Abstract
We summarize a recently developed microtechnology for printing biomaterials on biological surfaces. The technique is based on the use of immiscible aqueous solutions of two biopolymers and allows spatially defined placement of cells and biomolecules suspended in the denser aqueous phase on existing cell layers and extracellular matrix hydrogel surfaces maintained in the second phase. Printing takes place due to an extremely small interfacial tension and density difference between the two aqueous phases. The contact-free printing process ensures that both printed cells and the underlying cell monolayer maintain full viability and functionality. The technique accommodates both arbitrarily shaped patterns and microarrays of cells and bioreagents. The capability to print cells and small molecules on existing cell layers enables unique interrogations of the effects of cell-cell and cell-material interaction on cell fate and function. Furthermore, the very gentle conditions and the ability to directly pattern nongel embedded cells over cells make this technology appealing to tissue engineering applications where patterned multicellar organization with minimal scaffolding materials is needed, such as in dense tissues of the skeletal muscle and liver.
INTRODUCTION
Organ failure as a result of injury or disease affects the quality of life of millions of people worldwide. The past two decades have witnessed extensive research dedicated to the development of new strategies for restoring or creating new tissue constructs.1 Several approaches have resulted in the delivery of isolated cells to replace lost cells and restore the tissue function, the use of tissue-inducing factors such as growth- and differentiation-inducing molecules to a target site, and constructing tissue pieces in vitro using cells, hydrogels, and other factors as the building blocks.1 While the first two techniques yield promising results for small defects, extensive tissue damage in majority of patients leaves the last method as the only viable option.
Two major approaches have been developed for in vitro engineering of artificial tissue replacements: “top-down” and “bottom-up.”2 The former involves the fabrication of a porous scaffold of synthetic polymers (e.g., polyglycolide, polylactide, and polylactide coglycolide) and natural materials (e.g., fibrin and collagen) and subsequent injection of a cocktail of one or more cell types of the target tissue. The scaffold, which may also be chemically conjugated to various biomolecules, provides growth promoting cues to cells and guides their synthesis of extracellular matrix (ECM) proteins and other factors.3, 4, 5 The physicochemical and geometrical properties of the scaffold and the intrinsic propensity of cells to organize into three-dimensional structures are key determinants of the quality of tissue formation.6, 7 This approach has shown some promise;8, 9 nevertheless, there are several drawbacks inherent to the scaffold-based tissue engineering including nonuniform distribution of cells within the scaffold, lack of control over spatial placement of different cell types in a prearranged manner, and interference of the scaffold with direct cell-cell interactions. These can result in insufficient cellular organization and impaired formation and function of the construct. In addition, the scaffolding material may elicit adverse host responses.
The bottom-up approach can, in principle, overcome these limitations and accommodate placement of defined densities of various cell types and natural ECM materials in a spatially controlled manner to allow direct cell-cell and cell-ECM contact. The assembly of cell-laden microgels in two-phase media driven by energetic effects,2 printing of cell-loaded hydrogels through mechanical valves and needles,10, 11, 12 assembly of cellular spheroids onto templating agarose rods,13 rapid prototyping-based matrix fabrication,14, 15 micromolding of cells and hydrogels,16 inkjet printing of cells onto hydrogels,17, 18 and cell sheet engineering19, 20, 21 are major methods developed to control three-dimensional tissue structuring. Although the majority of these techniques are still in the research phase, the cell sheet engineering technology has already been successfully utilized in clinical settings for reconstruction of thin corneal tissue.22
For a bottom-up patterning∕printing approach to be useful for tissue engineering applications, a series of criteria should be met. The method should (i) be gentle to cells and avoid exerting large forces during preparation and patterning steps so that cells retain high viability and full functionality, (ii) be noncontact to allow placement of particulates on already existing cell layers, (iii) enable layered spatial patterning of multiple cell types specific to a tissue, (iv) be carried out in fully aqueous cell-friendly media to prevent drying or stressing of cells, (v) ensure fidelity of patterned∕printed biomaterials including cells, hydrogels, and small molecules, and (vi) avoid exposure to harmful mutation-inducing radiations (e.g., ultraviolet light) that may be used for cross-linking of hydrogels. Recently, we have developed a simple and efficient method for patterning biomolecules and layering of cells on living cell layers that meets these criteria. In the following, we summarize this approach and highlight its potential as a new tissue engineering technique.
DIRECT MICROPRINTING ON CELLS USING AQUEOUS TWO-PHASE SYSTEMS
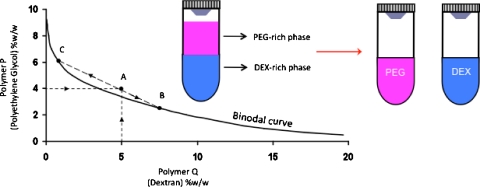
Aqueous mixtures of several polymers above certain concentrations segregate and give rise to two distinct phases.23 The resulting system is known as an aqueous two-phase system (ATPS) and is characterized by a unique phase diagram, which describes the composition of each phase and the range of concentrations that result in phase separation (Fig. 1). To form an ATPS, the polymers can first be dissolved separately in aqueous media at desired polymer concentrations (e.g., point A in Fig. 1). The two solutions are then thoroughly mixed and allowed to equilibrate. A sharp interface is formed after the equilibrium condition is established and two distinct phases with compositions of points B and C form (Fig. 1). Then, the two phases can be separated gently. This principle has been used in various macroscale bioseparation processes as a means of purifying biological particulates, including proteins and cells, based on the physicochemical properties of the phase forming polymers and biomolecules of ineterest.24, 25 ATPSs have also been used as microscale separation media to partition cells and proteins in microfluidic devices.26, 27 The nonionic polymers polyethylene glycol (PEG) and dextran (DEX) are most widely used in such applications and provide a mild bioseparation environment and act as the protectant of the particulates.25 This particular ATPS always gives rise to a top PEG phase and a bottom DEX phase due to a small density difference between them.
Figure 1.
Polymeric ATPS. A typical phase diagram of an ATPS comprising PEG (Mw: 8 K) and DEX (Mw: 500 K). Only those combinations of the two polymers P (PEG) and Q (DEX) above the binodal curve give an ATPS. Point A represents the initial concentration of each polymer in the entire solution, whereas points B and C describe the compositions of bottom and top phases in equilibrium, respectively. Line BC represents a tie line and is a unique property of the given ATPS.
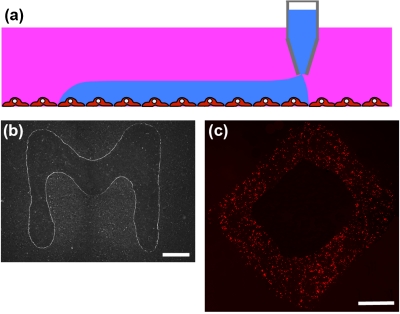
We have recently adapted this partitioning technology to develop a novel approach for direct contact-free printing of bioreagents on cell layers. The aqueous PEG phase prepared in culture media is used as the immersion phase to cover a monolayer of cells in culture. The DEX phase is used as the patterning phase and is loaded into a patterning tool such as an extruded pipette tip. The pipette tip is lowered in the vicinity of the cell layer. The denser DEX phase immediately dispenses out of the tip of the pipette to form a droplet. However, the extremely small interfacial tension between the two aqueous phases, usually of the order of 1–100 μJ∕m2, cannot support the weight of the drop, and thus, the DEX phase dispenses continuously. This autonomous dispensing combined with simultaneous horizontal movement of the patterning tool results in the formation of a linear pattern of the DEX phase on the cell monolayer maintained in the aqueous PEG phase [Figs. 2a, 2b].28 The ultralow PEG-DEX interfacial tension combined with the microscale roughness of the cell monolayer and the affinity of cell’s surface to the DEX phase help maintain the DEX phase patterns stable over extended periods of time (at least a day). Furthermore, bioreagents can be stably partitioned in the DEX phase and thus be localized for durations that are far beyond what can be expected if there was free diffusion of reagents and that are sufficient for bioreagents to exert their function. We confirmed this point, for example, by printing transfection complexes of nucleic acids (dsRed) on a monolayer of cells in an arbitrary shape. Cells were incubated for 8 h in the presence of ATPS media, which was then washed out, and the incubation continued for another 48 h with regular growth media to allow expression of the gene. This resulted in localized expression patterns of the gene that exactly mimicked the shape of the printed reagent [Fig. 2c].28 Thus, the ATPS microprinting technique enabled direct, gentle, contact-free, and stable spatial patterning of reagents on living cell monolayers in fully aqueous cell-friendly media. The resolution of printed patterns, that is, how much the patterned DEX phase spreads on the cell layer covered in the PEG phase, is determined by a force balance between the three interfacial tensions, i.e., PEG phase-cell monolayer (γPEG-cell), DEX phase-cell monolayer (γDEX-cell), and PEG phase-DEX phase (γPEG-DEX), where the three phases meet as well as the diameter and volume of the dispensing mechanism.28 With an extruded pipette tip of ∼150 μm in diameter, patterns as thin as 300 μm could be generated. Better resolutions on the scale of a few cells may be achievable using dispensing mechanisms of a few nanoliters in volume.
Figure 2.
Reagent microprinting on cells. (a) The denser DEX phase is used as the patterning phase and printed on cells immersed in the PEG phase. (b) The ATPS microprinting technique allows generating user-defined stable patterns of reagents on cells. (c) Long-term stability of patterns is demonstrated by including liposomal transfection complexes of a dsRed plasmid in the patterning DEX phase and printing them in the shape of a diamond. The resulting gene expression pattern resembles the printed diamond shape. Scale bar 1 mm in (b) and (c). [Panels (b) and (c) adapted with permission from NPG (Ref. 28)].
We have also evaluated the compatibility of the ATPS media with cells. Marinating cells in PEG- and DEX-containing culture media for 24 h as well as transfection of cells in ATPS media resulted in a high viability of over 95%, similar to that observed in control experiments without any polymer addition to media.28 It should be noted that our PEG-media is formulated with low concentrations of a high molecular weight PEG to minimize adverse cellular effects. As with any new media formulation, however, it may be important to verify compatibility when working with different cell types due to variability of culture condition requirements.
CELL MICROPRINTING ON CELLS
The ATPS microprinting strategy can be used as a new contact-free cell printing technique when the patterning DEX phase contains a suspension of cells. The utility of aqueous biphasic systems for cell printing strongly depends on the partitioning behavior of cells in ATPS; that is, which phase cells favor during the equilibration stage. The partition of cells in an ATPS depends on the surface properties of cells and several phase factors including polymer molecular weight, ionic composition of phases, temperature, and electrostatic potential difference and the interfacial tension between phases.29 Elucidating the individual effect of each phase factor is often difficult because varying one physical property can alter others too. However, it has been shown that cell partition in ATPSs is a function of the interfacial tension between the top and bottom phases, γ12, and the electrostatic potential difference between the two aqueous phases, Δψ [i.e., K=f(γ12,Δψ)].30 Here K denotes the partition coefficient and is defined as the ratio of the number of cells in the bottom and top phases, respectively.31 When both phases are prepared in the same culture media, the effect of the electrostatic potential can safely be ignored if the phase forming polymers are nonionic, such as PEG and DEX, and do not further contribute to the electrostatic potential of the phases. Thus, the partition of cells becomes mainly a function of the interfacial tension [i.e., K=f(γ12)]. Under this condition, it was shown that partition coefficient follows the relation below,30
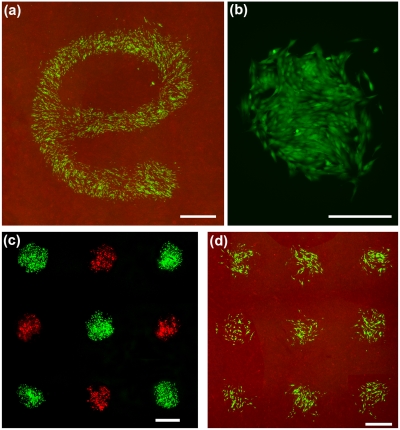
where α is a constant number. For the purpose of cell printing with the PEG-DEX two-phase system, cells should have favorable partitioning to the DEX phase (i.e., the patterning medium). According to the above equation for the K value to increase, the interfacial tension, γ12, should be minimized. The interfacial tension of aqueous biphasic systems is a function of the concentration of the phase forming polymers and for a given ATPS, it decreases by lowering the polymer concentrations.32 Decreasing γ12 also helps maintain physiologic levels of the tonicity of the phases and ensures that cells retain their normal morphology and function. We studied several two-phase systems and, based on independent measurements of the PEG-DEX interfacial tension and the partition coefficient, identified that the ATPS with 2.5%w∕w PEG (Mw of 35 K) and 3.2%w∕w DEX (Mw of 500 K) gives an extremely small interfacial tension of γ12=10 μJ∕m2 and a partition coefficient of K=78%.31 We used this system for cell printing on a cell monolayer and found that it results in uniform patterns of printed cells confirming the above principle about the relation between cell partition in ATPS and the interfacial tension [Fig. 3a]. Importantly, the ultralow interfacial tension-driven dispensing of the patterning aqueous phase is a real advantage over existing printing techniques that use damaging electrical and thermal energy or mechanical means to eject biomaterials. Using the ATPS microprinting approach, cells retain a full postprinting viability of ∼100% [Fig. 3b]. We have also adapted the technique to multiplexed microarray printing of cells both on hydrogel surfaces and cell layers. A series of nanoliter-volume dispensing pins residing on a fixture is loaded with the patterning DEX phase containing cells. The vertical movement of the pins, which is controlled using a micromanipulator, into the PEG phase results in the dispensing of the DEX phase and formation of droplets of cell suspension and subsequent attachment of cells to the underlying surface [Figs. 3c, 3d].
Figure 3.
Microprinting cells. (a) Green fluorescing cells printed on a monolayer of red fluorescing cells spelling “e.” (b) Printed cells retain a viability of ∼100%. (c) A duplex microarray of green and red fluorescent cells printed on a layer of porcine gel. (d) A microarray of green fluorescent cells printed on a monolayer of red fluorescent cells. Scale bar, 1 mm in (a), 400 μm in (b), and 800 μm in (c) and (d). [Panels (a)–(c) adapted with permission from Wiley-VCH (Ref. 31)].
APPLICATIONS TOWARD TISSUE ENGINEERING
Homotypic and heterotypic cell-cell interactions are known to have important regulatory effects on tissue formation and function.33 For example, embryonic fibroblast-endothelial cell interactions were shown to regulate vascularization of the engineered skeletal muscle and enhance the integrity of the resulting capillaries.8 Thus, the capability of direct printing of cells on living cell layers using the ATPS approach is an enabling tool for tissue engineering type applications.
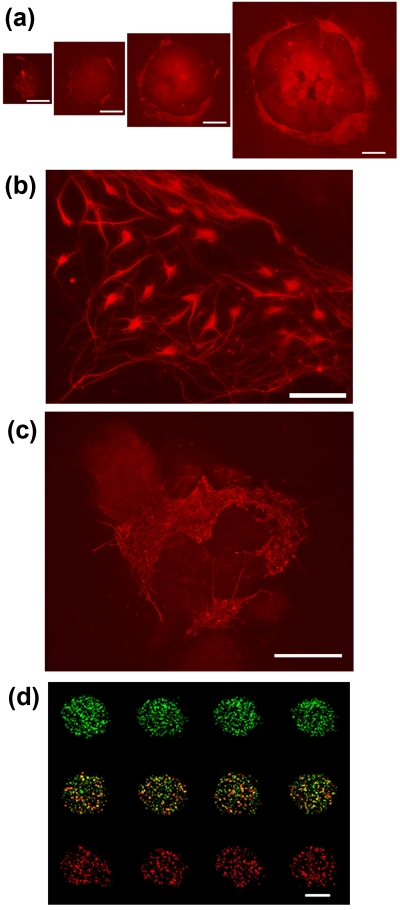
Over the past decade, there has been a growing interest in using stem cells as the cell source for engineering tissue constructs because of the potential of these cells to differentiate into various cell types of the body.34 The success of this approach depends on the ability to generate well-defined niche conditions, including cell-cell, cell-ECM, and cell-soluble factors interactions, and study the effect of these factors in directing stem cells’ fate decisions. The ATPS microprinting technique is a potent method to interrogate the influence of niche conditions on the fate of stem cells because it allows placement of cells on cell layers and ECM hydrogel surfaces and, in principle, can accommodate spatial control over localized delivery of biomolecules such as growth and differentiation factors to cells. We have conducted a series of cell and reagent printing studies to demonstrate this point. (i) Mouse embryonic stem cells (mESCs) were directly printed on a layer of support PA6 stromal cells to study direct heterocellular interactions effect on the differentiation of mESC to neurons. Selecting the printed mESC densities in the range of 20–500 cells per spot generated four different niche conditions. After 8 days of culture, mESCs grew to individual colonies of 0.9–5.8 mm2. Immunostaining of mESC for neuron-specific tubulin TuJ revealed neuronal differentiation of mESC at the periphery of colonies and a niche size-dependent differentiation phenotype where larger mESC colonies yielded higher levels of TuJ expression [Figs. 4a, 4b].31 This enhanced differentiation is not simply due to higher number of cells in larger colonies because normalizing the TuJ staining at the periphery of colonies with respect to the area of each colony still yielded an increasing trend with colony size.31 (ii) An array of two different types of feeder cells, mouse embryonic fibroblast (MEF) and PA6 cells, was printed on a porcine gel coated culture plate and overlaid with mESC.35 This created duplex heterocellular microarray of stem cell niches on a hydrogel surface. The MEF cell islands supported prolonged undifferentiated state of mESCs, whereas PA6 cells guided differentiation of mESCs to neurons. Interestingly on differentiation-inducing PA6 feeder cells, mESCs that grew to several small (100–300 μm in diameter) and interspaced (50–150 μm) colonies show very dense networks of neurons. This configuration resulted in one to two orders of magnitude enhancement in the differentiation efficiency compared to individual mESC colonies [Fig. 4c].31 Computational modeling suggested that observed differentiation patterns are due in part to the maintenance of concentration gradients of autocrine factors in the spacing among the colonies. (iii) As an example of localized delivery of small molecules, liposomal complexes of nucleic acids usually smaller than 100 nm36 were printed on defined clusters of cells from a monolayer of HEK293H cells. We obtained a multiplexed gene expression microarray for eGFP (top row), dsRed (bottom row), or cotransfection of both genes (middle row) within a lawn of nontransfected cells [Fig. 4d]. The absence of cross-contamination between the transfected cell clusters confirmed the confinement of genetic materials to the printed DEX phase droplets during the transfection period.28 Furthermore, other biomaterials including lentiviral vectors28 and suspensions of bacterial colonies37 have successfully been patterned. Altogether, these studies provide ample evidence that the ATPS microprinting is a versatile approach for spatially defined printing of biomaterials on delicate cell and ECM hydrogel surfaces.
Figure 4.
ATPS Microprinting for cell fate and function. (a) Size-dependent neuronal differentiation of individual mESC colonies resulting from printing different densities of mESC on feeder PA6 cells. (b) High magnification image of lower left corner of the third largest colony of panel (a). (c) Enhanced differentiation of interspaced small mESC colonies on a PA6 feeder island. (d) Multiplexed genetic materials printing on HEK293H cell clusters from a monolayer results in a spatially defined gene expression pattern. The concentration of GFP and dsRed plasmids in the clusters of the middle row is each half of that in the top and bottom rows. Scale bar 500 μm in (a), 100 μm in (b), 250 μm in (c), and 700 μm in (d). [Panel d adapted with permission from NPG (Ref. 28)].
CONCLUSION
We summarized a new microtechnology that utilizes two immiscible aqueous solutions to print biomaterials on various substrates. This strategy offers direct and contact-free placement of predefined patterns of cells and reagents on living cell monolayers. The printing process is extremely gentle to cells and carried out in fully aqueous cell-compatible media to accommodate full cell viability. These features make it an appealing technology for bottom-up tissue engineering applications. Interfacing this ATPS microprinting with robotic systems will facilitate automated layered printing of cells and hydrogels and enable constructing tissues using the required materials. This approach may help address the long-recognized challenge of constructing thick tissues of the skeletal muscle, liver, and heart that are multicellular, contain minimal scaffolding matrix within a dense mass of the cells, and are highly organized into three-dimensional architectures that serve tissue function.38, 39 The capability of organized placement of a particular cell type within a matrix of a second cell type using the ATPS microprinting may result in more efficient construction of organized tissue structures and faster tissue vascularization compared to scaffold-based methods, which rely on random seeding of cells. Nevertheless, a key challenge will be ensuring that cells embedded within previously printed layers of cells remain viable and functional. A combination of ATPS microprinting technology and other tissue engineering approaches may be required to tackle this problem. In the context of utility of stem cells for tissue engineering,40 the ATPS printing approach will be useful to determine niche conditions that result in optimum differentiation levels of stem cells toward a particular lineage with minimal usage of precious stem cells. The identification of niche size and colony interspacing effect on the efficiency of neuronal differentiation of mESCs, which was discussed above, is an elucidating example.
References
- Langer R. and Vacanti J. P., Science 260, 920 (1993). 10.1126/science.8493529 [DOI] [PubMed] [Google Scholar]
- Du Y., Lo E., Ali S., and Khademhosseini A., Proc. Natl. Acad. Sci. U.S.A. 105, 9522 (2008). 10.1073/pnas.0801866105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith L. G., Acta Mater. 48, 263 (2000). 10.1016/S1359-6454(99)00299-2 [DOI] [Google Scholar]
- Mahoney M. J. and Saltzman W. M., Nat. Biotechnol. 19, 934 (2001). 10.1038/nbt1001-934 [DOI] [PubMed] [Google Scholar]
- Bonadio J., Smiley E., Patil P., and Goldstein S., Nat. Med. 5, 753 (1999). 10.1038/10473 [DOI] [PubMed] [Google Scholar]
- Oberpenning F., Meng J., Yoo J. J., and Atala A., Nat. Biotechnol. 17, 149 (1999). 10.1038/6146 [DOI] [PubMed] [Google Scholar]
- Scott R., Marquardt L., and Willits R. K., J. Biomed. Mater. Res. Part A 93A, 817 (2010). [DOI] [PubMed] [Google Scholar]
- Levenberg S., Rouwkema J., Macdonald M., Garfein E. S., Kohane D. S., Darland D. C., Marini R., van Blitterswijk C. A., Mulligan R. C., D’Amore P. A., and Langer R., Nat. Biotechnol. 23, 879 (2005). 10.1038/nbt1109 [DOI] [PubMed] [Google Scholar]
- Weinberg C. B. and Bell E., Science 231, 397 (1986). 10.1126/science.2934816 [DOI] [PubMed] [Google Scholar]
- Moon S., Hasan S. K., Song Y. S., Xu F., Keles H. O., Manzur F., Mikkilineni S., Hong J. W., Nagatomi J., Haeggstrom E., Khademhosseini A., and Demirci U., Tissue Eng. 16, 157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorovich N. E., Wijn J. R. D., Verbout A. J., Alblas J., and Dhert W. J. A., Tissue Eng. 14, 127 (2008). 10.1089/ten.2007.0158 [DOI] [PubMed] [Google Scholar]
- Xu F., Moon S. J., Emre A. E., Turali E. S., Song Y. S., Hacking S. A., Nagatomi J., and Demirci U., Biofab. 2, 014105 (2010). 10.1088/1758-5082/2/1/014105 [DOI] [PubMed] [Google Scholar]
- Norotte C., Marga F. S., Niklason L. E., and Forgacs G., Biomater. 30, 5910 (2009). 10.1016/j.biomaterials.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers R., Hübner U., Schmelzeisen R., and Mülhaupt R., Biomaterials 23, 4437 (2002). 10.1016/S0142-9612(02)00139-4 [DOI] [PubMed] [Google Scholar]
- Sachlos E., Reis N., Ainsley C., Derby B., and Czernuszka J. T., Biomaterials 24, 1487 (2003). 10.1016/S0142-9612(02)00528-8 [DOI] [PubMed] [Google Scholar]
- Bian W. and Bursac N., Biomaterials 30, 1401 (2009). 10.1016/j.biomaterials.2008.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Jin J., Gregory C., Hickman J. J., and Boland T., Biomaterials 26, 93 (2005). 10.1016/j.biomaterials.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Cui X. and Boland T., Biomaterials 30, 6221 (2009). 10.1016/j.biomaterials.2009.07.056 [DOI] [PubMed] [Google Scholar]
- Yang J., Yamato M., Sekine H., Sekiya S., Tsuda Y., Ohashi K., Shimizu T., and Okano T., Adv. Mater. (Weinheim, Ger.) 21, 3404 (2009). 10.1002/adma.200801990 [DOI] [PubMed] [Google Scholar]
- Asakawa N., Shimizu T., Tsuda Y. Sekiya S., Sasagawa T., Yamato M., Fukai F., and Okano T., Biomaterials 31, 3903 (2010). 10.1016/j.biomaterials.2010.01.105 [DOI] [PubMed] [Google Scholar]
- Hannachi I. E., Yamato M., and Okano T., Biofab. 1, 022002 (2009). 10.1088/1758-5082/1/2/022002 [DOI] [PubMed] [Google Scholar]
- Nishida K., Yamato M., Hayashida Y., Watanabe K., Yamamoto K., Adachi E., Nagai S., Kikuchi A., Maeda N., Watanabe H., Okano T., and Tano Y., N. Engl. J. Med. 351, 1187 (2004). 10.1056/NEJMoa040455 [DOI] [PubMed] [Google Scholar]
- Albertsson P.-A. and Tjerneld F., Methods Enzymol. 228, 3 (1994). 10.1016/0076-6879(94)28003-7 [DOI] [PubMed] [Google Scholar]
- Johansson G., Methods Enzymol. 228, 28 (1994). 10.1016/0076-6879(94)28005-3 [DOI] [PubMed] [Google Scholar]
- Walter H. and Larsson C., Methods Enzymol. 228, 42 (1994). 10.1016/0076-6879(94)28006-1 [DOI] [PubMed] [Google Scholar]
- Yamada M., Kasim V., Nakashima M., Edahiro J., and Seki M., Biotechnol. Bioeng. 88, 489 (2004). 10.1002/bit.20276 [DOI] [PubMed] [Google Scholar]
- Münchow G., Hardt S., Kutter J. P., and Drese S., Lab Chip 7, 98 (2007). 10.1039/b612669n [DOI] [PubMed] [Google Scholar]
- Tavana H., Jovic A., Mosadegh B., Lee Q. Y., Liu X., Luker K. E., Luker G. D., Weiss S. J., and Takayama S., Nature Mater. 8, 736 (2009). 10.1038/nmat2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsson P.-A., Partition of Cell Particles and Macromolecules (Wiley, New York, 1986). [Google Scholar]
- Gerson D. F., Biochim. Biophys. Acta 602, 269 (1980). 10.1016/0005-2736(80)90310-7 [DOI] [PubMed] [Google Scholar]
- Tavana H., Mosadegh B., and Takayama S., Adv. Mater. (Weinheim, Ger.) 22, 2628 (2010). 10.1002/adma.200904271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberger S., Seaman G. V. F., Sharp K. A., and Brooks D. E., J. Colloid Interface Sci. 99, 194 (1984). 10.1016/0021-9797(84)90100-0 [DOI] [Google Scholar]
- Koike N., Fukumura D., Gralla O., Au P., Schechner J. S., and Jain R. K., Nature (London) 428, 138 (2004). 10.1038/428138a [DOI] [PubMed] [Google Scholar]
- Passier R., van Laake L. W., and Mummery C. L., Nature (London) 453, 322 (2008). 10.1038/nature07040 [DOI] [PubMed] [Google Scholar]
- Tavana H., Mosadegh B., Zamankhan P., Grotberg J. B., and Takayama S. (2010) (unpublished). [DOI] [PMC free article] [PubMed]
- Gonçalves E., Debs R. J., and Heath T. D., Biophys. J. 86, 1554 (2004). 10.1016/S0006-3495(04)74223-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi T., Lee S., Choi W. S., Kim D., Kim T., Mitchell R. J., and Takayama S., Analyst 135, 2848 (2010). 10.1039/c0an00464b [DOI] [PubMed] [Google Scholar]
- Vander A., Sherman J., and Luciano D., Human Physiology (McGraw-Hill, New York, 2001), pp. 292–323. [Google Scholar]
- Lam M. T., Huang Y.-C., Birla R. K., and Takayama S., Biomaterials 30, 1150 (2009). 10.1016/j.biomaterials.2008.11.014 [DOI] [PubMed] [Google Scholar]
- Bianco P. and Robey P. G., Nature (London) 414, 118 (2001). 10.1038/35102181 [DOI] [PubMed] [Google Scholar]