Abstract
Neural tube closure is a critical morphogenetic event that is regulated by dynamic changes in cell shape and behavior. Although previous studies have uncovered a central role for the non-canonical Wnt signaling pathway in neural tube closure, the underlying mechanism remains poorly resolved. Here, we show that the missing in metastasis (MIM; Mtss1) protein, previously identified as a Hedgehog response gene and actin and membrane remodeling protein, specifically binds to Daam1 and couples non-canonical Wnt signaling to neural tube closure. MIM binds to a conserved domain within Daam1, and this interaction is positively regulated by Wnt stimulation. Spatial expression of MIM is enriched in the anterior neural plate and neural folds, and depletion of MIM specifically inhibits anterior neural fold closure without affecting convergent extension movements or mesoderm cell fate specification. Particularly, we find that MIM is required for neural fold elevation and apical constriction along with cell polarization and elongation in both the superficial and deep layers of the anterior neural plate. The function of MIM during neural tube closure requires both its membrane-remodeling domain and its actin-binding domain. Finally, we show that the effect of MIM on neural tube closure is not due to modulation of Hedgehog signaling in the Xenopus embryo. Together, our studies define a morphogenetic pathway involving Daam1 and MIM that transduces non-canonical Wnt signaling for the cytoskeletal changes and membrane dynamics required for vertebrate neural tube closure.
Keywords: Wnt, Daam1, Neural tube closure, MIM, Mtss1, Xenopus
INTRODUCTION
Neural tube defects (NTDs) remain a prevalent human birth defect, occurring in ~1 in 1000 pregnancies in the United States and in an estimated 300,000 newborns per year worldwide (Copp et al., 2003; Ueno and Greene, 2003). Although the signaling pathways regulating neural fold closure remain to be fully delineated, several genes required for neural tube closure have been identified, including members of the non-canonical Wnt pathway previously identified to play significant roles in convergent extension (CE) and neural tube closure, such as Strabismus, Dishevelled and Celsr1 (Curtin et al., 2003; Jenny et al., 2003; Wallingford and Harland, 2002; Ybot-Gonzalez et al., 2007). Convergence (narrowing) and extension (lengthening) of tissues occurs by the active intercalation of cells along the axis of convergence (Keller, 2002). This dynamic process drives neural tube closure in vertebrates, and failure of CE results in neural tube closure defects (Wallingford, 2005; Wallingford and Harland, 2002).
Cell behaviors underlying neurulation have been extensively characterized in the mouse, chicken and Xenopus, and previous studies found that apical constriction and hinge point formation promote neural plate bending (Davidson and Keller, 1999; Haigo et al., 2003; Hildebrand and Soriano, 1999). Moreover, disruption of actin microfilaments within the neural tube inhibits neurulation, indicating a crucial role for the actin cytoskeleton during neural tube closure (Schoenwolf et al., 1988; Ybot-Gonzalez and Copp, 1999). However, the link between a signaling pathway that specifically regulates neural tube closure along the anteroposterior axis and actin cytoskeletal regulation remains unclear. We report that missing in metastasis (MIM; also known as Mtss1), an actin monomer-binding and membrane curvature-inducing protein, is a central factor for anterior neural tube closure.
MIM was originally isolated as a gene downregulated in bladder cancer (Lee et al., 2002) and encodes a multi-domain protein that is suggested to have a scaffolding function, although its in vivo role is not fully established (Machesky and Johnston, 2007). The N-terminal IRSp53/MIM homology domain (IMD) of MIM harbors F-actin bundling activity and is highly conserved across species (Machesky and Johnston, 2007; Quinones et al., 2010). The IMD has been shown to bind membranes, induce negative membrane curvature opposite to F-BAR domain-containing proteins, interact with the small GTPase Rac and mediate MIM self-association (Bompard et al., 2005; Disanza et al., 2006; Lee et al., 2007). The serine-rich domain (SRD) contains two tyrosine phosphorylation sites that can be phosphorylated by Src kinase, whereas the proline-rich domain (PRD) binds cortactin (Lin et al., 2005). The C-terminal WASP homology 2 (WH2) domain binds actin monomers (Lee et al., 2007; Mattila et al., 2003).
In this study, we have identified MIM as a Daam1 binding partner and characterized its in vivo role during vertebrate development. We demonstrate that a MIM-Daam1 protein complex is induced by Wnt signaling and that MIM regulates cytoskeletal changes and membrane dynamics required for anterior neural fold closure. Our studies suggest that MIM provides a direct link between Daam1-mediated non-canonical Wnt signaling and the remodeling of the actin cytoskeleton and changes in membrane dynamics required for neural tube closure.
MATERIALS AND METHODS
Antibodies, stains and recombinant protein
Monoclonal antibodies against Myc (9E10), GFP (sc-9996) and actin (sc-8432) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA), anti-HA was from Roche (Indianapolis, IN, USA) and anti-β-catenin from Transduction Laboratories (San Diego, CA, USA). Alexa Fluor-conjugated anti-mouse and anti-rabbit antibodies, Texas Red-Phalloidin and Oregon Green-Phalloidin were from Molecular Probes (Eugene, OR, USA). Rabbit anti-MIM antibodies were generated against a GST-fusion protein spanning the SRD and PRD of MIM and affinity purified. Recombinant Wnt3a was from R&D Systems (Minneapolis, MN, USA) and used at 200 ng/ml.
Plasmids and oligonucleotides
Xenopus MIM (XMIM) and MIM fragments generated by restriction digestion or PCR were subcloned into pCS2+MT, pcDNA-HA or pCS2+GFP vectors. XMIM morpholino oligonucleotides (MOs) complementary to the 5′UTR region (5′-CGGGAGATAGACGGTGCTTGAGTTC-3′) and the translational initiation site (5′-ATGGATACGAACATGGAGCGGGAGT-3′) were synthesized by Gene Tools (Philomath, OR, USA). An MO of similar length with a random sequence provided the negative control.
Yeast two-hybrid screen
A rat brain cDNA library (Clontech, Mountain View, CA, USA) was screened using C-Daam1 (see Fig. 1A) as bait. In total 3.9 million independent clones were screened, and three overlapping MIM fragments, in addition to other positives, were obtained (Khadka et al., 2009; Sato et al., 2006).
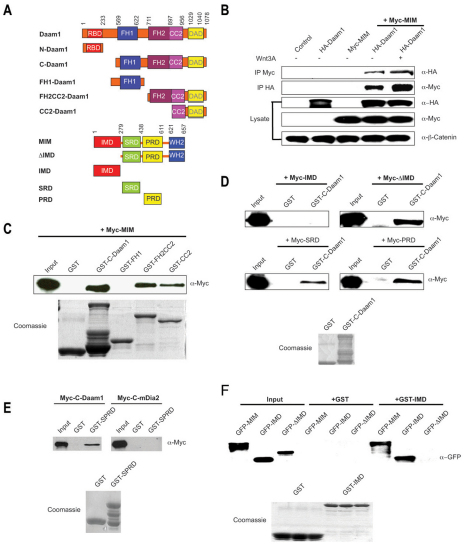
Fig. 1.
MIM is a Daam1-interacting protein. (A) Domain structure of Daam1 and MIM constructs. Numbers indicate amino acid positions. RBD, Rho-binding domain; DAD, diaphanous autoregulatory domain. (B) Full-length Daam1 interacts with full-length MIM and is positively regulated by Wnt3a (3 hours of treatment). HA-Daam1 and Myc-MIM constructs were co-transfected into human HEK293T cells and lysates were immunoprecipitated with the indicated antibodies. (C) MIM interacts with C-Daam1, the FH2-CC2 and CC2 domains, but not the FH1 domain, in GST pull-down assays. (D) The ΔIMD, SRD and PRD, but not the IMD, interact with Daam1. (E) MIM specifically interacts with Daam1 but not with the Formin protein mDia2. (F) MIM self-associates through its IMD. GST pull-down shows that full-length MIM and IMD, but not the ΔIMD, bind to recombinant IMD protein. For pull-downs, Myc-MIM, IMD, ΔIMD, SRD and PRD constructs were transfected into HEK293T cells and lysates were isolated. GST and GST-tagged C-Daam1, FH1, FH2-CC2, CC2, SPRD and IMD recombinant proteins were prepared from transfected E. coli BL21 cells. The integrity of GST-tagged proteins is shown by Coomassie staining.
Transfections and luciferase assays
Mammalian cell culture studies were performed using HEK293T cells or NIH3T3 cells. Cells were transfected using Polyfect reagent (Qiagen, Valencia, CA, USA) with 1-2 μg of the indicated plasmid. The total amount of transfected DNA was equalized by supplementation with vector DNA without inserts. Luciferase assays were performed as previously described (Asaoka et al., 2010).
GST pull-down assays and immunoprecipitation
Myc-tagged and HA-tagged proteins were generated by TNT Quick-Coupled Transcription/Translation systems (Promega, Madison, WI, USA). GST pull-down and immunoprecipitation assays were performed as previously described (Liu et al., 2008; Sato et al., 2006). We determined the increase in complex formation between Daam1 and MIM by normalizing to the input of HA-Daam1 and Myc-MIM and to β-catenin levels in the lysate.
Immunocytochemistry
Immunocytochemistry was performed as previously described (Khadka et al., 2009; Liu et al., 2008; Sato et al., 2006). Images were obtained using an Olympus IX70 fluorescence microscope with a 100× objective lens or a Zeiss Axiovert 100 confocal microscope. For quantification of effects on actin fibers, a baseline of ten actin fibers/cell was used. Cells containing more or fewer than ten fibers were scored as an increase or decrease, respectively. For quantification of membrane protrusions, cells with visible actin protrusions were counted.
Embryo manipulations, in situ hybridizations and explant assays
Embryo manipulations and explant assays were performed as described (Khadka et al., 2009; Liu et al., 2008; Sato et al., 2006). Embryo injections were performed with in vitro transcribed RNAs. Convergent extension assays in explants were performed as described (Khadka et al., 2009; Liu et al., 2008; Sato et al., 2006).
Embryo sectioning and in vivo imaging
RNAs (0.5-2 ng) encoding GFP-CAAX and membrane-tethered Cherry were microinjected separately into the dorsal blastomeres of four-cell stage Xenopus embryos, alone or together with Xdd1 RNA or MOs for Daam1 (Habas et al., 2001) and MIM. Embryos were fixed in 4% paraformaldehyde in PBS for 4 hours at room temperature and then washed with PBS. Embryos were embedded in 4% agarose, incubated with PBS overnight and then microtome-sectioned at 100 μm. Samples were washed in PBS containing 0.1% Triton X-100 and incubated with 4 U/ml Oregon Green-Phalloidin (Invitrogen, Carlsbad, CA, USA) for 6 hours. Samples were washed with PBS containing 0.1% Triton X-100 and mounted on glass slides with Fluoro Cel mounting medium (Electron Microscopy Sciences, Hatfield, PA, USA).
RESULTS
MIM is a Daam1-interacting protein
We previously reported that the C-terminal fragment of human Daam1 (C-Daam1) induces Rho activation and actin fiber formation, although the full spectrum of effectors downstream of Daam1 regulating these processes remains unknown (Habas et al., 2001). To identify these effectors, we performed a yeast two-hybrid screen using C-Daam1 as bait, and identified MIM as a binding partner amongst others (Khadka et al., 2009; Sato et al., 2006).
To study MIM function, we first cloned the Xenopus homolog of MIM (XMIM). XMIM is composed of 656 amino acids and shares 46% and 50% identity with the mouse and human homologs, respectively (see Fig. S1A in the supplementary material). The IMD and the WH2 domain are highly conserved across vertebrate species and Drosophila (see Fig. S1B in the supplementary material). Daam1-MIM interaction was confirmed by co-immunoprecipitation assays using epitope-tagged cDNAs (Fig. 1A,B). Full-length MIM interacted with full-length Daam1 and this interaction was enhanced ~3-fold upon Wnt3a stimulation (Fig. 1B). To delineate the domain(s) required for MIM-Daam1 interaction, we performed in vitro GST pull-down assays. MIM bound to C-Daam1 (Fig. 1A,C). Using smaller fragments harboring the FH1, FH2-coil-coiled (FH2-CC2) or CC2 domains separately, we determined that MIM specifically binds to the CC2 fragment of Daam1 (Fig. 1A,C). We then determined that it is either the SRD or PRD of MIM that interacts with Daam1 (Fig. 1A,D).
As Daam1 is a Formin protein, we considered whether MIM also interacts with other Formin proteins. Using GST pull-down assays, we found that MIM specifically binds to Daam1, but not to the well-characterized Formin protein mDia2 (Diap3), suggesting that MIM is not a general binding partner for Formin proteins (Fig. 1E).
Previous studies have shown that murine MIM can self-associate through its N-terminal IMD, so we tested whether XMIM self-associates in a similar manner. Using GST pull-down assays, we found that both XMIM and its IMD fragment bind to GST-tagged IMD protein, indicating that XMIM self-associates through its IMD (Fig. 1A,F) and suggesting an ability to oligomerize.
MIM co-localizes with Daam1 and modulates the actin cytoskeleton
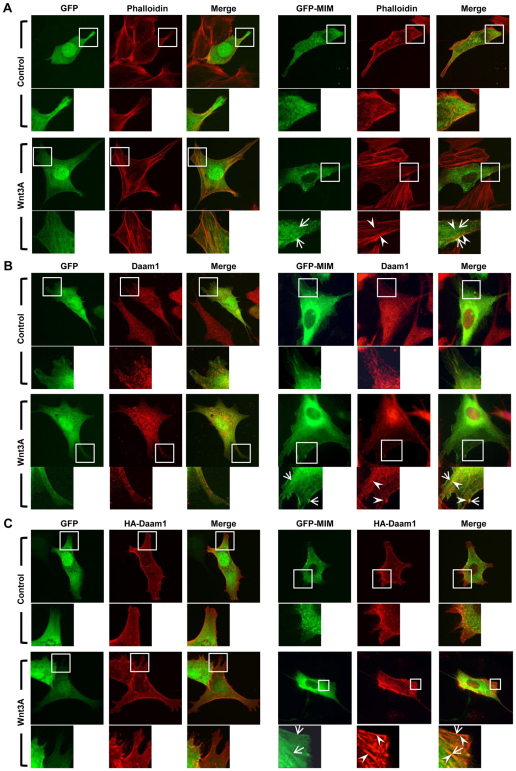
We next examined the subcellular localization of MIM and endogenous Daam1 in mouse NIH3T3 cells transfected with GFP-MIM. Immunofluorescence showed that MIM localized to the plasma membrane but was observed on actin fibers and at the plasma membrane in response to Wnt3a stimulation (Fig. 2A and see Fig. S2A in the supplementary material). Affinity-purified Daam1 polyclonal antibody (Sato et al., 2006) revealed that Daam1 was diffusely localized to the cytoplasm of NIH3T3 cells. Upon Wnt3a stimulation, endogenous Daam1 localized to structures resembling actin fibers and to the plasma membrane and co-localized with GFP-MIM (Fig. 2B and see Fig. S2B in the supplementary material). Since resolving the subcellular localization of endogenous Daam1 was difficult, we examined the co-localization of HA-Daam1 and GFP-MIM. Overexpressed MIM and Daam1 partially co-localized to structures resembling actin fibers and to the plasma membrane and this co-localization was enhanced in response to Wnt3a stimulation (Fig. 2C and see Fig. S2C in the supplementary material). The enhanced co-localization of MIM and Daam1 in response to Wnt stimulation, along with our protein interaction data, suggest that MIM and Daam1 might function in a common pathway for regulating the actin cytoskeleton.
Fig. 2.
Wnt stimulation induces co-localization of Daam1 and MIM. (A) GFP-MIM, but not GFP, is localized to actin fibers and the plasma membrane upon Wnt3a stimulation. (B,C) Endogenous Daam1 (B) or HA-Daam1 (C) (red) co-localizes with GFP-MIM, but not GFP, to structures resembling actin fibers (arrowheads) and at the plasma membrane (arrows) upon Wnt3a stimulation. Boxes indicate the regions magnified beneath. Mouse NIH3T3 cells were cultured in the presence of 10% serum and Wnt3a stimulation was for 3 hours.
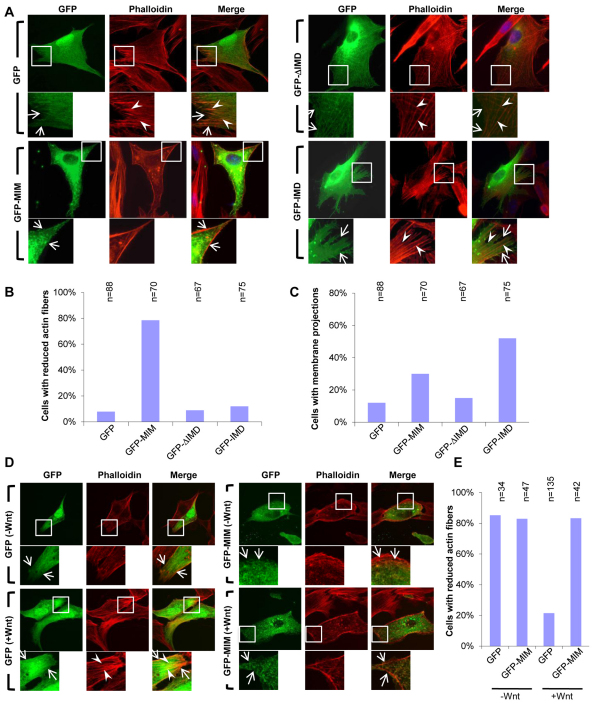
Previous reports showed that overexpression of murine or human MIM depletes actin fibers in cultured cells, possibly owing to its ability to bind to and sequester actin monomers (Gonzalez-Quevedo et al., 2005; Mattila et al., 2003). We tested whether XMIM had similar effects in cultured cells. Transfected NIH3T3 cells showed a dramatically decreased amount of actin fibers but had increased membranous filopodia-like projections compared with untransfected cells (Fig. 3A-C). When the C-terminus fragment ΔIMD was transfected (Fig. 1A) depletion of actin fibers was not observed, although ΔIMD co-localized with actin fibers (Fig. 3A,B). Overexpression of the N-terminal IMD fragment induced membranous filopodia-like projections (Fig. 3A,C), consistent with previous reports (Yamagishi et al., 2004).
Fig. 3.
MIM regulates cellular actin fiber integrity and the formation of membrane protrusions. (A) GFP-MIM, especially GFP-ΔIMD, strongly co-localizes with Phalloidin-stained actin fibers (red). However, GFP-IMD only partially localized to, or decorated, actin fibers (arrowheads), but induced numerous membranous protrusions (arrows). Cells transfected with GFP were used as controls and transfected cells were cultured in the presence of 10% serum. (B) Quantification of the effects of GFP-MIM, GFP-ΔIMD and GFP-IMD on the amount of actin fibers within transfected cells. (C) Quantification of the ability of GFP-MIM, GFP-ΔIMD and GFP-IMD to induce membrane projections within transfected cells. (D) Transfection of GFP-MIM but not GFP inhibits Wnt3a-mediated actin fiber induction. cDNAs were transfected into serum-starved NIH3T3 cells for 24 hours with 3 hours Wnt3a stimulation. (E) Quantification of the results of D. The number of cells analyzed in B, C and E are shown above each bar.
We next examined whether MIM is required for Wnt3a-mediated cytoskeletal reorganization using serum-starved NIH3T3 cells, which have reduced actin fibers in the absence of Wnt3a stimulation. Cells transfected with GFP had reduced numbers of actin fibers in the absence of Wnt3a, but in the presence of Wnt3a contained actin fibers. Overexpression of MIM strongly suppressed Wnt3a-induced actin fiber formation in transfected cells (Fig. 3D,E). These studies suggest that MIM might function in the regulation of changes in the actin cytoskeleton that are mediated by Wnt signaling.
Expression of MIM during embryogenesis
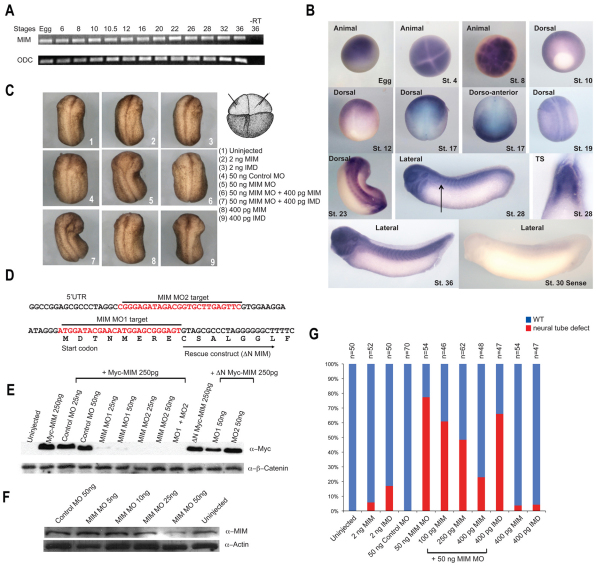
To elucidate the in vivo role of MIM during Xenopus development, we examined the temporal expression pattern of XMIM. RT-PCR uncovered that XMIM is a maternal gene that is expressed throughout development (Fig. 4A). In situ hybridization revealed a broad spatial expression pattern of XMIM in the developing embryo throughout the gastrula stages (Fig. 4B). During the neurula stage, expression of XMIM became refined in the neural folds and anterior neural plate (Fig. 4B). During later stages of development, XMIM was expressed at higher levels in the brain, eyes and spinal cord. This spatial expression of MIM overlaps with that of Daam1 during the gastrula and neurula stages (Nakaya et al., 2004).
Fig. 4.
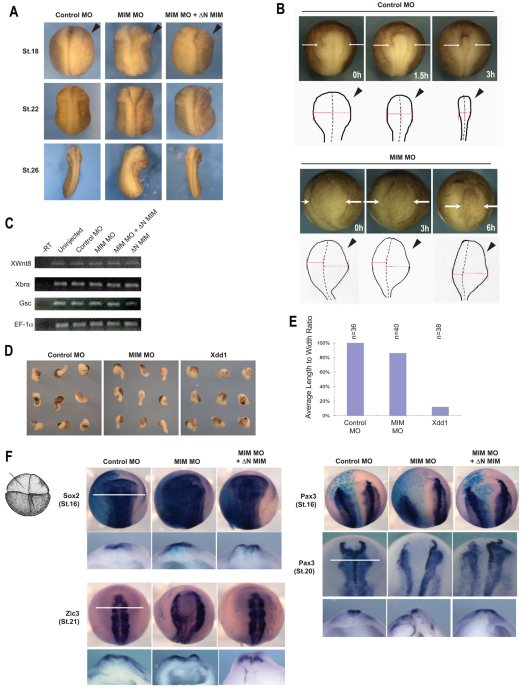
Temporal and spatial expression pattern of MIM and regulation of anterior neural fold closure by MIM. (A) Xenopus MIM (XMIM) is expressed throughout development as monitored by RT-PCR analysis. ODC is used as a loading control; −RT, without reverse transcriptase. (B) The spatial expression pattern of XMIM is dynamic, with highest expression observed in the neural folds of the neurula stage embryo. Stages and views of the embryos are indicated. No signal is detected using a XMIM sense probe. Arrow indicates the plane of the transverse section (TS). (C) Injection of MIM or IMD RNA produces no morphological defects in the Xenopus embryo, whereas injection of MIM MO (50 ng) dorsally inhibits anterior neural fold closure. The MIM MO phenotype can be effectively rescued by co-injection of 400 pg ΔN MIM RNA. (D) Location of the MIM MO target sequences; these are absent from the MIM rescue construct (ΔN MIM), which only includes sequence downstream of the MO1 target. (E) Injection of MIM MO1 or MO2 inhibits translation of Myc-MIM but not of endogenous β-catenin. (F) Injection of MIM MO1 inhibits translation of endogenous MIM but not of endogenous actin in the Xenopus embryo. (G) Quantification of the results of C. The number of injected embryos analyzed is shown above each bar.
Function of MIM during development
To delineate the role of MIM, we performed gain-of-function and loss-of-function studies in the Xenopus embryo. Dorsal or ventral injection of RNA encoding full-length MIM or just the IMD into both blastomeres at the four-cell stage resulted in no phenotype at the highest dose of 2 ng (Fig. 4C,G). However, injection of ΔIMD RNA dorsally or ventrally into both blastomeres at the four-cell stage blocked cytokinesis at the lowest dose of 5 pg, precluding functional analysis of this fragment.
To further investigate the in vivo function of MIM, we examined the effects of knockdown of MIM in the Xenopus embryo. We designed two antisense morpholino oligonucleotides (MO1 and MO2) to deplete the XMIM protein (Fig. 4D) and tested their efficiency in inhibiting translation of a Myc-MIM cDNA containing the 5′UTR and MO binding sequences. Both MIM MOs, but not the control MO, effectively inhibited translation of Myc-MIM in Xenopus embryos but did not affect the endogenous levels of β-catenin (Fig. 4E). We also generated a rescue construct of MIM (ΔN MIM) that lacked the MO binding sequences, and observed that translation of ΔN MIM was unaffected by the MOs (Fig. 4D,E). Additionally, we generated a MIM-specific polyclonal antibody and observed that injection of MIM MO1 or MO2 into the Xenopus embryo caused a reduction of endogenous MIM protein but not of actin (Fig. 4F).
For loss-of-function analyses, we injected 25 ng of each MIM MO (50 ng total) into each dorsal marginal blastomere of the four-cell stage embryo and compared the whole embryo phenotype of the morphants with that of control MO-injected siblings. Both the MIM MO-injected and control MO-injected embryos developed normally through early gastrulation. However, from early neurulation onwards, the neural plate was wider, and narrowing toward the dorsal midline was delayed, in MIM morphant embryos as compared with control MO-injected siblings. During the late neurula stage, the neural tube in MIM morphants failed to close. Notably, anterior neural tube closure was more severely affected than that of the posterior neural tube, as the posterior neural tube eventually closed (Fig. 4C,G). Co-injection of ΔN-MIM, which lacks the MO binding sequence, with the MIM MOs effectively rescued the defects in anterior neural fold closure induced by the MIM MOs (Fig. 4C,G).
To elucidate whether the MIM MO-induced anterior neural tube closure defects were cell-autonomous, we injected 25 ng of each MIM MO into one dorsal marginal blastomere of the four-cell embryo and used time-lapse imaging to follow the whole embryo phenotype (Fig. 5A). Depletion of MIM caused anterior neural fold closure defects only on the injected side (Fig. 5A). We further monitored anterior neural plate elevation and midline convergence by time-lapse imaging, and observed that the anterior neural fold on the MIM MO-injected side never apposed as prominently as on the uninjected side. Additionally, the medial convergence of the elevated fold for closure was severely delayed on the MIM MO-injected side as compared with the uninjected side or with control MO-injected embryos (Fig. 5B). These results demonstrate that MIM regulates neural tube closure during development.
Fig. 5.
Depletion of MIM specifically blocks anterior neural fold closure but does not affect convergent extension movements or the expression of mesodermal and anterior marker genes. (A) Time-lapse images of Xenopus embryos injected with control MO (25 ng), MIM MO (25 ng) and MIM MO + ΔN MIM RNA (25 ng + 400 pg) illustrate that the neural fold on the injected side (arrowheads) fails to close. (B) Measurement of the failure of anterior neural fold closure in MIM MO-injected embryos. Arrows point to the elevated neural fold region (solid black outline in schematic), which moves towards the midline (dashed black line). The solid red line marks the distance between the elevated neural fold region of the uninjected side and the midline, and the dashed red line marks the distance between the elevated neural fold region of the injected side (arrowheads) and the midline. (C) Depletion of MIM does not affect the expression of the mesodermal marker genes XWnt8, Xbra and Gsc in stage 10.5 embryos as analyzed by RT-PCR. EF-1α is used as loading control. (D) Depletion of MIM does not inhibit elongation of Keller explants, whereas dominant-negative Dishevelled (Xdd1, 2 ng) does inhibit elongation. (E) Quantification of Keller explant elongation in D by analysis of the length-to-width ratio of explants. The number of explants analyzed is shown above each bar. (F) Embryos injected dorsally with control MO (50 ng), MIM MO (50 ng) and MIM MO + ΔN MIM RNA (50 ng + 400 pg) show normal mesodermal and neural gene induction but abnormal tissue localization owing to neural fold elevation and closure defects, as examined by whole-mount in situ hybridization. The MIM MO-injected side is indicated in the accompanying diagram for Sox2, Pax3 and Zic3 staining. The MIM MO phenotype is rescued by co-injection of ΔN MIM RNA. Sox2- and Pax3-stained embryos were co-injected with lacZ RNA and stained for β-galactosidase (blue) to label the injected side. Sox2 was expressed in a wider region on the injected than the uninjected side (stage 16). Zic3 expression was only affected in the anterior. Pax3 expression was shifted laterally at stages 16 and 19. Solid white lines indicate the position at which embryos were sectioned.
MIM does not regulate mesodermal cell fate specification or convergent extension movements
As components of the non-canonical pathway, including Daam1, have been shown to regulate CE independent of mesoderm specification (Keller et al., 2003; Wallingford et al., 2002), we investigated the role of MIM in these processes. RT-PCR showed that knockdown and overexpression of MIM had no effects on the expression of the mesodermal genes XWnt8, Xbra and Gsc (Fig. 5C). For CE, Keller explants showed that overexpression or depletion of MIM had no significant effect on explant elongation, whereas the dominant-negative form of Dishevelled, Xdd1, potently inhibited elongation (Fig. 5D,E; data not shown). These results demonstrate that MIM is not required for CE or mesoderm specification during Xenopus development.
Neural fold closure defects induced by loss of MIM are independent of anterior gene expression
We next examined the expression and localization of mesodermal and neural markers by in situ hybridization to ascertain whether depletion of MIM affected neural and anterior gene expression. Embryos were injected in one dorsal marginal blastomere at the four-cell stage with 25 ng control MO or MIM MOs or with MIM MOs plus 400 pg ΔN MIM RNA, along with a lacZ tracer RNA to label the injected side. Sox2 is a pan-neural marker that is expressed in the neural plate at stage 16 (Mizuseki et al., 1998). In MIM MO-injected embryos, Sox2 expression was observed within the region that marked a wider neural plate in the MO-injected side as compared with the uninjected side (Fig. 5F). Pax3 is a segmental mesodermal marker that is expressed in dorsal neural fold cells (Bang et al., 1997). Localization of Pax3 was shifted laterally on the MIM MO-injected side at stages 16 and 19 (Fig. 5F). Zic3 is crucial for both neural and neural crest development and is expressed in the neural plate during neurula stages (Nakata et al., 1997). Zic3 expression was also shifted laterally on the MIM MO-injected side (Fig. 5F). Thus, MIM does not regulate the expression of these marker genes as their induction is normal, despite the abnormal tissue localization.
MIM and Hedgehog signaling
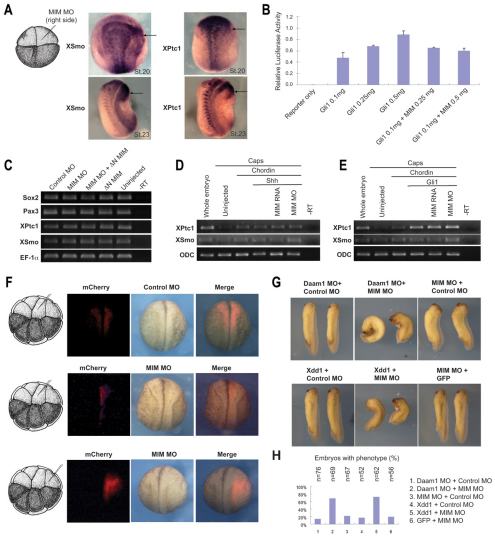
The failure of anterior neural tube closure that results from depletion of MIM could be due to defects in Hedgehog signaling (Eggenschwiler and Anderson, 2007). MIM has been reported to be a Hedgehog target gene in epidermal cells that can synergize with Gli1 to mediate transcription of Hedgehog target genes, including keratin 17 (Callahan et al., 2004). Depletion of MIM can suppress Hedgehog-mediated induction of target genes, including patched 1 and Gli1 (Bershteyn et al., 2010; Callahan et al., 2004; Gonzalez-Quevedo et al., 2005). We tested whether Hedgehog signaling in the MIM MO-injected embryos was affected. Expression of Xenopus Patched1 or Smoothened was unaffected in MIM-depleted embryos, although a failure of anterior neural tube closure was apparent along with somitogenesis defects (Fig. 6A). This prompted us to examine whether MIM can synergize with Gli1 to mediate transcription by employing a Gli-responsive reporter (Sasaki et al., 1999) that is responsive to Hedgehog signaling, transfected into human HEK293T cells. Transfected Gli1 dose-dependently activated the reporter, and co-expression of MIM did not synergize with Gli1-dependent reporter activation (Fig. 6B). A similar lack of effect of MIM on Gli1-dependent reporter activation was also seen in NIH3T3 and HeLa cells (not shown).
Fig. 6.
Depletion of MIM does not affect Hedgehog signaling and MIM synergizes with components of non-canonical Wnt signaling. (A) Depletion of MIM does not inhibit expression of the Hedgehog target genes Patched1 and Smoothened as detected by whole-mount in situ hybridization. Arrows indicate the MIM MO-injected side. (B) Luciferase assays show that transfection of Gli1 into human HEK293T cells dose-dependently activates reporter expression, and that co-expression of MIM with Gli1 does not affect reporter activation. (C) Depletion of MIM does not affect expression of the marker genes Sox2, Pax3, Patched1 (XPtc1) or Smoothened (XSmo) in stage 20 Xenopus embryos as seen by RT-PCR. EF-1α is used as loading control. (D,E) Neuralized Xenopus animal caps (chordin-injected) in the presence of Sonic hedgehog (Shh) (D) or Gli1 (E) express the Hedgehog target genes XPtc1 and XSmo as detected by RT-PCR. Co-injection of MIM RNA or MIM MO does not affect the expression levels of XPtc1 or XSmo. ODC is used as a loading control. (F) Targeted injection was performed into Xenopus dorsal medial animal blastomeres at the 16-cell stage. mCherry RNA (red) was co-injected with control MO, MIM MO and MIM MO + ΔN MIM RNA as a lineage tracer. Medial injection of MIM MO, but not of control MO, results in an anterior neural fold closure defect and this defect is rescued by co-injection of ΔN MIM RNA. (G) MIM synergizes with components of the non-canonical Wnt pathway. Ten nanograms of control MO, Daam1 MO or MIM MO or 250 ng of Xdd1 RNA was injected, respectively. Suboptimal doses of Daam1 MO and Xdd1 RNA co-injected with control MO only induced occasional, mild phenotypes, but co-injection of MIM MO with either Daam1 MO or Xdd1 RNA synergistically enhanced both the severity and the frequency of phenotypes. (H) Quantification of the severity of the results illustrated in G. The number of embryos examined is shown above each bar.
We next tested whether MIM affected Hedgehog signaling and anterior gene expression in the Xenopus embryo using RT-PCR analysis, and observed no effects on Sox2, Pax3, Patched1 or Smoothened expression (Fig. 6C). To further examine whether MIM regulates Hedgehog signaling, we employed animal cap assays. Neuralized animal caps injected with either Sonic hedgehog or Gli1 RNA induced the Hedgehog target genes Patched1 and Smoothened. However, overexpression or depletion of MIM in these animal caps had no effect on the expression levels of Patched1 or Smoothened (Fig. 6D,E). This suggests that the anterior neural fold closure defect caused by the loss of MIM was independent of Hedgehog signaling.
MIM function is required in the medial neural plate
In the early neurula stages, the medial neural plate involutes to form the ventral neural tube, whereas the lateral neural plate forms the dorsal neural tube (Bronner-Fraser and Fraser, 1997). To test whether MIM function is broadly required throughout the neural plate or specifically within the medial or lateral neural plate, we performed targeted injections of MIM MO at the 16-cell stage using membrane-tethered Cherry (mCherry) protein as a tracer to label injected tissue. In medially injected embryos, mCherry-labeled cells formed a single stripe in the dorsal midline. By contrast, in laterally injected embryos, mCherry-labeled cells were located outside of the dorsal midline border (Fig. 6F). Interestingly, open neural tubes were only observed in medially injected MIM MO embryos, whereas laterally injected MIM MO embryos or medially injected control MO embryos had normal neural tube closure (Fig. 6F). Therefore, the function of MIM is required specifically in the midline during neural fold closure.
MIM synergizes with non-canonical Wnt signaling components
As genetic approaches are not amenable in Xenopus laevis, we tested whether simultaneous depletion of MIM and Daam1 leads to synergistic phenotypic effects, which would be expected if two factors function in the same pathway. When a low dose of Daam1 MO or MIM MO was co-injected with control MO, only 15% of injected embryos showed mild gastrulation defects as hallmarked by mild dorsal curvature (Fig. 6G,H). When low doses of MIM MO and control MO were co-injected, less than 5% of embryos displayed any defects. However, co-injection of the low dose of Daam1 MO and MIM MOs resulted in severe defects in more than 60% of the injected embryos (Fig. 6G,H). To test whether MIM functionally interacts with other non-canonical Wnt signaling components, we co-injected a low dose of MIM MOs with Xdd1 RNA, a dominant-negative form of Dishevelled that inhibits non-canonical Wnt signaling (Wallingford and Habas, 2005). Co-injection of low doses of Xdd1 plus control MO only caused mild body axis curvature in the injected embryos. However, when low doses of Xdd1 and MIM MO were combined and co-injected, ~70% of injected embryos exhibited gastrulation defects (Fig. 6G,H). This indicates that MIM functionally interacts with the non-canonical Wnt signaling cascade.
MIM regulates anterior neural fold elevation, apical constriction and hinge point formation
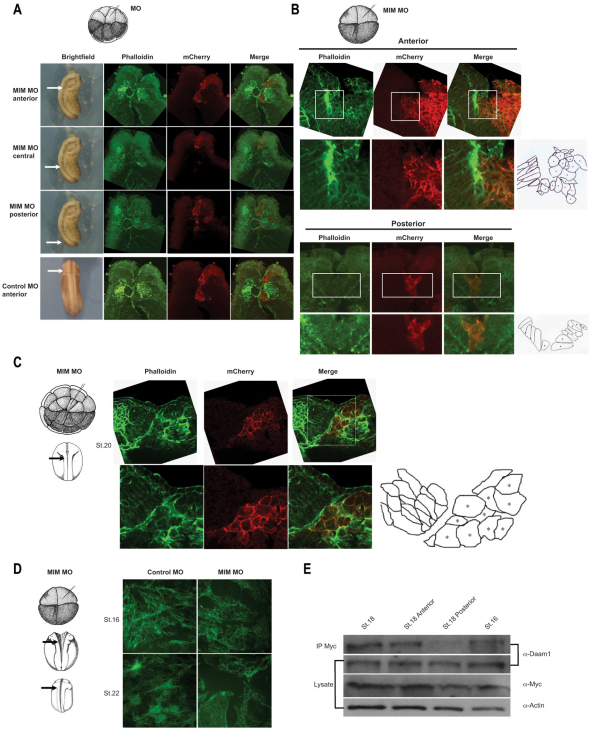
Neural tube closure requires the coordination of several morphogenetic events, including neural plate bending, neural fold elevation and CE, that require cell shape changes and polarized cell migration (Wallingford, 2005; Ybot-Gonzalez et al., 2007). We explored whether MIM is required for these processes. We injected MIM MO and mCherry RNA into one midline dorsal marginal blastomere at the 16-cell stage; vertical transverse sectioning was performed on stage 22 embryos and the sectioned tissues were stained with Oregon Green-Phalloidin. Serial sectioning along the anterior-posterior axis revealed that depletion of MIM led to a failure of elevation in the anterior neural plate, whereas the medial and posterior neural fold elevated normally (Fig. 7A).
Fig. 7.
Depletion of MIM affects anterior neural fold elevation, hinge point formation and apical constriction. (A) MIM depletion inhibits neural fold elevation and closure only in the anterior of the Xenopus embryo. Arrows indicate where sectioning was performed. Transverse sections of a stage 22 embryo show the actin cytoskeleton stained with Phalloidin (green), and mCherry (red) labels cells injected with control MO or MIM MO. Neural fold elevation is only affected in the anterior of the embryo and not in the medial or posterior regions. (B) MIM is required for hinge point formation. In control MO-injected embryos, the anterior neural plate bends at two paired lateral hinge points (not shown). In embryos injected unilaterally with MIM MO, no hinge point forms on the injected (right-hand) side. Actin staining confirms the absence of a hinge point on the MIM MO-injected side (boxed). Drawings illustrate the cell shape on the control MO-injected and MIM MO-injected sides. Asterisks indicate injected side. (C) Strong actin staining is shown on the uninjected side, but no actin accumulation is found on the MIM MO-injected side (boxed). Drawing illustrates the cell shape on the control MO-injected and MIM MO-injected sides. Uninjected cells adopt elongated, polarized morphologies, whereas MIM MO-injected cells are short, round and disoriented. (D) Examination of cells within the anterior neural fold reveals that depletion of MIM inhibits actin fibers, resulting in disrupted morphology. (E) Western blotting shows that MIM preferentially interacts with endogenous Daam1 in the anterior of the embryo. Myc-MIM RNA was injected dorsally and whole embryo or sectioned embryo lysates were isolated at stage 16 and stage 18.
Cell shape change during neural fold elevation is most pronounced in the initial bending regions or ‘hinge points’. In neurula stages, apical constriction occurs predominantly in two paired hinge points that exhibit accumulated apical actin, as visualized by Oregon Green-Phalloidin staining (Haigo et al., 2003). To test whether MIM is required for hinge point formation, we injected MIM MO and mCherry RNA into one dorsal marginal blastomere at the four-cell stage. In contrast to the paired hinge points in control embryos, no accumulated actin staining or hinge point was found on the injected side of MIM MO-injected embryos (Fig. 7A,B).
Failure of hinge point formation and of apical constriction were directly revealed in transverse sections of unilaterally MIM MO-injected embryos. Apical F-actin accumulation is the hallmark of apical constriction (Haigo et al., 2003). Phalloidin staining showed that apical actin failed to accumulate on the MIM MO-injected side, but accumulated normally on the uninjected and elevating side of the neural plate (Fig. 7B). The defects were evident in both the superficial and deep layer neural plate cells. Therefore, we propose that depletion of MIM blocks hinge point formation and apical constriction during neural plate elevation.
One of the cell behaviors underlying apical constriction is the polarization and elongation of neuroepithelial cells along their apicobasal axis. We observed that cells on the uninjected side elongated and adopted cell furrowing and wedging, which are required for neural plate bending and elevation (Fig. 7C). By contrast, injection of MIM MO (mCherry-labeled cells) inhibited both cell shape changes and apicobasal axis orientation (Fig. 7C). Thus, at the cellular level, depletion of MIM disrupts neuroepithelial cell elongation and polarization.
We then examined the organization of actin fibers in the neuroepithelial cells. In control MO-injected cells, numerous actin fibers were present traversing the cell interior; however, in MIM MO-injected cells these fibers were absent and the remaining fibers were highly disorganized (Fig. 7D). These findings suggest a key role for MIM in modulating the actin cytoskeleton rearrangements that underlie neural fold elevation, apical constriction and hinge point formation.
Preferential interaction between MIM and Daam1 in the anterior embryo
To examine how depletion of MIM specifically affects anterior neural fold closure, we performed co-immunoprecipitation of MIM and Daam1 from Xenopus embryo lysates. Full-length MIM specifically associated with endogenous Daam1 in stage 16 and 18 embryos, when neural fold closure occurs. Interestingly, we found that MIM was preferentially associated with Daam1 in the anterior portion of the embryo, even though the level of Daam1 protein was equivalent in anterior and posterior (Fig. 7E). Thus, the regional complex observed in vivo between Daam1 and MIM strongly reflects the neural tube region that is affected upon MIM depletion.
DISCUSSION
The signaling pathways and molecular players that regulate neural tube closure remain an intensive area of investigation. Studies have uncovered a role for the non-canonical Wnt pathway in regulating neural tube closure, but whether individual components of this pathway are required for specific aspects of neural tube closure along the anterior-posterior axis remained undeciphered. We report a central role for the MIM protein in regulating neural fold elevation and closure of the anterior neural tube.
MIM was originally isolated as a cDNA fragment that is present in non-metastatic, but absent in metastatic bladder cancer cell lines (Lee et al., 2002; Loberg et al., 2005). MIM was postulated to be involved in regulation of the actin cytoskeleton as it contains proline-rich sequences and a WH2 actin-binding motif (Lee et al., 2002) that in theory can provide actin monomers for actin fiber growth involving molecules such as cortactin (Lee et al., 2007; Lin et al., 2005). However, the in vivo role of the WH2 domain is not resolved, but it is proposed to regulate recruitment of MIM and its binding partners to regulate cytoskeletal changes (Lee et al., 2007).
Here we show that MIM plays a central role in transducing non-canonical Wnt signaling to regulate anterior neural tube closure. We have identified MIM as a binding partner for Daam1 that specifically binds to a coil-coiled region downstream of the FH2 domain (Fig. 1C). Recent structural studies have shown that the conformation of the Daam1 FH2 domain is stabilized by the β-sheet-like structure within this coil-coiled region, suggesting a regulatory role for this linker region (Lu et al., 2007; Yamashita et al., 2007). Molecules that bind this FH2 linker region might regulate the ability of the FH2 domain of Daam1 to mediate actin nucleation. In addition, it is possible that MIM targets Daam1 subcellularly to modulate changes to the actin cytoskeleton. Indeed, we found that MIM preferentially interacts with Daam1 in the anterior of the embryo, suggesting that their interaction is required for anterior neural tube closure (Fig. 7E). MIM can therefore provide a key route linking non-canonical Wnt signaling via Daam1 to the actin cytoskeleton.
Neural tube closure requires a coordinated series of cell polarization and constriction events for elevation and subsequent closure of the neural folds; the full plethora of factors that regulate this process remain unresolved. Although CE is required during gastrulation for axial extension and neural tube closure, defects in CE result in neural tube closure defects that are centered from the midbrain through to the lower spine region, but not in the anterior region (Wallingford, 2005; Wallingford and Harland, 2002). We show that in the Xenopus embryo, MIM is highly expressed within the anterior neural folds (Fig. 4B) and knockdown of MIM specifically impairs the elevation and closure of the anterior neural tube without affecting CE (Fig. 4F,G and Fig. 5A,B). Thus, the expression pattern of MIM correlates with its function within the anterior neural tube.
A key process that contributes to neural tube closure is apical constriction, which regulates neural fold elevation and apposition. In mammals there are two central bending points termed the medial and paired dorsolateral hinge points (Copp et al., 2003). The actin cytoskeleton also plays a crucial role during elevation of the neural folds and, interestingly, closure of the neural folds in the cranial region is more susceptible to molecules that affect the actin cytoskeleton (Copp et al., 2003; Ybot-Gonzalez and Copp, 1999). Here, we find that knockdown of MIM specifically disrupts hinge point formation and apical constriction (Fig. 7A-D) and we propose that MIM regulates hinge point formation via its ability to influence the actin cytoskeleton.
MIM possesses two key domains required for anterior neural tube closure. The N-terminal IMD has been shown to bind lipid membranes and induce negative curvature (Bompard et al., 2005; Disanza et al., 2006; Lee et al., 2007), and such membrane curvature is required for neural fold apposition. Second, the WH2 domain can bind to actin and provide actin monomers for the Daam1-mediated actin fiber polymerization required for neural fold apposition and hinge point formation (Lee et al., 2007; Lin et al., 2005). MIM-Daam1 interaction can therefore govern two key features: membrane curvature and remodeling along with effects on the actin cytoskeleton. These two features are crucial for neural tube apposition, hinge point formation and neural tube closure. Indeed, the WH2-containing fragment of MIM localizes to actin fibers and membranous protrusions are observed in cells expressing the IMD (Fig. 3A-C). Notably, expression of full-length MIM, but not the IMD fragment, effectively rescues neural tube closure defects caused by the MIM MOs (Fig. 4F,G). We were unable to test for rescue using the ΔIMD-MIM fragment as injection of this fragment interfered with cell division (Fig. 4C,G). This suggests that the IMD is not sufficient for MIM function during anterior neural tube closure.
Hedgehog signaling also plays important roles during neural tube closure, and disrupted Sonic hedgehog signaling can result in neural tube closure defects, whereas excessive signaling can abrogate the formation of paired hinge points (Goodrich et al., 1997; Ybot-Gonzalez et al., 2002). The finding that MIM is a Hedgehog target gene that cooperates with the Gli1 transcription factor to potentiate Hedgehog signaling (Callahan et al., 2004; Gonzalez-Quevedo et al., 2005) provides the intriguing possibility that MIM might be a key cross-talk factor between the non-canonical Wnt and Hedgehog signaling pathways. MIM was recently shown to modulate Drosophila border cell migration via endocytic regulation (Quinones et al., 2010; Quinones and Oro, 2010) and to regulate ciliogenesis and Hedgehog signaling in dermal cells (Bershteyn et al., 2010). It is noteworthy that defects in Hedgehog signaling also result in anterior neural tube closure defects (Copp and Greene, 2010). However, we did not observe any defects in the expression of a number of Hedgehog target genes upon MIM depletion in the Xenopus embryo, nor did we observe transcriptional synergy between MIM and Gli1 using a transcriptional reporter-based assay in culture cells (Fig. 5A-E). Thus, we favor the possibility that the defects observed in anterior neural tube closure are not due to dysregulated Hedgehog signaling, but rather to an effect on Daam1-mediated non-canonical Wnt signaling.
In summary, we propose a model of non-canonical Wnt signaling-mediated neural tube closure in which MIM is recruited into a complex with Daam1 in the anterior neural folds. This Daam1-MIM complex mediates actin polymerization and hinge point formation. MIM, via its IMD, regulates the negative curvature of the plasma membrane required for neural fold apposition and closure. As Daam1 can regulate distinct aspects of gastrulation and neural tube closure via a cohort of interacting proteins, including Profilin1 for blastopore closure (Sato et al., 2006), Profilin2 for CE movements (Khadka et al., 2009) and now MIM for anterior neural tube closure, we propose that non-canonical Wnt-mediated control of gastrulation via Daam1 is mediated via distinct and/or overlapping functions of its interacting partners. Identification of the full plethora of Daam1 binding partners and elucidation of their discrete functions remain important future goals.
Supplementary Material
Acknowledgements
We thank members of the L.R. and R.H. laboratories for discussion and critical comments; and Drs Jeffrey Miller, Michael Matisse, Jing Yang, Jean-Pierre Saint-Jeannet, Chenbei Chang and Karen Symes for reagents. This work is supported by grants from the March of Dimes (1-FY07-681), NSF (#0544061) and NIH (GM078172) to R.H. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.058800/-/DC1
References
- Asaoka Y., Kanai F., Ichimura T., Tateishi K., Tanaka Y., Ohta M., Seto M., Tada M., Ijichi H., Ikenoue T., et al. (2010). Identification of a suppressive mechanism for Hedgehog signaling through a novel interaction of Gli with 14-3-3. J. Biol. Chem. 285, 4185-4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang A. G., Papalopulu N., Kintner C., Goulding M. D. (1997). Expression of Pax-3 is initiated in the early neural plate by posteriorizing signals produced by the organizer and by posterior non-axial mesoderm. Development 124, 2075-2085 [DOI] [PubMed] [Google Scholar]
- Bershteyn M., Atwood S. X., Woo W. M., Li M., Oro A. E. (2010). MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev. Cell 19, 270-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bompard G., Sharp S. J., Freiss G., Machesky L. M. (2005). Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J. Cell Sci. 118, 5393-5403 [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M., Fraser S. E. (1997). Differentiation of the vertebrate neural tube. Curr. Opin. Cell Biol. 9, 885-891 [DOI] [PubMed] [Google Scholar]
- Callahan C. A., Ofstad T., Horng L., Wang J. K., Zhen H. H., Coulombe P. A., Oro A. E. (2004). MIM/BEG4, a Sonic hedgehog-responsive gene that potentiates Gli-dependent transcription. Genes Dev. 18, 2724-2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp A. J., Greene N. D. (2010). Genetics and development of neural tube defects. J. Pathol. 220, 217-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp A. J., Greene N. D., Murdoch J. N. (2003). The genetic basis of mammalian neurulation. Nat. Rev. Genet. 4, 784-793 [DOI] [PubMed] [Google Scholar]
- Curtin J. A., Quint E., Tsipouri V., Arkell R. M., Cattanach B., Copp A. J., Henderson D. J., Spurr N., Stanier P., Fisher E. M., et al. (2003). Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr. Biol. 13, 1129-1133 [DOI] [PubMed] [Google Scholar]
- Davidson L. A., Keller R. E. (1999). Neural tube closure in Xenopus laevis involves medial migration, directed protrusive activity, cell intercalation and convergent extension. Development 126, 4547-4556 [DOI] [PubMed] [Google Scholar]
- Disanza A., Mantoani S., Hertzog M., Gerboth S., Frittoli E., Steffen A., Berhoerster K., Kreienkamp H. J., Milanesi F., Di Fiore P. P., et al. (2006). Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat. Cell Biol. 8, 1337-1347 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler J. T., Anderson K. V. (2007). Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Quevedo R., Shoffer M., Horng L., Oro A. E. (2005). Receptor tyrosine phosphatase-dependent cytoskeletal remodeling by the hedgehog-responsive gene MIM/BEG4. J. Cell Biol. 168, 453-463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich L. V., Milenkovic L., Higgins K. M., Scott M. P. (1997). Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109-1113 [DOI] [PubMed] [Google Scholar]
- Habas R., Kato Y., He X. (2001). Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell 107, 843-854 [DOI] [PubMed] [Google Scholar]
- Haigo S. L., Hildebrand J. D., Harland R. M., Wallingford J. B. (2003). Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13, 2125-2137 [DOI] [PubMed] [Google Scholar]
- Hildebrand J. D., Soriano P. (1999). Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell 99, 485-497 [DOI] [PubMed] [Google Scholar]
- Jenny A., Darken R. S., Wilson P. A., Mlodzik M. (2003). Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 22, 4409-4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. (2002). Shaping the vertebrate body plan by polarized embryonic cell movements. Science 298, 1950-1954 [DOI] [PubMed] [Google Scholar]
- Keller R., Davidson L. A., Shook D. R. (2003). How we are shaped: the biomechanics of gastrulation. Differentiation 71, 171-205 [DOI] [PubMed] [Google Scholar]
- Khadka D. K., Liu W., Habas R. (2009). Non-redundant roles for Profilin2 and Profilin1 during vertebrate gastrulation. Dev. Biol. 332, 396-406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Kerff F., Chereau D., Ferron F., Klug A., Dominguez R. (2007). Structural basis for the actin-binding function of missing-in-metastasis. Structure 15, 145-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. G., Macoska J. A., Korenchuk S., Pienta K. J. (2002). MIM, a potential metastasis suppressor gene in bladder cancer. Neoplasia 4, 291-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Liu J., Wang Y., Zhu J., Zhou K., Smith N., Zhan X. (2005). Differential regulation of cortactin and N-WASP-mediated actin polymerization by missing in metastasis (MIM) protein. Oncogene 24, 2059-2066 [DOI] [PubMed] [Google Scholar]
- Liu W., Sato A., Khadka D., Bharti R., Diaz H., Runnels L. W., Habas R. (2008). Mechanism of activation of the Formin protein Daam1. Proc. Natl. Acad. Sci. USA 105, 210-215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loberg R. D., Neeley C. K., Adam-Day L. L., Fridman Y., St John L. N., Nixdorf S., Jackson P., Kalikin L. M., Pienta K. J. (2005). Differential expression analysis of MIM (MTSS1) splice variants and a functional role of MIM in prostate cancer cell biology. Int. J. Oncol. 26, 1699-1705 [PubMed] [Google Scholar]
- Lu J., Meng W., Poy F., Maiti S., Goode B. L., Eck M. J. (2007). Structure of the FH2 domain of Daam1: implications for formin regulation of actin assembly. J. Mol. Biol. 369, 1258-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M., Johnston S. A. (2007). MIM: a multifunctional scaffold protein. J. Mol. Med. 85, 569-576 [DOI] [PubMed] [Google Scholar]
- Mattila P. K., Salminen M., Yamashiro T., Lappalainen P. (2003). Mouse MIM, a tissue-specific regulator of cytoskeletal dynamics, interacts with ATP-actin monomers through its C-terminal WH2 domain. J. Biol. Chem. 278, 8452-8459 [DOI] [PubMed] [Google Scholar]
- Mizuseki K., Kishi M., Matsui M., Nakanishi S., Sasai Y. (1998). Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development 125, 579-587 [DOI] [PubMed] [Google Scholar]
- Nakata K., Nagai T., Aruga J., Mikoshiba K. (1997). Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc. Natl. Acad. Sci. USA 94, 11980-11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M. A., Habas R., Biris K., Dunty W. C., Jr, Kato Y., He X., Yamaguchi T. P. (2004). Identification and comparative expression analyses of Daam genes in mouse and Xenopus. Gene Expr. Patterns 5, 97-105 [DOI] [PubMed] [Google Scholar]
- Quinones G. A., Oro A. E. (2010). BAR domain competition during directional cellular migration. Cell Cycle 9 (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinones G. A., Jin J., Oro A. E. (2010). I-BAR protein antagonism of endocytosis mediates directional sensing during guided cell migration. J. Cell Biol. 189, 353-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H., Nishizaki Y., Hui C., Nakafuku M., Kondoh H. (1999). Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126, 3915-3924 [DOI] [PubMed] [Google Scholar]
- Sato A., Khadka D. K., Liu W., Bharti R., Runnels L. W., Dawid I. B., Habas R. (2006). Profilin is an effector for Daam1 in non-canonical Wnt signaling and is required for vertebrate gastrulation. Development 133, 4219-4231 [DOI] [PubMed] [Google Scholar]
- Schoenwolf G. C., Folsom D., Moe A. (1988). A reexamination of the role of microfilaments in neurulation in the chick embryo. Anat. Rec. 220, 87-102 [DOI] [PubMed] [Google Scholar]
- Ueno N., Greene N. D. (2003). Planar cell polarity genes and neural tube closure. Birth Defects Res. C Embryo Today 69, 318-324 [DOI] [PubMed] [Google Scholar]
- Wallingford J. B. (2005). Neural tube closure and neural tube defects: studies in animal models reveal known knowns and known unknowns. Am. J. Med. Genet. 135C, 59-68 [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., Harland R. M. (2002). Neural tube closure requires Dishevelled-dependent convergent extension of the midline. Development 129, 5815-5825 [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., Habas R. (2005). The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development 132, 4421-4436 [DOI] [PubMed] [Google Scholar]
- Wallingford J. B., Fraser S. E., Harland R. M. (2002). Convergent extension: the molecular control of polarized cell movement during embryonic development. Dev. Cell 2, 695-706 [DOI] [PubMed] [Google Scholar]
- Yamagishi A., Masuda M., Ohki T., Onishi H., Mochizuki N. (2004). A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 279, 14929-14936 [DOI] [PubMed] [Google Scholar]
- Yamashita M., Higashi T., Suetsugu S., Sato Y., Ikeda T., Shirakawa R., Kita T., Takenawa T., Horiuchi H., Fukai S., et al. (2007). Crystal structure of human DAAM1 formin homology 2 domain. Genes Cells 12, 1255-1265 [DOI] [PubMed] [Google Scholar]
- Ybot-Gonzalez P., Copp A. J. (1999). Bending of the neural plate during mouse spinal neurulation is independent of actin microfilaments. Dev. Dyn. 215, 273-283 [DOI] [PubMed] [Google Scholar]
- Ybot-Gonzalez P., Cogram P., Gerrelli D., Copp A. J. (2002). Sonic hedgehog and the molecular regulation of mouse neural tube closure. Development 129, 2507-2517 [DOI] [PubMed] [Google Scholar]
- Ybot-Gonzalez P., Savery D., Gerrelli D., Signore M., Mitchell C. E., Faux C. H., Greene N. D., Copp A. J. (2007). Convergent extension, planar-cell-polarity signalling and initiation of mouse neural tube closure. Development 134, 789-799 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.