This study defines a previously unknown combinatorial interaction between transcription factors in plants and provides a better understanding of how plants integrate signals to regulate petal development. It also highlights the role of a motif in the bHLH transcription factor that resembles a motif that was previously shown to be important in auxin signaling.
Abstract
Plant organ growth and final size are determined by coordinated cell proliferation and expansion. The BIGPETALp (BPEp) basic helix-loop-helix (bHLH) transcription factor was shown to limit Arabidopsis thaliana petal growth by influencing cell expansion. We demonstrate here that BPEp interacts with AUXIN RESPONSE FACTOR8 (ARF8) to affect petal growth. This interaction is mediated through the BPEp C-terminal domain (SDBPEp) and the C-terminal domain of ARF8. Site-directed mutagenesis identified an amino acid consensus motif in SDBPEp that is critical for mediating BPEp-ARF8 interaction. This motif shares sequence similarity with motif III of ARF and AUXIN/INDOLE-3-ACETIC ACID proteins. Petals of arf8 mutants are significantly larger than those of the wild type due to increased cell number and increased cell expansion. bpe arf8 double mutant analyses show that during early petal development stages, ARF8 and BPEp work synergistically to limit mitotic growth. During late stages, ARF8 and BPEp interact to limit cell expansion. The alterations in cell division and cell expansion observed in arf8 and/or bpe mutants are associated with a change in expression of early auxin-responsive genes. The data provide evidence of an interaction between an ARF and a bHLH transcription factor and of its biological significance in regulating petal growth, with local auxin levels likely influencing such a biological function.
INTRODUCTION
The growth rate and final size of plant organs are determined by two distinct yet integrated processes: cell proliferation, with concomitant accumulation of biomass, and cell expansion. These processes are highly reproducible when genetically identical individuals are grown in the same environment (Menand et al., 2004; Ingram and Waites, 2006). The genetic and molecular mechanisms that coordinate these two processes are beginning to be elucidated in plants and in animals (Anastasiou and Lenhard, 2007; Bögre et al., 2008; Busov et al., 2008; Krizek, 2009).
In plants, the physical dimensions of leaves, roots, and stems can vary significantly depending on the amount of light, temperature variations, or soil conditions. By contrast, flower organ size is remarkably constant within a given species, indicative of strong endogenous control during morphogenesis. Maintaining correct growth patterns to generate petal organs with characteristic shapes and sizes is important for the positioning of the sexual organs to ensure successful pollination and/or pollen dispersal. During the past decade, a number of studies have focused on how petal organ identity is defined (Krizek and Fletcher, 2005; Irish, 2010). For example, in Arabidopsis thaliana, the transcription factors PISTILLATA, APETALA3 (AP3), SEPALLATA (SEP1 to SEP4), and AP1 are responsible for the specification of petal organ identity (Honma and Goto, 2001; Theissen and Saedler, 2001; Krizek and Fletcher, 2005; Irish, 2010). However, how a particular flower organ such as the petal attains its final characteristic shape and size based on a specific growth pattern remains unclear. The discovery of genes regulating floral organ growth downstream of the floral organ identity genes has contributed to the understanding of the molecular and genetic mechanisms that underlie flower development (Zik and Irish, 2003; Wellmer et al., 2004; Zimmermann et al., 2004; Nakayama et al., 2005; Schmid et al., 2005; Szécsi et al., 2006; Brioudes et al., 2009, 2010). Nevertheless, there are still many unanswered questions. To move beyond the floral organ identity specification toward determining the downstream regulatory interactions and characterizing the biological functions of the effector genes, one ambitious task is to integrate the various pathways that specify petal organ patterning, size, shape, and symmetry into a comprehensive network. Such a task requires the discovery and the fine characterization of the underlying pathways involved in petal development.
Petal growth depends on cell division during early development stages but is controlled by cell expansion during late flower development (Hill and Lord, 1989; Smyth et al., 1990). Several genes were shown to regulate petal growth by affecting cell proliferation and/or cell expansion in an organ-specific manner (Anastasiou and Lenhard, 2007). Some of these genes (e.g., JAGGED, AINTEGUMENTA, and ARGOS) were shown to affect petal growth by positively regulating cell proliferation (Mizukami and Fischer, 2000; Hu et al., 2003; Dinneny et al., 2004), whereas other genes (BigBrother, KLU, and DA1) were shown to affect final organ size by negatively regulating the duration/period of cell proliferation (Disch et al., 2006; Anastasiou et al., 2007; Li et al., 2008). FRILL1 is implicated in maintaining the mitotic state and subsequently affects cell expansion during the late stage of petal lamina formation (Hase et al., 2000). BIGPETALp (BPEp) is a basic helix-loop-helix (bHLH) transcription factor preferentially expressed in petals. It was shown to regulate petal growth by restricting cell expansion (Szécsi et al., 2006; Brioudes et al., 2009). Interestingly, BPEp mRNA originates from an alternative splicing event in the ubiquitously expressed gene BPE regulated by phytohormone jasmonate signaling (Brioudes et al., 2009).
Here, we demonstrate the physical and genetic interactions between BPEp and the AUXIN RESPONSE FACTOR8 (ARF8). We report the biological significance of this interaction in negatively regulating mitotic growth during early petal development stages and postmitotic growth during late petal development stages, thus influencing organ growth in plants.
RESULTS
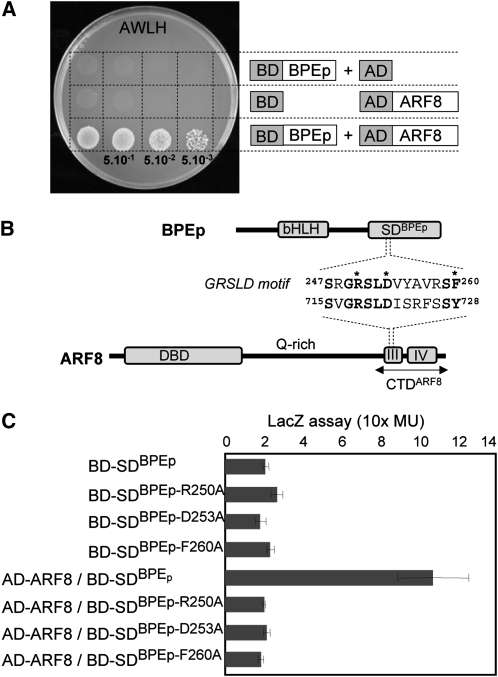
BPEp Interacts with ARF8 in Yeast and in Planta through Their SDBPEp and C-Terminal Domain, Respectively
We performed a yeast two-hybrid screen to identify putative BPEp binding proteins. BPEp full-length protein or its C-terminal specific domain (SDBPEp) (Szécsi et al., 2006) were fused to the GAL4 binding domain and used as baits. The SDBPEp domain is translated from the fifth intron that is retained in BPEp through an alternative splicing event. Since the fifth intron is not retained in BPEub, the other transcript expressed from the BPE gene (Szécsi et al., 2006; Brioudes et al., 2009), the SDBPEp domain is unique to BPEp. Two million clones of the floral cDNA library CD4-30 (Fan et al., 1997) were screened using each of the BPEp constructs. Out of the 40 positive clones, 20 harbored cDNA inserts in the correct reading frame and corresponded to known or putative proteins in Arabidopsis. One target binding partner corresponded to the ARF8 transcription factor (Figure 1A). Different clones corresponding to ARF8 were identified in two independent screens, suggesting that ARF8 represents a good BPEp interacting protein candidate. ARF8 was also isolated when using the SDBPEp as bait, suggesting that the BPEp interacts with ARF8 through its SDBPEp C-terminal domain. Moreover, all clones of ARF8 isolated in the yeast two-hybrid screens contained the C-terminal domain (CTDARF8) that harbors the motifs III and IV. We conducted bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaf cells using SDBPEp and CTDARF8 (see Supplemental Figure 1 online). Fluorescence was observed in cells in which SDBPEp and CTDARF8 individually fused to parts of yellow fluorescent protein (YFP) were expressed, indicating reconstitution of YFP (see Supplemental Figure 1A online). We observed the physical interaction in the cytoplasm since both constructs lack nuclear localization signals. These data are in agreement with the yeast-two hybrid results and suggest that that BPEp and ARF8 interact in planta through their C-terminal domains SDBPEp and CTDARF8, respectively (see Supplemental Figure 1A online).
Figure 1.
BPEp and ARF8 Proteins Interact in Yeast.
(A) Interaction between BPEp and ARF8 in yeast two-hybrid assays. Yeast cotransformed with pAS2.1-BPEp (BD-BPEp) as bait and pAD-GAL4-2.1-ARF8 (AD-ARF8) as prey or with plasmid vectors without inserts for control of self-activation was diluted three times, and drops were deposited on selective medium A−W−L−and H− (AWLH).
(B) Schematic representation of the BPEp and the ARF8 proteins. The GRSLD motif in SDBPEp that shares sequence similarity with motif III of ARF8 C-terminal domain (CTDARF8) is shown. Conserved amino acids are shown in bold. Key amino acids in the GRSLD motif of SDBPEp that were mutagenized are indicated with asterisks. DBD, DNA binding domain.
(C) Interactions between ARF8 and SDBPEp wild type or mutated at amino acids Arg-250, Asp-253, or Phe-260 were analyzed using β-galactosidase assays in yeast. Mutations of the key amino acids in the GRSLD motif interfere with BPEp and ARF8 interaction. Values are means of three biological replicates. Error bars indicate sd.
ARF C-terminal motifs are known to be required for homodimerization and also for heterodimerization with AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) proteins through shared III and IV motifs (Ulmasov et al., 1997). Alignments between SDBPEp and CTDARF8 highlighted an amino acid sequence referred to as the GRSLD motif (Figure 1B) in the SDBPEp that shares high sequence similarity with motif III of ARF proteins (see Supplemental Figure 2 online). The GRSLD motif is composed of a conserved stretch of five amino acids, Gly249-Arg-Ser-Leu-Asp253, a conserved Ser-259, and two conserved aromatic amino acids Phe-260 and Phe-271 (Figure1B; see Supplemental Figure 2 online). Site-directed mutagenesis was performed to mutate Arg-250, Asp-253, or Phe-260 of the GRSLD motif into Ala. Using quantitative β-galactosidase assays, we analyzed whether the SDBPEp domains containing the point mutations retained the ability to interact with ARF8 and found that all mutated SDBPEp proteins lost their ability to interact with ARF8 (Figure 1C). Similarly, a considerable reduction in fluorescent complementation in BiFC assays was observed when using mutated SDBPEp R250A, D253A, F260A, or R250A/S251A/D253A (see Supplemental Figures 1B to 1E online). Together, the data demonstrate that the GRSLD motif is important for mediating BPEp and ARF8 interaction in vivo.
BPEp and ARF8 were fused with green fluorescent protein (GFP) and mOrange, respectively, and then transiently expressed in N. benthamiana leaf cells (see Supplemental Figures 1F to 1H online). To prevent the fifth intron from being spliced off in BPEp, we used a version of BPEp sequence in which the splicing donor and acceptor sequences of intron 5 were mutated (see Methods). The ARF8 and BPEp proteins colocalized in the nucleus of the tobacco cells. These data together with that from the BiFC assays suggest that interaction between ARF8 and BPEp likely occurs in the nucleus.
BPEp and ARF8 Show Similar Expression Profiles during Petal Development
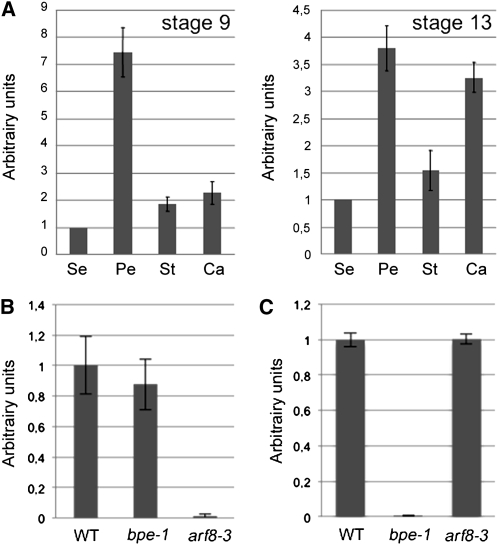
Szécsi et al. (2006) reported that BPEp is preferentially expressed in petals and that its expression is highly induced at late flower development stages where petals differentiate and expand. Analysis of ARF8 expression using the Genevestigator gene Atlas (Zimmermann et al., 2004) showed that ARF8 is ubiquitously expressed, and its expression level is also the highest at late flower development during petal differentiation and expansion stages. Nagpal et al. (2005) showed using the promoterARF8:β-glucuronidase fusion that ARF8 is expressed in the petals of stage 9 and 10 flowers. Real-time quantitative RT-PCR (qPCR) experiments showed that like BPEp, ARF8 is preferentially expressed in petals (at flower development stages 9 and 13) compared with the other flower organs (Figure 2A). Therefore, BPEp and ARF8 have overlapping expression patterns during petal development.
Figure 2.
ARF8 Exhibits High Expression Levels in Petals.
(A) qPCR analyses of ARF8 mRNA in sepals (Se), petals (Pe), stamens (St), and carpels (Ca) at flower development stages 9 and 13. Expression value of ARF8 in sepals was set to 1, and the relative ARF8 expression in the different flower organs is presented. Each value represents the mean of three biological replicates ± se.
(B) and (C) qPCR analyses of ARF8 mRNA accumulation (B) and of BPEp mRNA accumulation (C) in bpe-1 and arf8-3 inflorescences. WT, wild type.
qPCR analyses of BPEp expression in an arf8 loss-of-function mutant background, and vice versa, showed that the accumulation of BPEp is not affected in arf8-3 and similarly ARF8 expression is not affected in bpe-1 (Figures 2B and 2C), demonstrating that BPEp and ARF8 do not influence each other’s expression in an epistatic manner.
Loss of Function of ARF8 Results in Petals with Increased Surface Area and Modified Vein Patterning
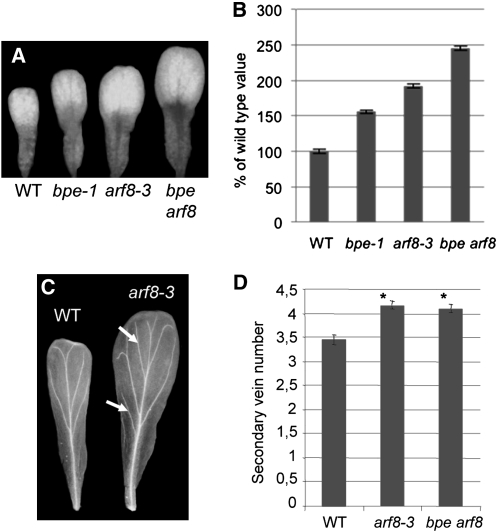
Previously, the ARF8 loss-of-function line arf8-3 was shown to have normal roots and shoots and to have flowers with normal organ numbers and positions compared with the wild-type plants (Nagpal et al., 2005). We investigated arf8-3 mature flower organs for size and shape modifications. At development stage 14, no size modification for sepals was observed, compared with the wild type (see Supplemental Figure 3 online). Consistent with a previous report (Nagpal et al., 2005), stamens of arf8-3 were shorter, whereas carpels were longer compared with the wild type (see Supplemental Figure 3 online). Measurements of the petal surface showed that petals of arf8-3 were significantly larger in size (P value < 0.05; t test) compared with the wild-type plants (~1.8-fold) (Figures 3A and 3B). These data suggest that, like BPEp, ARF8 is a negative regulator of petal growth. Moreover, petals of arf8-3 mutants at flower development stages 12 and beyond displayed a more branched arrangement of vein bundles (Figure 3C). Statistical studies revealed that arf8-3 mature petals have a significantly increased number of secondary vein loops compared with the wild type (Figure 3D); hence, arf8-3 exhibits vein patterning modifications similar to those previously observed in the petals of bpe-1 (Brioudes et al., 2009).
Figure 3.
Loss of Function of ARF8 Affects Petal Organ Growth.
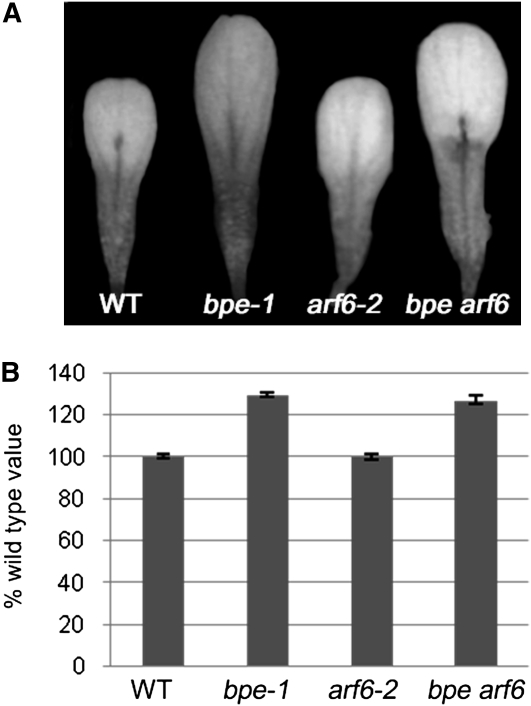
(A) Fully expanded petals at flower development stage 14 in bpe-1, arf8-3, bpe arf8, and wild-type Arabidopsis (WT).
(B) bpe-1, arf8-3, bpe arf8, and wild-type mature (development stage 14) petal surface area. Values are given as mean ± se relative to the wild-type value, set at 100%.
(C) Vein patterning in cleared petals of arf8-3 and wild-type plants. Arrows indicate extra secondary veins in petals of arf8-3.
(D) Number of secondary veins in petals of arf8-3 and bpe arf8 mutants compared with the wild type. Asterisk indicates significant difference from the wild type at the 5% significance level (t test).
Petal Size Increase in arf8-3 Is Associated with Modifications in Both Cell Proliferation and Cell Expansion
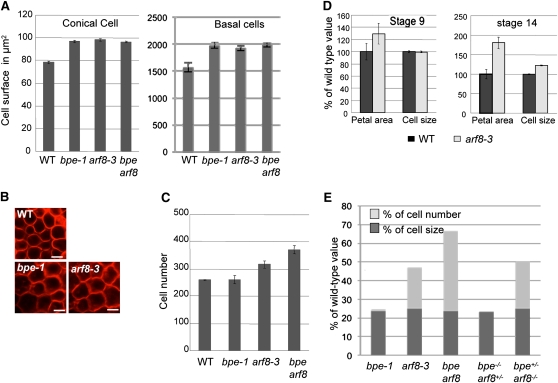
In an earlier study, we showed that the increase in petal surface area in bpe-1 correlates with increased cell expansion (Szécsi et al., 2006). To examine if the petal overgrowth observed in arf8-3 was associated with increased cell proliferation and/or cell expansion, we analyzed cell size and cell number in mature arf8-3 petals (stage 14) and compared the results to the wild type and bpe-1. Conical and basal cell size measurements showed that in mature petals of arf8-3, cells were ~23% larger in size compared with the wild type (Figures 4A and 4B). However, this increase in cell size observed in arf8-3 does not explain the total overgrowth of petals (90% increase compared with the wild type). Moreover, mature petals of arf8-3 flowers at development stage 14 were ~1.5-fold bigger compared with bpe-1 (Figures 3A and 3B), but the cell size increase was similar in arf8-3 and bpe-1 petals (Figures 4A and 4B), thus suggesting a modification in cell number in petals of arf8-3 as well. Consistent with this suggestion, cell number in mature arf8-3 petals at stage 14 was significantly increased (P value < 0.05; t test) compared with that of the wild type or bpe-1 (Figure 4C). In summary, the analyses of petal cell number and size suggest that petal overgrowth in arf8-3 correlates with higher cell proliferation as well as cell overexpansion.
Figure 4.
Petal Overgrowth in ARF8 Loss-of-Function Plants Is Associated with Increased Cell Number and Cell Size.
(A) Cell size measurements (conical cells left and basal cells right) in mature petals of bpe-1, arf8-3, and bpe arf8 show that loss of function of ARF8 or BPE results in similar increased cell size compared with the wild type (WT).
(B) Images of FM4-64–stained epidermal cells from the adaxial distal region of mature petals (stage 14) in wild-type, arf8-3, and bpe-1 plants. Bars = 10 μm.
(C) Cell number measurements in mature petals of arf8-3, bpe-1, and bpe arf8. Values are given as mean ± se.
(D) Petal surface area and cell size measurements at petal development stages 9 and 14. Note that petal overgrowth is observed in arf8-3 before stage 9 and that this petal overgrowth is not associated with increased cell size. Values are given as mean ± se relative to the wild-type value, set at 100%.
(E) Diagram showing the petal overgrowth associated with cell size (dark gray) and cell number (light gray) in single mutants bpe-1, arf8-3, double mutant bpe arf8, and in sesquimutants bpe−/− arf8+/− and bpe+/− arf8−/−. Results are given as percentages of the wild type, whose overgrowth is set at 0.
[See online article for color version of this figure.]
In Arabidopsis, petal development is characterized by a modular growth pattern that consists of early phases of growth (stages 5 to 9) controlled by cell division and later stages (stage 9 up to stage 13) controlled predominantly by cell expansion (Hill and Lord, 1989; Smyth et al., 1990). To examine if the petal overgrowth observed in arf8-3 occurs at early and/or at late development stages, petal surface area was measured at flower development stage 9 in arf8-3 and wild-type plants (Figure 4D). Interestingly, at stage 9, petal cell size was identical in arf8-3 and wild-type plants, but the petals of arf8-3 were around 1.3-fold larger than those of the wild type. We conclude that petal overgrowth in arf8-3 at stage 9 was due to increased cell production leading to an increase in cell number and hence an overall increase in petal size. Taken together, these data show that petal overgrowth in arf8-3 is a composite result of a higher cell proliferation at early development stages (before flower development stage 9) and increased cell expansion at late development stages.
It has been reported that increased cell expansion, especially in leaves and in petals, can be associated with increased endoreduplication (Melaragno et al., 1993; Hase et al., 2005). Analysis of the DNA content of petal cell nuclei by flow cytometry confirmed a low level of endoreduplication in fully developed wild-type petals with a maximum of 8c content for <10% of the nuclei (see Supplemental Figure 4 online). A similar spectrum of DNA content per nucleus was observed in arf8-3 petals, whereas a slight but significant decrease of 4c nuclei was measured in bpe-1. Hence, no enhanced endoreduplication was observed in arf8-3 and bpe-1 petals compared with the wild type, demonstrating that the cell size increase observed in petals of the arf8-3 and bpe-1 lines was not resulting from enhanced endoreduplication, a common landmark of cell expansion (Melaragno et al., 1993; Sugimoto-Shirasu and Roberts, 2003).
ARF8 and BPE Genetically Interact to Influence Petal Development
To test the genetic interaction between BPE and ARF8 further, double mutants of bpe-1 and arf8-3 were generated (bpe arf8). Petal surface area was measured in the bpe arf8 line and compared with those of the wild type, bpe-1, and arf8-3. In bpe arf8 double mutants, petals were significantly increased (t test P value <0.05) in size compared with those of the wild type (almost 2.5-fold), bpe-1 (~1.6-fold), or arf8-3 (~1.3-fold) (Figures 3A and 3B). To examine to what extent cell proliferation and cell expansion contribute to the observed petal overgrowth in bpe arf8, cell number and size were measured and compared with those of the wild type, bpe-1, and arf8-3 (Figures 4A and 4C). In bpe arf8, the petal cell size increase was comparable to that of bpe-1 and arf8-3 single mutants, demonstrating that BPEp and ARF8 likely act in the same pathway to influence cell expansion during petal development. However, a higher cell number count was observed in petals of bpe arf8 compared with the arf8-3 single mutant, suggesting a higher cell proliferation rate in petals of bpe arf8 double mutants (Figure 4C). This result confirms that ARF8 affects cell proliferation during petal development and further suggests a synergistic effect between bpe-1 and arf8-3 losses of function on cell proliferation but not on cell expansion. Analyses of DNA content of petal-cell nuclei by flow cytometry showed that petals of bpe arf8 exhibited slightly higher numbers of 4c nuclei compared with the wild type (see Supplemental Figure 4 online). However, no higher numbers of 8C, 16C, 32C, or 64C nuclei were observed in bpe arf8 petals compared with the wild type, hence suggesting that as for bpe-1 and arf8-3 single mutants, there was no enhanced endoreduplication in bpe arf8 petals compared with the wild type.
To characterize further the synergistic interaction between BPE and ARF8, we generated the sesquimutants bpe−/− arf8+/−bpe+/− arf8−/− (Latin sesqui meaning one and a half). The bpe−/− arf8+/− plants are homozygous for the bpe-1 mutation but heterozygous for the arf8-3 mutation, whereas the bpe+/− arf8−/− plants are heterozygous for bpe-1 but homozygous for arf8-3. Similar to the bpe-1 single mutant, petals of bpe−/− arf8+/− were ~23% larger in size compared with the wild type as result of an increase in cell expansion (Figure 4E), indicating that the phenotype of bpe-1 is not enhanced by a mutation in one copy of ARF8. By contrast, bpe+/− arf8−/− petals were larger in size compared with arf8-3 (Figure 4E), suggesting that the phenotype associated with arf8 loss of function is enhanced by removing a copy of BPE. It should be noted that plants heterozygous for arf8 or for bpe have petals with a wild-type phenotype.
We examined if the petal overgrowth phenotype in bpe+/− arf8−/− was due to a modification in cell number and/or cell expansion (Figure 4E). Cell size in bpe+/− arf8−/− was similar to that in arf8-3, bpe-1, or bpe arf8 mutants. By contrast, cell number was higher in bpe+/− arf8−/− compared with arf8-3 but lower than in bpe arf8. Therefore, the overgrowth in bpe+/− arf8−/− is associated with higher cell proliferation, but not cell expansion, thus corroborating the synergistic effect between BPE and ARF8 on cell proliferation. These data demonstrate that ARF8 is involved in the control of cell proliferation in petals during early flower development stages, whereas at late flower development stages both ARF8 and BPEp are involved in the control of petal growth by limiting cell expansion. Furthermore, BPEp works synergistically with ARF8 to regulate cell proliferation.
Nagpal et al. (2005) reported that ARF6 and ARF8 are partially functionally redundant. Therefore, we analyzed the putative genetic interaction of ARF6 and BPE. Petals in arf6-2 single mutants have wild-type size (Figures 5A and 5B). Surface measurements showed that petal size in bpe arf6 double mutants is similar to that in the bpe-1 single mutant (Figures 5A and 5B). This result suggests that there is no genetic interaction between BPE and ARF6 and that ARF6 is not implicated in regulating cell proliferation and cell expansion during petal development.
Figure 5.
Regulation of Petal Growth by BPEp Is Independent of ARF6.
(A) Fully expanded petals at flower development stage 14 in wild-type (WT), bpe-1, arf6-2, and bpe arf6 plants.
(B) Surface area measurements of mature petals (stage 14) in bpe-1, arf6-2, bpe arf6, and wild-type plants. Values are given as mean ± se relative to the wild-type value, set at 100%.
Loss of Function of BPEp Leads to Changes in Expression of Auxin Response Genes
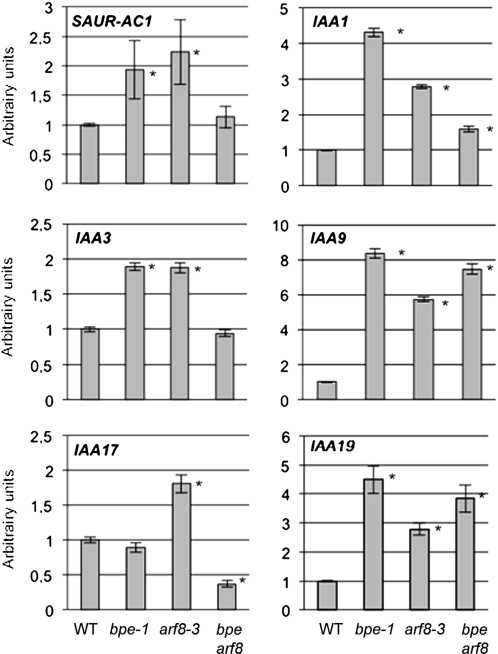
Transcription factors of the ARF family contribute to the regulation of expression of various genes in response to auxin. The physical and genetic interactions between BPEp and ARF8 suggest that BPEp function likely affects auxin-mediated gene responses. To test this possibility, we investigated the transcriptional responses of genes known to be regulated by the auxin signaling pathway in petals of bpe-1, arf8-3, and bpe arf8 mutants. We examined the expression of six auxin-responsive genes, five Aux/IAAs, and SMALL AUXIN-UP RNA (SAUR) (Paponov et al., 2008). The results shown in Figure 6 indicate that bpe-1 and arf8-3 plants display altered expression of auxin-regulated genes. The loss of function of BPE led to a high and significant increase in the accumulation of all analyzed genes in petal cells, except for IAA17, which showed no modification of expression in bpe-1 but higher accumulation in petal cells of arf8-3. These data suggest that BPE and ARF8 are likely involved in maintaining appropriate repression of some auxin response genes in petals. Analyses of expression of these auxin-responsive genes in the double mutant bpe arf8 showed that IAA1, IAA9, and IAA19 had increased expression similar to that in the single mutants, thus with no increased effect over that observed in arf8-3 or bpe-1. SAUR-AC1 and IAA3 RNA levels in petal cells of the double mutant were similar to those in the wild type, and IAA17 RNA levels were reduced in the double mutant plants (Figure 6). These discrepancies in the accumulation of the auxin-regulated genes in bpe arf8 reflect the complex regulation of expression of auxin-regulated genes, which are subject to various combinations of transcriptional regulators, some of these being modulated by cell-specific, developmental stage, or various abiotic signals (Paponov et al., 2008). Together, the data suggest that BPE and ARF8 are likely involved in maintaining appropriate expression of auxin response genes in petals and hence suggest that the interaction between BPEp and ARF8 and their roles in regulating petal growth are likely related to auxin signaling.
Figure 6.
qPCR Analysis of the Response of Aux/IAA and SAUR-AC1 Transcript Levels to Loss of Function of BPE, ARF8, or BPE and ARF8 in Arabidopsis Petals at Development Stages 7 to 9.
qPCR data were normalized with respect to mRNA of the genes ACTIN8 and TUBULIN4, and se was calculated for each sample. Expression value of wild type (WT) was set to 1, and the relative expression of Aux/IAAs and SAUR-AC1 is presented. The analysis was performed using three biological repeats with similar results. Asterisks indicate significant difference from the wild type with P value < 0.001 (t test).
DISCUSSION
The bHLH BPEp Interacts with an ARF in Vivo
We identified ARF8 as an interaction partner of BPEp and demonstrated the function of ARF8 in regulating mitotic growth at early development stages and postmitotic expansion at late petal development stages.
BPEp is a member of the large bHLH transcription factor family (Toledo-Ortiz et al., 2003). It has been shown that bHLH transcription factors interact with other bHLH and/or with proteins of other families (e.g., MYB and WD40) to form multimeric protein complexes that function in multiple pathways leading to diverse cell fates (Bernhardt et al., 2003; Ramsay and Glover, 2005). Our data describe a previously unknown interaction between a bHLH and an ARF transcription factor. Interestingly, the interaction between BPEp and ARF8 is mediated by SDBPEp, the C-terminal domain unique to BPEp, and the CTDARF8, which contains Aux/IAA motifs III and IV. An alternative splicing event gives rise to the specific domain SDBPEp (Szécsi et al., 2006; Brioudes et al., 2009). Amino acid sequence analysis and site-directed mutagenesis identified the GRSLD motif in SDBPEp as mediating BPEp-ARF8 interaction. The GRSLD motif shares sequence similarity with motif III of the CTDs of ARF and Aux/IAA proteins (see Supplemental Figure 2 online). CTDs of ARFs are required for homodimerization or heterodimerization with Aux/IAA proteins through shared III and IV motifs. These interactions modulate auxin response genes that affect cellular responses to auxin and various developmental processes (reviewed in Chapman and Estelle, 2009; Perrot-Rechenmann, 2010). Auxin stimulates growth at low concentrations and restricts growth at higher concentrations (Teale et al., 2006). Our data suggest a previously unknown link between the BPEp bHLH transcription factor and auxin signaling during petal development. This is supported by the fact that BPEp interacts with ARF8 through the same domain that was previously shown to be involved in the interaction with Aux/IAA proteins (Tatematsu et al., 2004). Therefore, auxin likely influences the function of BPEp in influencing cell expansion via interaction with ARF8. This is further supported by the fact that expression of auxin response genes, such as Aux/IAA and SAUR-AC1, is modified in petals of bpe-1 and arf8-3 loss-of-function mutants. Interestingly, loss of function of BPE results in derepression of some early auxin response genes, suggesting that BPEp acts as a repressor. ARF8 belongs to a subgroup of five ARF proteins that possess a Glu-rich middle domain. These ARFs were shown to activate auxin-induced genes in transient expression assays (Ulmasov et al., 1999; Wilmoth et al., 2005) and are thus considered to be transcriptional activators. However, the transcriptional activity might depend on the organ or cell type and interacting proteins, as our data suggest that in petals, BPEp and ARF8 together likely contribute to decreasing the auxin response. Furthermore, similar to arf8-3, petals of the bpe-1 loss-of-function line show a modified venation pattern (Brioudes et al., 2009), thus providing more evidence suggesting that auxin may influence petal development via BPEp. In 2007, Shin et al. (2007) reported the interaction of MYB77 and ARF7. Our data provide further evidence that ARF transcription factors interact with proteins outside the ARF and Aux/IAA families.
ARF8 Influences Cell Expansion and Proliferation during Petal Growth
Several ARF transcription factors have been shown to play roles in different aspects of plant organ development (Liscum and Reed, 2002; Guilfoyle and Hagen, 2007). For example, ARF2 has been reported to affect seed size by negatively regulating cell division and cell expansion; ARF3 was shown to be involved in the development of gynoecium, ARF5 was implicated in embryo development, and ARF7 and ARF19 act in hypocotyl and root development (Sessions et al., 1997; Hardtke and Berleth, 1998; Harper et al., 2000; Tatematsu et al., 2004; Schruff et al., 2006; Shin et al., 2007). In our work, the analysis of arf8-3 suggests that ARF8 is involved in the negative regulation of mitotic growth and postmitotic expansion during petal morphogenesis (Figure 7). Recently, Tabata et al. (2010) reported that loss of function of ARF8 resulted in mature petals being shorter in length compared with the wild type, whereas Nagpal et al. (2005) showed that loss of function of ARF8 resulted in petals that were not shorter than wild-type petals. Our petal surface area measurement data show that petals of arf8 are larger than those of the wild type and thus suggest ARF8 as a negative regulator of petal growth. This role of ARF8 is further supported by cell number and cell size measurements demonstrating that the larger petal surface in arf8-3 is a sum of increased cell number and larger cell size.
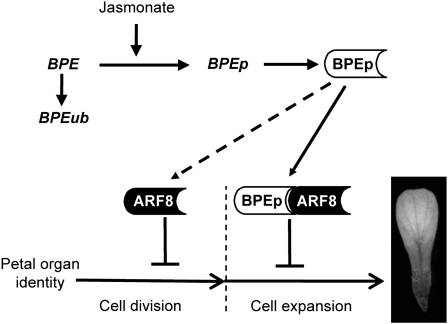
Figure 7.
Model Summarizing the Roles of ARF8 and BPEp in the Regulation of Petal Development.
This model shows the action of ARF8 in restricting cell proliferation at early flower development stages and of ARF8 and BPEp in limiting cell expansion at late development stages. The synergistic interaction between BPEp and ARF8 in early petal development is indicated with a dashed arrow. The role of jasmonate signaling in the regulation of BPEp expression has been reported (Brioudes et al., 2009). The second transcript BPEub originating from the BPE gene (Szécsi et al., 2006) is shown. BPEp and ARF8 proteins are presented as boxes.
The petal overgrowth phenotypes associated with cell expansion are strikingly similar in single arf8-3 or bpe-1 mutants, the bpe arf8 double mutant, and combinations of sesquimutants (bpe−/− arf8+/− or bpe+/− arf8−/−), suggesting that BPEp and ARF8 are involved in the same pathway required for the regulation of cell expansion during petal growth. Therefore, it is likely that these two transcription factors interact to form a complex that is essential to limit cell expansion and petal growth at late flower development stages. Unlike BPEp, ARF8 also regulates cell proliferation in petals at early flower development stages, thus explaining the even larger size of petals in arf8-3 compared with bpe-1. This role of ARF8 in regulating cell proliferation is likely independent of the function of BPEp. However, the analyses of bpe arf8 and of the sesquimutants bpe−/−/arf8+/− and bpe+/−/arf8−/− suggest a synergistic interaction between BPEp and ARF8 in cell proliferation in petals at early flower development stages. A similar synergistic interaction that involves ARF7 and MYB77 was recently shown to be involved in the regulation of lateral root density and growth in Arabidopsis (Shin et al., 2007). Moreover, gene dosage effects on ARF8 function in regulating cell division were observed in the bpe-1 mutant background, such that bpe−/− arf8+/− showed an intermediate phenotype between arf8-3 and the bpe arf8 double mutant. A similar gene dosage-dependent function has been reported for ARF6 and ARF8 during adventitious root formation and flower development (Nagpal et al., 2005; Gutierrez et al., 2009). Interestingly, ARF8 and ARF6 have been shown to promote jasmonate production in flowers, and this regulation is likely related to auxin signaling (Nagpal et al., 2005; Tabata et al., 2010). The fact that BPEp expression is regulated by jasmonate (Brioudes et al., 2009) suggests that there may be a feedback loop between auxin and jasmonate signaling to regulate petal growth and that BPEp and ARF8 are part of such a regulatory loop.
Previous studies suggested that in leaves, a compensation mechanism is activated to maintain normal plant organ sizes when aberrant or deficient cell divisions occur (Truernit and Haseloff, 2008; Tsukaya, 2008). The result of the compensation mechanism is increased cell volume in the presence of decreased cell number (Horiguchi et al., 2006; Ferjani et al., 2007). Compensation was found to occur in lateral organs with a determinate fate, such as leaves and petals, and is usually associated with endoreduplication (Hase et al., 2000, 2005; Ferjani et al., 2007). Here, we show that the increased cell expansion in petals is associated with an increase in cell number and no or only subtle changes in the ploidy level. Therefore, we observe no such compensation mechanism at play for the mutant plant lines bpe-1, arf8-3, bpe arf8, bpe−/− arf8+/−, and bpe+/− arf8−/−. Petals were proposed not to endoreduplicate naturally. In fact, suppression of endoreduplication is thought to be important for petal morphogenesis (Hase et al., 2000, 2005). However, it has been shown that mutations (e.g., in FRILL) negatively affecting cell division enhance cell expansion (compensation) and also cause abnormal endoreduplication in the distal part of petals. The fact that the increase in petal cell expansion in bpe-1 and arf8-3 was not associated with endoreduplication is another argument that ARF8 and BPEp must affect cell expansion in a pathway that does not involve the compensation mechanism. Therefore, our study suggests that cell expansion can be uncoupled from cell division during petal organ growth.
ARF8 has pleiotropic functions. For example, it was shown that ARF8 is involved in stamen filament elongation, anther dehiscence, restricting hypocotyl elongation in response to changes in auxin level, positive regulation of adventitious rooting, and negative regulation of fruit initiation by acting on cell division and cell expansion (Vivian-Smith et al., 2001; Tian et al., 2004; Nagpal et al., 2005; Goetz et al., 2006; Gifford et al., 2008; Gutierrez et al., 2009). Our data demonstrate that ARF8 is also a negative regulator of petal growth by restricting cell division at early development stages and limiting cell expansion through an interaction with BPEp at late development stages. These multiple roles of ARF8 are consistent with its patterns of expression in seedlings, developing flowers, and fruits (Tian et al., 2004; Nagpal et al., 2005; Goetz et al., 2006; Wu et al., 2006). A similar pleiotropic function has been demonstrated for the tomato (Solanum lycopersicum) ARF DR12, the putative homolog of Arabidopsis ARF4 (Jones et al., 2002). Loss of function of DR12 caused a pleiotropic phenotype that included dark-green immature fruit, unusual cell division in the fruit pericarp, blotchy ripening, enhanced fruit firmness, upward curling leaves, and increased hypocotyl and cotyledon growth. Nagpal et al. (2005) reported that ARF6 and ARF8 are partially functionally redundant. However, petals of arf6 have wild-type size and petals of the bpe arf6 double mutant exhibit a bpe-1 phenotype (Figure 5), thus suggesting that the regulation of petal growth by BPEp is independent from ARF6. This is consistent with the fact that no interaction between BPEp and ARF6 was observed in yeast. The double mutant arf6 arf8 was shown to exhibit petals with reduced size compared with the wild type, a phenotype that is opposite to that observed in the single mutant arf8 (Nagpal et al., 2005). Flowers of arf6 arf8 were shown to arrest in development at stages 10 to 12 (Nagpal et al., 2005), thus explaining the small size of petals in the double mutant arf6 arf8. Moreover, arf6 arf8 plants exhibited severe dwarfing and organ size reduction, pleiotropic phenotypes that were related to hormonal defects (related to auxin and jasmonates). The expression of many other genes that have been shown to modulate growth, including transcription factors, kinases, cell wall synthesis genes, and those in multiple hormonal pathways (including auxin, gibberellins, and jasmonate responses and signaling) is modified in arf6 arf8. Therefore, arf6 arf8 developmental defects are likely to be independent of the regulation asserted by the ARF8/BPEp complex.
In summary, the data here provide evidence of interaction between a bHLH transcription factor (BPEp) and ARF8 and unravels the specificity and the biological significance of such interaction in influencing petal organ growth by regulating cell proliferation and cell expansion (Figure 7), two of the many processes regulated by auxin. This defines a previously unknown combinatorial interaction between transcription factors in plants and provides a better understanding of how plants integrate signals to regulate petal development. To characterize further the biological significance of the interaction between BPEp and ARF8, one of the future tasks will be to identify the downstream target genes to understand better its biological functions and to elucidate the systemic signaling that is triggered during Arabidopsis petal morphogenesis.
METHODS
Plant Lines
The arf8-3 and arf6-2, the sesquimutants arf8-3 +/− arf6-2 (Nagpal et al., 2005), and the bigpetal-1 (bpe-1) (Szécsi et al., 2006) knockout lines have been described. Wild-type (Columbia-0 accession) and mutant Arabidopsis thaliana plants were kept in growth chambers with conditions of 16 h/8 h day/night at 22°C and 100 μE/m2/s light. Nicotiana benthamiana plants were grown in greenhouse with conditions of 16 h/8 h day/night and 24°C/22°C day/night.
Two-Hybrid Screen
Full-length BPEp open reading frame (ORF) or BPEp C-terminal specific domains SDBPEp (Szécsi et al., 2006) were PCR amplified (see Supplemental Table 1 online) and then cloned into plasmid pAS2.1 (Clontech) in frame with the BD-GAL4 as NcoI-EcoRI or EcoRI-EcoRI fragments, respectively. The two-hybrid screen was performed by mating as described (Fromont-Racine et al., 1997) using yeast strains AH109, Y187 (Clontech), and the CD4-30 two-hybrid library from Arabidopsis inflorescences in pADGal4-2.1 (Fan et al., 1997). Cells were selected on medium lacking Leu (L−), Trp (W−), His (H−), and adenine (A−). Yeast clones that survived after three rounds of streaking on the above medium were grown on selective medium lacking Leu and Trp, harvested, and then disrupted by vortexing in presence of glass beads. Recombinant pAD-GAL4 2.1 plasmids were purified from yeast lysate, and cDNA inserts were sequenced. cDNA clones with an ORF in frame with the GAL4-AD domain were selected and the corresponding genes in the Arabidopsis genome were identified. The interaction of BPEp with the identified proteins was then confirmed by cotransformation in AH109 yeast strain followed by selection for expression of ADE2 and HIS3 reporters.
Yeast drops were prepared using AH109 cotransformed with pAS2.1-BPEp (BD-BPEp) as bait and pAD-GAL4-2.1-ARF8 (AD-ARF8) as prey. Double transformed yeast cells were selected on selective medium lacking Trp and Leu, resuspended in TE, and then diluted three times (5·10−1, 5·10−2, and 5·10−3). Yeast drops were then deposited on selective medium A−W−L−and H−. β-Galactosidase activity was quantified from liquid cultures of yeast as previously described (Schneider et al., 1996).
Yeast strains were cultured and handled according to the Yeast Protocol Handbook (Clontech).
BiFC Assays and BPEp/ARF8 Coexpression in Tobacco Cells
The DNA fragments coding for SDBPEp (last 122 amino acids in the BPEp sequence) and CTDARF8 (last 127 amino acids in the ARF8 sequence) were cloned in the pBiFP2 and pBiFP3 vectors (Desprez et al., 2007), respectively. The resulting clones harbor SDBPEp fused to the N-terminal domain of the YFP and CTDARF8 fused to the C-terminal of the YFP, respectively, expressed under the control of the constitutive 35S promoter. Split-YFP fused SDBPEp and CTDARF8 were transiently expressed in N. benthamiana leaf cells as previously described (Tai et al., 1999). An Agrobacterium tumefaciens strain carrying the 35S:p19 construct (Voinnet et al., 2003) was coinfiltrated to achieve maximum level of protein expression. Four days after infiltration, tobacco leaf cells were analyzed on an Axiovert100M LSM-510 confocal laser scanning microscope (Zeiss).
The BPEp ORF was cloned in frame with the GFP in the vector pK7FWG2 using the Multisite Gateway cloning technology (Invitrogen). The resulting clone harbors the BPEp-GFP fusion expressed under the control of the 35S promoter. To prevent the fifth intron from splicing off in BPEp, the splice donor site was mutated without changing the amino acid content (Glu-221 [GAG] → Glu-221 [GAA] and Val-222 [GTG] → Val-222 [GTA]). Similarly, ARF8 was fused to the mOrange and expressed under the control of the 35S promoter in the destination vector pB7m34GW. BPEp-GFP and mOrange-ARF8 fusion proteins were transiently expressed in N. benthamiana as above, and their cellular localization was analyzed using confocal microscopy.
Gene Expression Analyses
Total RNA was extracted from stage 9 or stage 13 floral buds using the Spectrum Plant Total RNA kit (Sigma-Aldrich) or from dissected sepals, petals, stamens, and carpels using TRI Reagent according to the manufacturer’s instructions (Molecular Research Center). Contaminating DNA was removed using the DNA-freeTM kit (Ambion). One microgram of total RNA was used in a reverse transcription assay with RevertAid M-MuLV Reverse Transcriptase (Fermentas). Target cDNAs were quantified by qPCR using FastStart universal SYBR green master (Roche) on a Step-One Plus Real-Time PCR system (Applied Biosystems). Primer sequences are available in Supplemental Table 1 online. Expression levels were normalized with ACTIN8 and TUBULIN4 reference genes. These genes were chosen using the GeNorm application (Vandesompele et al., 2002). The calculated M value for the two reference genes was 0.102. Three biological replicates were used for each experiment, and two qPCR technical replicates were performed for each biological replicate.
Petal Size and Petal Cell Size and Number Measurements
Petals from flowers at stage 14-15 (maximum expansion) (Smyth et al., 1990) were carefully dissected and then cleared overnight in a solution containing 86% ethanol and 14% acetic acid followed by two incubations of 4 h each in 70% ethanol (Szécsi et al., 2006). For these experiments, we used young plants (mutants and wild type with no mature siliques) of the same age and grown under the same conditions. Petal organ length, width (distal region of petal blade), and area, as well as the adaxial epidermis cell size and number, were measured from digital images (UTHSCSA Image Tool version 3.0, D. Wilcox, B. Dove, D. Mc David, and D. Greer). Cell number per 0.045 mm2 surface was counted for each mutant and then compared with that in wild-type petals. For all mutants and the wild type, this measurement was performed in the distal part of the petal (conical cells). The surface areas of at least 60 petals from at least eight plants of the wild type, arf8-3, arf6-2, bpe-1, bpe arf6, and bpe arf8 double mutants were measured. Cell size determination was performed by measuring at least 400 cells.
For FM4-64 staining, petals were removed and kept alive in Murashige and Skoog (Duchefa) liquid medium for one night to wet their surfaces. FM4-64 was directly applied on petal adaxial surface. Digital images were taken with an Axiovert100M LSM-510 confocal laser scanning microscope (Zeiss).
Measurements of DNA Content of Petal Cell Nuclei
Nuclei were extracted from fresh petal tissue by thin chopping in Galbraith’s medium (Galbraith et al., 1983). Nuclei were stained with 50 mg/mL propidium iodide before analysis of DNA content on an EPICS Elite ESP flow cytometer (Beckman-Coulter).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BPE, At1g59640; ARF8, At5g37020; ARF6, At1g30330; ACTIN8, At1g49240); TUBULIN4, At5g44340; IAA1, NP_193192; IAA2, NP_188943; IAA3, NP_171920; IAA4, NP_199183; IAA5, NP_173011; IAA6, NP_175692; IAA7, NP_974355; IAA8, NP_850028; IAA9, NP_851275; IAA17, NP_171921; IAA19, NP_188173; IAA27, NP_194637; ARF1, NP_001031208; ARF2, NP_974980; ARF4, NP_200853; ARF5, NP_173414; ARF6, NP_001031115; ARF7, NP_568400; ARF8, NP_198518; ARF9, NP_001031706; ARF10, NP_180402; ARF11, NP_001031548; ARF12, NP_174691; ARF14, NP_174786; ARF15, NP_174784; ARF16, NP_567841; ARF17, NP_565161; ARF18, NP_567119; ARF19, NP_173356; ARF20, NP_174758; ARF21, NP_174701; and ARF22, NP_174699.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ARF8 and BPEp Proteins Interact in Vivo and Colocalize in the Nucleus.
Supplemental Figure 2. Alignment of Motif III of ARF and Aux/IAA C-Terminal Domains with the GRSLD Motif of SDBPEp.
Supplemental Figure 3. Sepal, Stamen, and Carpel Size in arf8-3 and Wild-Type Plants.
Supplemental Figure 4. DNA Content of Petal Cell Nuclei.
Supplemental Table 1. Sequences of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Dali Ma, Annick Dubois, and Olivier Hamant (Ecole Normale Supérieure, Lyon, France) for critical reading of the manuscript and Jason W. Reed (University of North Carolina, Chapel Hill, NC) for kindly providing arf8-3 and arf6-2 mutant lines. This work was funded by the Action Concertee Initiative-Young Investigator Program, by the Agence Nationale de Recherche (ANR-09-BLAN-0006), and by the Biology Department of the French National Institute for Agronomic Research (Institut National de la Recherche Agronomique).
References
- Anastasiou E., Kenz S., Gerstung M., MacLean D., Timmer J., Fleck C., Lenhard M. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13: 843–856 [DOI] [PubMed] [Google Scholar]
- Anastasiou E., Lenhard M. (2007). Growing up to one’s standard. Curr. Opin. Plant Biol. 10: 63–69 [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bögre L., Magyar Z., López-Juez E. (2008). New clues to organ size control in plants. Genome Biol. 9: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brioudes F., Joly C., Szécsi J., Varaud E., Leroux J., Bellvert F., Bertrand C., Bendahmane M. (2009). Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J. 60: 1070–1080 [DOI] [PubMed] [Google Scholar]
- Brioudes F., Thierry A.M., Chambrier P., Mollereau B., Bendahmane M. (2010). Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc. Natl. Acad. Sci. USA 107: 16384–16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busov V.B., Brunner A.M., Strauss S.H. (2008). Genes for control of plant stature and form. New Phytol. 177: 589–607 [DOI] [PubMed] [Google Scholar]
- Chapman E.J., Estelle M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S. (2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Yadegari R., Fischer R.L., Yanofsky M.F., Weigel D. (2004). The role of JAGGED in shaping lateral organs. Development 131: 1101–1110 [DOI] [PubMed] [Google Scholar]
- Disch S., Anastasiou E., Sharma V.K., Laux T., Fletcher J.C., Lenhard M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16: 272–279 [DOI] [PubMed] [Google Scholar]
- Fan H.Y., Hu Y., Tudor M., Ma H. (1997). Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12: 999–1010 [DOI] [PubMed] [Google Scholar]
- Ferjani A., Horiguchi G., Yano S., Tsukaya H. (2007). Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 144: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain J.C., Legrain P. (1997). Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16: 277–282 [DOI] [PubMed] [Google Scholar]
- Galbraith D.W., Harkins K.R., Maddox J.M., Ayres N.M., Sharma D.P., Firoozabady E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220: 1049–1051 [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M., Vivian-Smith A., Johnson S.D., Koltunow A.M. (2006). AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460 [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Bussell J.D., Pacurar D.I., Schwambach J., Pacurar M., Bellini C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S., Berleth T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17: 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper R.M., Stowe-Evans E.L., Luesse D.R., Muto H., Tatematsu K., Watahiki M.K., Yamamoto K., Liscum E. (2000). The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase Y., Fujioka S., Yoshida S., Sun G., Umeda M., Tanaka A. (2005). Ectopic endoreduplication caused by sterol alteration results in serrated petals in Arabidopsis. J. Exp. Bot. 56: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Hase Y., Tanaka A., Baba T., Watanabe H. (2000). FRL1 is required for petal and sepal development in Arabidopsis. Plant J. 24: 21–32 [DOI] [PubMed] [Google Scholar]
- Hill J.P., Lord E.M. (1989). Floral development in Arabidopsis thaliana: A comparison of the wild type and the homeotic pistillata mutant. Can. J. Bot. 67: 2922–2936 [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Horiguchi G., Ferjani A., Fujikura U., Tsukaya H. (2006). Coordination of cell proliferation and cell expansion in the control of leaf size in Arabidopsis thaliana. J. Plant Res. 119: 37–42 [DOI] [PubMed] [Google Scholar]
- Hu Y., Xie Q., Chua N.H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15: 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram G.C., Waites R. (2006). Keeping it together: Co-ordinating plant growth. Curr. Opin. Plant Biol. 9: 12–20 [DOI] [PubMed] [Google Scholar]
- Irish V.F. (2010). The flowering of Arabidopsis flower development. Plant J. 61: 1014–1028 [DOI] [PubMed] [Google Scholar]
- Jones B., Frasse P., Olmos E., Zegzouti H., Li Z.G., Latché A., Pech J.C., Bouzayen M. (2002). Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J. 32: 603–613 [DOI] [PubMed] [Google Scholar]
- Krizek B.A. (2009). Making bigger plants: Key regulators of final organ size. Curr. Opin. Plant Biol. 12: 17–22 [DOI] [PubMed] [Google Scholar]
- Krizek B.A., Fletcher J.C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6: 688–698 [DOI] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M.W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 22: 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E., Reed J.W. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49: 387–400 [PubMed] [Google Scholar]
- Melaragno J.E., Mehrotra B., Coleman A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menand B., Meyer C., Robaglia C. (2004). Plant growth and the TOR pathway. Curr. Top. Microbiol. Immunol. 279: 97–113 [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P., Ellis C.M., Weber H., Ploense S.E., Barkawi L.S., Guilfoyle T.J., Hagen G., Alonso J.M., Cohen J.D., Farmer E.E., Ecker J.R., Reed J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118 [DOI] [PubMed] [Google Scholar]
- Nakayama N., Arroyo J.M., Simorowski J., May B., Martienssen R., Irish V.F. (2005). Gene trap lines define domains of gene regulation in Arabidopsis petals and stamens. Plant Cell 17: 2486–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov I.A., Paponov M., Teale W., Menges M., Chakrabortee S., Murray J.A., Palme K. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1: 321–337 [DOI] [PubMed] [Google Scholar]
- Perrot-Rechenmann C. (2010). Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schneider S., Buchert M., Hovens C.M. (1996). An in vitro assay of beta-galactosidase from yeast. Biotechniques 20: 960–962 [DOI] [PubMed] [Google Scholar]
- Schruff M.C., Spielman M., Tiwari S., Adams S., Fenby N., Scott R.J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Sessions A., Nemhauser J.L., McColl A., Roe J.L., Feldmann K.A., Zambryski P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491 [DOI] [PubMed] [Google Scholar]
- Shin R., Burch A.Y., Huppert K.A., Tiwari S.B., Murphy A.S., Guilfoyle T.J., Schachtman D.P. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Szécsi J., Joly C., Bordji K., Varaud E., Cock J.M., Dumas C., Bendahmane M. (2006). BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 25: 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R., Ikezaki M., Fujibe T., Aida M., Tian C.E., Ueno Y., Yamamoto K.T., Machida Y., Nakamura K., Ishiguro S. (2010). Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 51: 164–175 [DOI] [PubMed] [Google Scholar]
- Tai T.H., Dahlbeck D., Clark E.T., Gajiwala P., Pasion R., Whalen M.C., Stall R.E., Staskawicz B.J. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 96: 14153–14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K., Kumagai S., Muto H., Sato A., Watahiki M.K., Harper R.M., Liscum E., Yamamoto K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W.D., Paponov I.A., Palme K. (2006). Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471 [DOI] [PubMed] [Google Scholar]
- Tian C.E., Muto H., Higuchi K., Matamura T., Tatematsu K., Koshiba T., Yamamoto K.T. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 40: 333–343 [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Haseloff J. (2008). Arabidopsis thaliana outer ovule integument morphogenesis: Ectopic expression of KNAT1 reveals a compensation mechanism. BMC Plant Biol. 8: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2008). Controlling size in multicellular organs: Focus on the leaf. PLoS Biol. 6: e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian-Smith A., Luo M., Chaudhury A., Koltunow A. (2001). Fruit development is actively restricted in the absence of fertilization in Arabidopsis. Development 128: 2321–2331 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wellmer F., Riechmann J.L., Alves-Ferreira M., Meyerowitz E.M. (2004). Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 16: 1314–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth J.C., Wang S., Tiwari S.B., Joshi A.D., Hagen G., Guilfoyle T.J., Alonso J.M., Ecker J.R., Reed J.W. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43: 118–130 [DOI] [PubMed] [Google Scholar]
- Wu M.F., Tian Q., Reed J.W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218 [DOI] [PubMed] [Google Scholar]
- Zik M., Irish V.F. (2003). Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell 15: 207–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.