescence. Genome Biol. 5: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh S., Siddiqui H., Allu A.D., Matallana-Ramirez L.P., Caldana C., Mehrnia M., Zanor M.I., Köhler B., Mueller-Roeber B. (2010). A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J. 62: 250–264 [DOI] [PubMed] [Google Scholar]
- Beal M.J., Falciani F., Ghahramani Z., Rangel C., Wild D.L. (2005). A Bayesian approach to reconstructing genetic regulatory networks with hidden factors. Bioinformatics 21: 349–356 [DOI] [PubMed] [Google Scholar]
- Bellman R. (1961). Adaptive Control Processes: A Guided Tour. (Princeton, NJ: Princeton University Press; ). [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57: 289–300 [Google Scholar]
- Benjamini Y., Liu W. (1999). A step-down multiple testing procedure that controls the false discovery rate under independence. J. Stat. Planning Inference 82: 163–170 [Google Scholar]
- Breeze E., Harrison E., Page T., Warner N., Shen C., Zhang C., Buchanan-Wollaston V. (2008). Transcriptional regulation of plant senescence: From functional genomics to systems biology. Plant Biol. (Stuttg.) 10(Suppl. 1), 99–109 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P.O., Nam H.G., Lin J.F., Wu S.H., Swidzinski J., Ishizaki K., Leaver C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signaling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Choudhury A., Lahiri A. (2011). Comparative analysis of abscisic acid-regulated transcriptomes in Arabidopsis. Plant Biol. (Stuttg.) 13: 28–35 [DOI] [PubMed] [Google Scholar]
- Churchill G.A. (2004). Using ANOVA to analyze microarray data. Biotechniques 37: 173–175, 177 [DOI] [PubMed] [Google Scholar]
- Coll N.S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J.L., Epple P. (2010). Arabidopsis type I metacaspases control cell death. Science 330: 1393–1397 [DOI] [PubMed] [Google Scholar]
- Demidov D., Hesse S., Tewes A., Rutten T., Fuchs J., Ashtiyani R.K., Lein S., Fischer A., Reuter G., Houben A. (2009). Aurora1 phosphorylation activity on histone H3 and its cross-talk with other post-translational histone modifications in Arabidopsis. Plant J. 59: 221–230 [DOI] [PubMed] [Google Scholar]
- Demidov D., Van Damme D., Geelen D., Blattner F.R., Houben A. (2005). Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dóczi R., Brader G., Pettkó-Szandtner A., Rajh I., Djamei A., Pitzschke A., Teige M., Hirt H. (2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H., Walker J.M., Friedman E.M., Thompson A.R., Vierstra R.D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277: 33105–33114 [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood D.C., Mead A., Sergeant M.J., Burton K.S. (2008). Statistical modelling of transcript profiles of differentially regulated genes. BMC Mol. Biol. 9: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P., Mack A.A., Morris V.R., Dangl J.L. (2003). Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc. Natl. Acad. Sci. USA 100: 6831–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Somssich I.E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10: 366–371 [DOI] [PubMed] [Google Scholar]
- Forcat S., Bennett M.H., Mansfield J.W., Grant M.R. (2008). A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Fulda M., Shockey J., Werber M., Wolter F.P., Heinz E. (2002). Two long-chain acyl-CoA synthetases from Arabidopsis thaliana involved in peroxisomal fatty acid β-oxidation. Plant J. 32: 93–103 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194 [DOI] [PubMed] [Google Scholar]
- Grbic V., Bleecker A.B. (1995). Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 8: 595–602 [Google Scholar]
- Guo Y., Gan S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Gutterson N., Reuber T.L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377 [DOI] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Fukushige H., Hildebrand D.F., Gan S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard N.A. (2011). Iterative reclassification in agglomerative clustering. J. Comput. Graph. Stat. http://dx.doi.org/10.1198/jcgs.2011.09111
- Heard N.A., Holmes C.C., Stephens D.A. (2006). A quantitative study of gene regulation involved in the immune response of anopheline mosquitoes: An application of Bayesian hierarchical clustering of curves. JASA 101: 18–29 [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S., Feller U. (2002). Nitrogen metabolism and remobilization during senescence. J. Exp. Bot. 53: 927–937 [DOI] [PubMed] [Google Scholar]
- Hwang I., Sheen J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Dröge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., Parcy F.; bZIP Research Group (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Kel A.E., Gossling E., Reuter I., Cheremushkin E., Kel-Margoulis O.V., Wingender E. (2003). MATCHTM: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 13: 3576–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr M.K., Churchill G.A. (2001). Experimental design for gene expression microarrays. Biostatistics 2: 183–201 [DOI] [PubMed] [Google Scholar]
- Ketelaar T., Voss C., Dimmock S.A., Thumm M., Hussey P.J. (2004). Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 567: 302–306 [DOI] [PubMed] [Google Scholar]
- Kizis D., Lumbreras V., Pagès M. (2001). Role of AP2/EREBP transcription factors in gene regulation during abiotic stress. FEBS Lett. 498: 187–189 [DOI] [PubMed] [Google Scholar]
- Kikis E.A., Khanna R., Quail P.H. (2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44: 300–313 [DOI] [PubMed] [Google Scholar]
- Kim H.J., Ryu H., Hong S.H., Woo H.R., Lim P.O., Lee I.C., Sheen J., Nam H.G., Hwang I. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Woo H.R., Kim J., Lim P.O., Lee I.C., Choi S.H., Hwang D., Nam H.G. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kim J.M., DellaPenna D. (2006). Defining the primary route for lutein synthesis in plants: The role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. USA 103: 3474–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., To T.K., Nishioka T., Seki M. (2010). Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 33: 604–611 [DOI] [PubMed] [Google Scholar]
- Lee E.J., Matsumura Y., Soga K., Hoson T., Koizumi N. (2007). Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 48: 405–413 [DOI] [PubMed] [Google Scholar]
- Lenhard B., Wasserman W.W. (2002). TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics 18: 1135–1136 [DOI] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colón-Carmona A., Gutiérrez R.A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Martínez D.E., Costa M.L., Guiamet J.J. (2008). Senescence-associated degradation of chloroplast proteins inside and outside the organelle. Plant Biol. (Stuttg.) 10(Suppl. 1), 15–22 [DOI] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Matys V., et al. (2006). TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 34(Database issue): D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. (2003). HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15: 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Meierhoff K., Westhoff P. (1996). Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Miao Y., Laun T., Zimmermann P., Zentgraf U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55: 853–867 [DOI] [PubMed] [Google Scholar]
- Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K. (2006). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580: 6537–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., Rieter E., Klionsky D.J., Reggiori F. (2009). Multiple roles of the cytoskeleton in autophagy. Biol. Rev. Camb. Philos. Soc. 84: 431–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K.A.H., MacKerness S.A., Page T., John C.F., Murphy A.M., Carr J.P., Buchanan-Wollaston V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23: 677–685 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Katsura K., Maruyama K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60: 51–68 [DOI] [PubMed] [Google Scholar]
- Noodén L.D., Singh S., Letham D.S. (1990). Correlation of xylem sap cytokinin levels with monocarpic senescence in soybean. Plant Physiol. 93: 33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ongaro V., Leyser O. (2008). Hormonal control of shoot branching. J. Exp. Bot. 59: 67–74 [DOI] [PubMed] [Google Scholar]
- Palaniswamy S.K., James S., Sun H., Lamb R.S., Davuluri R.V., Grotewold E. (2006). AGRIS and AtRegNet. A platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent C., Dimitrov S. (2003). Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 116: 3677–3685 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Fründt C., Gutierrez C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33: 801–811 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Somssich I.E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28: 123–133 [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J.M., Fulda M.S., Browse J.A. (2002). Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129: 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol A., Kwiatkowska A., Jerzmanowski A., Prymakowska-Bosak M. (2007). Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227: 245–254 [DOI] [PubMed] [Google Scholar]
- Staswick P.E. (1994). Storage proteins of vegetative plant tissues. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45: 303–322 [Google Scholar]
- Staswick P.E. (2008). JAZing up jasmonate signaling. Trends Plant Sci. 13: 66–71 [DOI] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58: 321–346 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B., Somssich I.E. (2004). WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 7: 491–498 [DOI] [PubMed] [Google Scholar]
- van der Graaff E., Schwacke R., Schneider A., Desimone M., Flügge U.I., Kunze R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141: 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley J.W., Coughlan S., Hudson M.E., Covington M.F., Kaspi R., Banu G., Harmer S.L., Dehesh K. (2007). Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 3: 1800–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Wang P., Korkaric M., Capper R.G., Saunders N.J., Langdale J.A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L.M., Gan S., Quirino B., Amasino R.M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Weisshaar B., Jenkins G.I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1: 251–257 [DOI] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall P.H., Krishen A., Young S.S. (1998). Using prior information to allocate significance levels for multiple endpoints. Stat. Med. 17: 2107–2119 [DOI] [PubMed] [Google Scholar]
- Woo H.R., Chung K.M., Park J.-H., Oh S.A., Ahn T., Hong S.H., Jang S.K., Nam H.G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13: 1779–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Kerr K., Cui X., Churchill G. (2003). MAANOVA: A software package for the analysis of spotted cDNA microarray experiments. The Analysis of Gene Expression Data: Methods and Software, Parmigiani G., Garett E., Irizarry R., Zeger S., (New York: Springer; ), pp. 313–341 [Google Scholar]
- Yang T., Poovaiah B.W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277: 45049–45058 [DOI] [PubMed] [Google Scholar]
- Yoo J.H., et al. (2005). Direct interaction of a divergent CaM isoform and the transcription factor, MYB2, enhances salt tolerance in Arabidopsis. J. Biol. Chem. 280: 3697–3706 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Hanaoka H., Sato S., Kato T., Tabata S., Noda T., Ohsumi Y. (2004). Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.