Abstract
Microfluidic devices allow for precise control of the cellular and noncellular microenvironment at physiologically relevant length- and time-scales. These devices have been shown to mimic the complex in vivo microenvironment better than conventional in vitro assays, and allow real-time monitoring of homotypic or heterotypic cellular interactions. Microfluidic culture platforms enable new assay designs for culturing multiple different cell populations and∕or tissue specimens under controlled user-defined conditions. Applications include fundamental studies of cell population behaviors, high-throughput drug screening, and tissue engineering. In this review, we summarize recent developments in this field along with studies of heterotypic cell-cell interactions and tissue specimen culture in microfluidic devices from our own laboratory.
INTRODUCTION
Cell-cell interactions regulate organ function in both tissue homeostasis and disease by controlling basic cellular functions such as survival∕apoptosis, migration, proliferation, and differentiation. These interactions can be homotypic or heterotypic, and depending on the intercellular distance, can occur via multiple mechanisms,1 such as direct physical contact,2 diffusion of soluble paracrine factors,3 electrical signal transmission,4, 5 and transduction of mechanical cues6 through the surrounding extracellular matrix (ECM). During tissue regeneration, interactions between stem cells and other cell types within the “niche” environment are essential to preserve stem cell proliferative potential and multipotency.7 Cellular interactions among small groups of cells have also been shown to control oocyte polarization through cell-cell receptor signaling, during the developmental processes of body axes formation in Drosophila.8 In diseased tissues, cell-cell interactions may become perturbed and this is crucial not only for disease initiation (e.g., neurodegenerative disorders9), but also during later stages of disease progression (e.g., cancer metastasis10, 11). In vitro culture platforms designed to model, image, and measure these interactions under healthy and diseased conditions will not only advance our fundamental knowledge of these phenomena, but also enable drug discovery and high-throughput screening with time and cost benefits.
Cells sense and adapt to a complex microenvironment in vivo, which consists of cellular and noncellular components (ECM, biochemical, bioelectrical, and biophysical factors).12 Although in vivo studies have been carried out to monitor cell-cell interactions and cell signaling within their native microenvironments, these studies are limited by expensive experimental manipulations (e.g., animal models), lack of control over local experimental conditions, and complex imaging setups.13 Study of cell-cell interactions in vitro is advantageous due to more tightly controlled experimental conditions, higher experimental throughput, and lower costs. Among in vitro approaches, microfluidics has proven to be a powerful technology to study cell biology over the past few years,14, 15, 16, 17 offering significant improvements over traditional cellular assays such as user-defined assay design and precise control of the cellular and noncellular microenvironment. In particular, microfluidic cell culture systems have been designed with a wide variety of microchannel dimensions and geometries18 for generating soluble and insoluble gradients, on-chip valving and pumping through multilayer fabrication,19 and incorporation of membranes20 and three-dimensional (3D) hydrogels21 for increased functionality. The first generation of microfluidic cell culture devices enabled the study of cellular behaviors of a single cell type under well-defined biochemical gradients,18 shear stress,20 and cell-ECM interactions in three dimensions.22
Developing next generation microfluidic devices to investigate cell-cell and cell-ECM signaling is highly desirable due to the ability to integrate multiple cell types and controlled microenvironments at physiologically relevant length- and time-scales. New microfluidic assays for cell-cell signaling should enable user-defined control of the distance and type of interaction between different cell populations, along with spatiotemporal delivery of biochemical and biophysical stimuli individually21 or combined20, 23, 24 to different cell populations. Furthermore, integration with quantitative assays for measuring the intracellular (e.g., fluorescent calcium dyes25) and extracellular [e.g., ELISA (Ref. 26)] signals will be critical for obtaining dose-response curves and validating the results against traditional assays. Advancing the state-of-the-art microfluidic platforms for modeling, measuring, and imaging cell-cell signaling will be important to improve our understanding of the effects of homotypic and heterotypic cell-cell interactions on fundamental cellular behavior, tissue morphogenesis, and disease. Furthermore, it may provide tools for the development of “surrogate organ” platforms for preclinical drug27, 28 and toxicity testing, and clinical diagnostics assays of individual patient samples, as an early step toward personalized medicine.
In the first section of this review, we present a brief overview of conventional and micropatterning approaches used to study cell-cell interactions. In the second part, we review the use of microfluidic devices to recreate and monitor heterotypic cell-cell interactions and culture tissue specimens, with applications in diverse areas such as vascular biology, tumor biology, liver and neural tissue engineering, along with some recent data from our own laboratory. Finally, we summarize the possible applications of the microfluidic cell-cell interaction devices and discuss future research directions.
CONVENTIONAL AND MICROPATTERNING APPROACHES
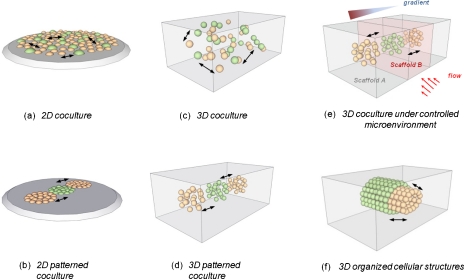
Cell-cell signaling is often studied using macroscale culture systems, such as Transwell assays, two-dimensional (2D) plating or 3D seeding of different cell populations, and addition of excised tissues into existing cultures.29 These assays are also termed as “macroscale culture assays” because of the inability to control cell plating and biomolecular transport at length-scales down to a cell diameter. The Transwell assay includes a porous filter to allow for biomolecular transport between two separate wells, where two different cell populations can be seeded. These assays can only facilitate paracrine interactions between two cell populations because of a stiff, nondegradable filter inhibiting direct contact, and have provided biological insight into the interactions of a large variety of cell types such as neuron-astrocytes,30 tumor-endothelial cells,31 fibroblasts-epithelial cells,32 and fibroblasts-tumor cells.33 Other macroscale coculture strategies have employed seeding different cell populations on a 2D substrate [Fig. 1a] (Ref. 33) or within a biologically degradable 3D ECM [Fig. 1c],34 and these platforms have enabled both paracrine signaling and direct physical interactions of cell populations.32, 35 Most macroscale coculture studies have focused on the interactions between two cell populations as mentioned above, although recently triculture systems have also been employed for the study of epithelial-fibroblast-adipocyte interactions during human breast development,34 hepatocyte-hepatic stellate cell-endothelial cell interactions recreating the liver microstructure,36 and myoblast-embryonic fibroblast-endothelial cells for skeletal muscle tissue vascularization.37
Figure 1.
Different configurations for studying paracrine and physical interactions between two different cell populations (green and orange). Conventional macroscale approaches without intercellular distance control: seeding in (a) 2D and (c) 3D. Micropatterning approaches with intercellular distance control: (b) 2D and (d) 3D. Microfluidic cell culture of different cell populations: (e) in 3D scaffolds under microenvironmental control and (f) for tissue specimen culture or forming organized cellular structures.
Although these macroscale coculture assays have provided significant insights into how interactions among different cell populations regulate cellular phenotype, a major limitation is the lack of control over cell-cell distance and the degree of cell-cell interaction because they are based on global seeding strategies. Furthermore, macroscale culture assays are usually not easily adaptable for live-cell imaging and do not offer the capability to create locally defined biochemical, bioelectrical, and biophysical stimuli conditions, such as growth-factor concentration gradients and shear stress imposed through blood or interstitial flow. In the following, we briefly discuss microfabrication approaches which can overcome these limitations by providing control over spatial distribution of cells in culture, resulting in better manipulation of homotypic and heterotypic cell-cell interactions.
Microfabrication technology has enabled the creation of patterned 2D surfaces [Fig. 1b] and 3D scaffolds [Fig. 1d] with improved spatial control over macroscale culture platforms. These systems, termed “microscale” culture platforms, have been utilized to explore the effects of homotypic and heterotypic cell-cell interactions on tissue functions with length-scales ranging from tens to hundreds of micrometers. For a more detailed discussion, the reader is referred to the papers by Bhatia et al.38 and Kaji et al.39 Hybrid coculture systems integrating both conventional and microscale approaches have also been developed. For example, Torisawa et al.40 developed coculture microarrays by using a polydimethylsiloxane (PDMS) stamp to create wells for seeding two different cell populations. In micropatterned systems, the arrays of different wells are usually fed by a single larger well, creating numerous challenges with respect to the cell seeding protocol and the establishment of different test conditions in different wells.
As shown in Fig. 1, the assay configuration and dimensionality influences intercellular distance, which will in turn determine the mode of cell-cell interaction (e.g., paracrine or physical contact) and the effective concentration of soluble factors sensed by both cell types.41 Compared with micropatterned systems, microfluidic devices incorporate flow control and allow for spatiotemporally user-defined delivery of cells within different channels or 3D scaffolds. Along with the capabilities to control and monitor biochemical and biophysical stimuli, microfluidic approaches offer a useful platform for developing new cell-cell interaction assays [Fig. 1e] and organized cellular structures∕tissue specimen culture [Fig. 1f] under controlled conditions, which more accurately mimic the complex in vivo microenvironment.
MICROFLUIDIC CELL CULTURE
Microfluidic cell culture: Advantages and challenges
The field of microfluidic cell culture has grown significantly over the past decade, and microfluidic platforms are becoming very attractive for developing new biological assays, ranging from single-molecule studies to tissue-level studies. Within the past three years, many new microfluidic platforms have been custom-designed to improve traditional cell biology assays. A number of advantages over traditional Petri dish culture make microfluidic platforms more attractive for cell biology: (i) user-defined assay design, (ii) control of cell patterning∕seeding and cellular microenvironmental factors at relevant length-scales, (iii) small reagent∕media volumes, (iv) excellent adaptability to imaging equipment, and (v) multiplexing capacities for applications in screening assays. One area of microfluidic cell culture that has received considerable interest is microfluidic gradient generators, which can expose cells to controlled biochemical stimuli. A wide variety of device designs have been developed to precisely control the spatiotemporal distribution of biochemical cues, soluble or patterned, on 2D surfaces to recreate biomolecular gradients with tunable concentration profiles and time-scales in 2D42, 43 or 3D environments.21, 44, 45, 46 In addition to biochemical factors, microfluidic devices integrating continuous flow conditions have been used to expose cells to biophysical stimuli, such as controlled levels of shear stress and interstitial flow through a 3D matrix.47
Despite these advantages, cell culture in microfluidic devices is associated with a number of new challenges that need to be taken into consideration. A majority of microfluidic devices are fabricated from PDMS which can have two potential side effects on cell behavior: adsorption of small hydrophobic molecules into the polymer and leaching of uncured PDMS oligomers into the channels. In a recent study, Regehr et al.48 showed that uncured PDMS oligomers could be detected on the cell membrane of cultured cells in the PDMS channels. Furthermore, these authors used quantitative ELISA assays to demonstrate that a small hydrophobic molecule estrogen (272 Da) partitioned into the surrounding PDMS, which could have resulted therefore in a decreased effective estrogen concentration available to the cells. Apart from the standard PDMS-based devices, the use of biocompatible materials such as alginate, collagen, chitosan, and other hydrogels has received considerable interest, as reviewed by Domachuk et al.49 Furthermore, frequent culture media change in microfluidic devices lacking continuous flow can become an issue, especially for large cell numbers or rapidly proliferating cells resulting in nutrients and oxygen depletion.16 Media depletion can be eliminated by establishing fluid flow in the device, on-chip by integrating a peristaltic pump, or by interfacing external flow devices.16 With respect to cell-cell interaction assays, care must be taken in selecting the appropriate media for cocultures, efficient cell seeding for optimizing cell viability, combining experimental results with computational models to predict and analyze transport of paracrine factors, and quantifying results with reproducible image∕data analysis.17, 50 Results from microfluidic assays must be carefully interpreted in comparison to macroscale culture data due to the inherently different transport phenomena in microfluidic and macroscale cultures.50, 51 In studies which require high-throughput screening of large cell numbers, the hydrodynamic environment needs to be considered as discussed by Yin et al.25, 52 In their studies, Yin et al. demonstrated that the shear stress levels need to be appropriately tuned in order to provide good quantitative agreement between intracellular calcium flux measurements in single cell microfluidic assays and traditional well-plate assays.
Microfluidic studies of cell-cell interactions
Most studies have addressed the effects of biochemical and biophysical microenvironments on a single cell type, referred here as “monoculture.” The flexibility of microfluidic device design enables multiple microchannels to be incorporated at user-defined physiologically relevant spacings, allowing for culture of multiple cell types. Furthermore, 3D ECM environments can be incorporated into these channels, while allowing for real-time imaging and control of the biochemical and biophysical factors through the 3D ECM. In Secs. 3B1, 3B2, 3B3, 3B4, 3C, we review research in microfluidic device design for studying heterotypic cell-cell interactions and tissue specimen culture, along with some relevant studies from our laboratory.
Mural-endothelial cell interactions
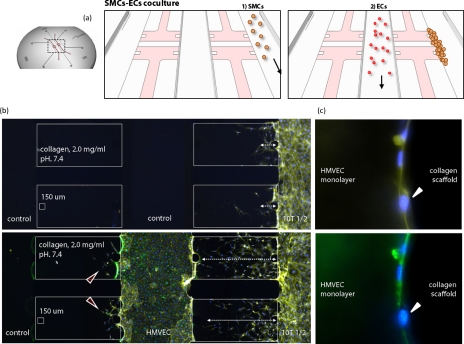
Mural cells refer to the pericytes and vascular smooth muscle cells (SMCs) which surround blood vessels in the formation of a normal, mature vasculature. Vessel maturation requires various angiogenic factors to assist endothelial cells (ECs) in forming cell-cell junctions and creating a vessel wall containing mural cells.53 Not only chemical factors (e.g., ANG-1, PDGF, TGF-β, S1P, PIGF) but also mechanical stimuli (e.g., interstitial flow, smooth muscle activation) play an important role in vessel maturation and remodeling. However, lack of concise knowledge of mural cells physical and paracrine interactions with ECs with respect to their matrix synthesis and deposition potential suggests the need for a mural cell-EC coculture model. Earlier coculture models between ECs and mural cells, which included direct coculture, coculture separated by extracellular matrix and spheroidal coculture, suggested that ECs modulate the proliferation of mural cells and that mural cells regulate EC quiescence.54, 55 A 3D microfluidic coculture model including ECs and SMCs was established by our group, enabling real-time monitoring and quantification of 3D cell migration under controlled biochemical and biophysical conditions.21 This coculture model enabled seeding of ECs and mural cells individually in different channels, allowing for the establishment of test and control conditions simultaneously in the same experiment [Fig. 2a]. The 3D collagen type I gel physically separates the initial populations of ECs and mural cells but allows for paracrine communication between the two cell types via diffusion. Cells are capable of migrating through the 3D gel and physically interacting with each other, while still maintaining their 3D morphology. Mack et al. used this platform to study the effect of endothelial Kruppel-like factor (KLF) expression on SMC migration, which has implications in the biomechanical remodeling of blood vessels in response to shear stress.21 Quantification of SMC migration demonstrated that KLF overexpression on ECs reduced the number of invading SMCs into the 3D gel compared to the control EC condition and SMC monoculture, demonstrating that paracrine signaling between EC∕SMC regulates SMC 3D invasion.21 We also investigated the effects of SMC∕EC interactions on EC monolayer stabilization and sprouting.21 Results [Figs. 2b, 2c] showed that (i) ECs formed a continuous monolayer in the microfluidic channel and could form vascular sprouts into the collagen type I gel, (ii) SMC actively migrated toward the endothelial monolayer [Fig. 2b], and (iii) SMC migration was enhanced by the presence of ECs. Interestingly, the presence of SMC precursor murine cells resulted in stabilized EC monolayers [Fig. 2c], while the control side of EC developed a small number of sprouts distally from the SMC. These two microfluidic studies suggest that bidirectional paracrine signaling controls not only SMC invasion but also EC phenotype (sprouting or stabilized). To further investigate paracrine signaling, an important extension of this work would be to study how SMC migration and EC sprouting depend on their intercellular distance, as defined by the width of the 3D collagen type I gel. Microfluidic systems integrating control over paracrine and direct physical interactions between ECs and mural cells under controlled microenvironmental (e.g., shear stress) conditions may also be utilized for improving scaffold vascularization, which is of critical importance in tissue engineering applications.56 Finally, systems need to be developed that allow for vascularization across the entire length of a 3D matrix so that the mural cell-EC interactions can be observed in a more natural setting.
Figure 2.
Microfluidic device for culturing mural cells and endothelial cells. (a) Coculture model including mural cells and EC (HMVEC). (b) SMCs are cultured in the right channel and ECs are cultured in the center channel. The presence of ECs increased 3D invasion of SMCs into the collagen type I ECM, while ECs become stabilized by the presence of SMC (Ref. 21). (c) Adhered SMCs on the HMVEC monolayer, shown by a white arrowhead. Nucleus (DAPI; blue) and actin filaments (rhodamine phalloidin; yellow). Green antibody staining indicates HMVECs.
Hepatocyte-endothelial cell interactions
Vascularization of tissue-engineered constructs is still a key challenge in the field of liver tissue engineering. Numerous 3D culture methods of hepatocytes have been previously reported,57, 58, 59, 60 which suggest that the supply of oxygen and nutrients by diffusion becomes limited as hepatocytes are cultured in 3D and the cultured hepatocyte tissues become thicker. Since the diffusion limit for oxygen is presumed to be ∼100 μm,56 cells in the tissues need vascular networks to transport oxygen, nutrients, and metabolites. A detailed understanding of the hepatocyte-endothelial cell interactions will have significant implications for developing improved liver tissue engineering approaches.
We recently developed a hepatocyte-EC coculture model in a microfluidic device to investigate the interactions between hepatocytes and ECs.24 The device has two microfluidic channels, one in which hepatocytes were seeded, and a second seeded with ECs, separated by a 3D gel region, which allowed for cell-cell communication and cell migration. First, hepatocytes were introduced into the top channel [Fig. 3a], and they formed 3D tissues when they were exposed to interstitial flow across the collagen gel between the top and the bottom channels. When ECs were introduced into the bottom channel to initiate coculture, they migrated into collagen gel and formed capillary networks extending toward the hepatocyte tissues in the top channel. This result shows that hepatocyte-EC interactions promote EC capillary morphogenesis, since EC monoculture under these conditions does not lead to sprout formation.24
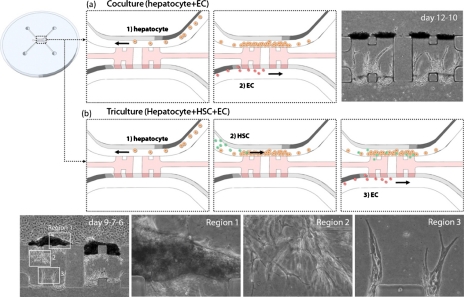
Figure 3.
Microfluidic device for integrating different liver cells. (a) Coculture model including hepatocytes and EC. Hepatocytes are cultured in the top channel in the figure and form 3D structures adjacent to the collagen type I gel between the top and the bottom channels. ECs are cultured in the bottom channel, and they migrate into the gel to form capillary networks. (b) Triculture model including hepatocytes, HSC, and EC. HSCs are introduced before adding ECs. Phase contrast images show that three cell types behave differently in the different regions 1–3.
The coculture model described above can be extended to a triculture model in which hepatocytes, hepatic stellate cells (HSCs), and ECs are sequentially introduced in a controlled fashion [Fig. 3b]. First, hepatocytes were seeded in one channel, and they formed 3D tissues similar to that observed in hepatocyte-EC coculture. When HSCs were also introduced into the same channel, they migrated across the hepatocyte tissues, and some cells migrated into the collagen type I gel. Finally, when ECs were seeded into the opposite channel to create the triculture model, they extended into the collagen matrix to form vascular sprouts which developed into capillary networks within the gel. Results showed that hepatocytes formed 3D tissues adjacent to collagen gel on day 9 [region 1, Fig. 3b], while HSCs extended cytoplasmic processes within the collagen gel and intervened between the hepatocyte tissues and ECs on day 7 [region 2, Fig. 3b], and ECs formed capillary networks extending toward HSCs and hepatocytes on day 6 [region 3, Fig. 3b]. Thus, it can be concluded that spatial and temporal control of cell seeding in a microfluidic device results in controlled cellular morphogenesis depending on each cell type. This triculture model could be further improved by the incorporation of 3D hydrogels which would provide for longer-term cultures, minimizing cell-mediated gel contraction, for investigating potential modulation of endothelial sprout formation by the hepatocytes and HSCs.
In a related study, a hepatocyte-HSC-EC triculture model was established using a conventional culture technique,36 in which sequential layers of hepatocytes, HSC, and EC were created with a microporous membrane. HSCs mediated the communication between hepatocytes and ECs in terms of EC capillary morphogenesis. However, the role of HSCs on EC capillaries is still unknown, and needs to be elucidated using methods such as the microfluidic triculture model described above. One of the factors that limit the detailed study of these heterotypical cell interactions is the difficulty in measuring the various factors being secreted from each cell type to mediate the signaling. Since the medium volumes are so small, standard biochemical assays are of limited value. The expression of factors with fluorescent labeling or scaffold-tethered probes would be advantageous, but the development of these can be an arduous process even for a small number of targets. Thus, identification of the critical signaling pathways remains a major hurdle.
Tumor-endothelial cell interactions
The interactions of tumor cells (TCs) with vascular ECs are critical for tumor progression as well as for later stages of cancer metastasis.61 These interactions include paracrine signaling of angiogenic factors secreted from the tumor cells, especially during the initial stages of tumor development, leading to the formation of new and leaky vessels.10 At later stages, once the TCs invade toward the ECs, contact interactions including receptor-mediated processes become important in heterotypic TC-EC adhesion and transendothelial migration.62, 63, 64 It is important to note that the TC-EC interactions occur within the tumor microenvironment, including other metastasis regulating factors such as cell-ECM contact, biophysical forces, and other host cells. Studying these processes in vivo is difficult, while conventional in vitro cultures do not mimic the in vivo conditions. Microfluidic devices allow for the integration of these critical microenvironmental factors and cell-cell interactions, offering new cellular assays which can provide insight into how cancer cell metastatic potential is influenced by these factors and their interactions.
Using controlled assembly of two complementary substrates,65 on which HeLa (cervical cancer) cells and human umbilical vein endothelial cells were seeded separately, Kaji et al.65 developed a coculture assay to study the effect of paracrine factors on cell migration of both types. In a recent study from the same group, the authors incorporated flow control to investigate the influence of paracrine signaling on cell behavior.23 The authors showed that flow from the ECs to the TCs had no effect on the net movement of either cell type, whereas by switching flow direction the ECs retracted and the TCs migrated into the empty space. Although the above work represents a simple platform to study paracrine interactions, the cells were seeded on a 2D substrate, which is a significant limitation for studying cell migration in the context of cancer invasion66 and cancer angiogenesis. By integrating flow-control and an endothelial monolayer on a microfluidic channel, Song et al.20 presented a novel system to model TC adhesion on ECs during blood-borne metastasis. Their system enables spatially restricted basal cytokine stimulation of the endothelial monolayer seeded on a porous membrane. The authors showed that either basal addition of CXCL12 or increasing shear stress from 0.5 to 2.5 dyn∕cm2 enhanced metastatic breast cancer cell adhesion compared to apical stimulation, demonstrating the synergistic effects of biochemical and biophysical activation on cell adhesion under flow.
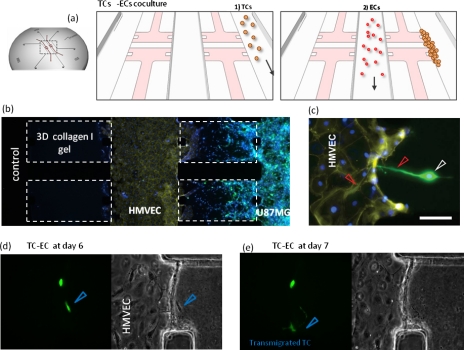
Our group recently developed microfluidic-based assays14, 21 to monitor TC-EC interactions in real-time, while enabling accurate control of biochemical and biophysical factors within a 3D matrix. An important advantage of this system [Fig. 4a] is its ability to recreate not only paracrine interactions [Fig. 4b] through soluble factor communication between both cell types across the ECM, but also physical contact interactions once the tumor cells adhere to the endothelial monolayer [Fig. 4c]. Furthermore, the system is capable of high-resolution time-lapse microscopy for visualizing the dynamics of TC-EC interactions [Figs. 4d, 4e]. The incorporated 3D collagen type I gel can be remodeled both by tumor and endothelial cells, allowing for the formation of new blood vessels and better mimicking in vivo conditions. One microenvironmental factor which was not considered in the coculture studies described by our group is fluid-flow mediated shear stress level on the EC, which could be integrated by establishment of continuous flow along the EC channel. This microfluidic system could also be used to study multiple steps of the metastatic cascade, while incorporating additional metastasis supporting cell types, such as fibroblasts or immune cells, and this represents an important direction for future work.
Figure 4.
Microfluidic device for recreating TC-EC interactions. (a) Schematic of the coculture model. (b) Brain cancer cells (U87MG) are cultured in the right channel and microvascular dermal endothelial cells are cultured in the center channel. U87MG invade into the collagen type I gel and approach the endothelial cells. (c) Protruding U87MG toward the endothelial monolayer. [(d) and (e)] Time-lapse images (fluorescent and phase-contrast) every 24 h of U87MG transmigrating into EC channel. Transmigrated cell marked with blue arrowhead. Nucleus (DAPI; blue) and actin filaments (rhodamine phalloidin; yellow). Green staining indicates GFP expressing brain cancer cells.
Other cell-cell interactions
Given the role of epithelial-stromal cell interactions in a wide array of diseases, a number of groups have developed microfluidic coculture systems for studying these interactions and understanding their biological behavior. These systems offer several major improvements over conventional Boyden-chamber based Transwell assays, such as real-time assessment of cell activity in 3D environment, and the ability to spatially pattern multiple cell types consistently. Bauer et al.67 used a simple array of 3D microchannels to coculture carcinoma cells and fibroblasts and compared it to traditional macroscale coculture. The authors observed that compared to monocultures of carcinoma cells, coculturing carcinoma cells with fibroblasts enhanced their proliferation and led to larger cell cluster formation. In another study of stromal-tumor cell interactions, Liu et al.68 showed that cancer associated fibroblasts (CAFs) promote gland adenoid cystic carcinoma cell invasion in 3D microfluidic coculture model. The authors found that the increase in cancer cell invasion was specific to the CAFs, since coculture with a normal fibroblast cell line did not promote cancer cell invasion. Furthermore, the authors showed that the increased invasion could be prohibited by using Matrix Metalloproteinase (MMP) inhibitors. In a similar study, Huang et al.69 developed a microfluidic device which allows cell seeding in multiple adjacent 3D gel channels to more accurately resemble the tumor microenvironment. When metastatic breast cancer cells and macrophages were cultured in 3D collagen and matrigel, respectively, they observed that macrophages invaded into the collagen gel only when cancer cells were present within the gel. Similarly, progressive macrophage invasion was observed over 12 days of culture, which suggests that the cancer cells may have been releasing signaling molecules that activated macrophage and invasion, consistent with other in vitro70 and in vivo71 studies. These studies shed light on the dynamic remodeling of ECM matrix by host cells and invading cells, the pattern of cell migration and the various roles of biochemical and biophysical stimuli within the complex tumor microenvironment.
During nervous system development, numerous guidance molecules play a critical role in directing the tip of growing neurites (growth cones) to their final destinations, resulting in the formation of a complex circuitry. Studying the precise mechanism by which these guidance molecules are presented to the growth cones (diffusion, binding to ECM, paracrine signaling) has been an intense area of research over the past few years and numerous in vitro systems have been employed for this purpose. However, traditional assays such as bath application of growth factors, substrate bound cues,72 tissue explant culture in vitro, pipette turning assay,73 and Dunn chamber assay are limited by their 2D cultures, difficulty in quantification, lack of spatiotemporal control on growth-factor delivery and sensing, and low reproducibility. We addressed these limitations by developing a microfluidic assay in which primary neurons could be seeded onto a three-dimensional collagen gel and cocultured in the presence of another cell type, transfected to release a guidance cue over the culture period.74 The device has a chamber for filling a 3D gel, three channels for supplying media and seeding cells, and a growth-factor gradient can be created across the 3D scaffold mimicking in vivo conditions. When mouse hippocampal neurons were cultured in these devices in the bottom channel, netrin-transfected fibroblasts in the right channel, and untransfected dermal fibroblasts in the left channel, so as to create a netrin gradient across the gel, it was observed that a significantly higher number of extending neurites were attracted toward netrin-transfected fibroblasts, compared to control cultures (with no fibroblasts) or cultures which received netrin exogenously instead. These results offer great insight into the functioning of guidance molecule sensing by growth cones in the presence of different microenvironmental conditions, and the utility of this device in drug screening and fundamental neurobiological assays. This device could be further improved by modifying the 3D gel chamber geometry to provide independent control of the chemoattractant absolute concentration and gradient strength for elucidating the growth-cone chemoattractant gradient sensitivity in a 3D matrix. Also, continuous flow of media and growth-factor gradients can be incorporated using a reservoir system, which eliminates the need for gradient reestablishment after regular intervals. The advantages of this device include the ease of handling and cell culture in 3D environment, rapid generation of pressure differentials and growth-factor gradients across a 3D gel on demand, provision to coculture up to three different cell types, live-cell imaging, precise quantification of axonal turning in 3D, high reproducibility of experimental observations, and ability to even culture tissue biopsies under controlled environments.
Periprosthetic osteolysis, a pathological condition after total hip replacement, occurs when the implant debris (usually polymeric or metallic) activates resident macrophages to release cytokines, which indirectly signal osteoblasts and osteoclasts, possibly resulting in bone resorption and decoupling of implant-tissue interface.75, 76 However, investigating these cell-cell interactions in cocultures without actual physical contact between the cell types has been a challenge with conventional techniques. Wei et al.77 developed a microfluidic system by laser direct-writing on polymethyl methacrylate (PMMA) substrates, which permits culturing two different cell types separately in upstream and downstream microwells separated by a microchannel that allows media exchange. An inherent advantage of this system is that it permits direct rapid media transfer with built-in linear concentration gradients from one cell type to the other, unlike conventional coculture systems which use transferred conditioned media.78 The authors found that inflammatory cytokines such as TNF-α and IL-1β were released by macrophages cultured in upstream wells upon exposure to 100 μg∕mL PMMA debris, which stimulated prostaglandin E2 release by osteoblasts cultured in downstream wells. This microfluidic coculture platform offers important advantages over Transwell assays, but it is limited to a 2D substrate and one-directional cell-cell communication under flow conditions because of the serial connections between upstream∕downstream wells, although bidirectional paracrine interactions could be achieved under static conditions.
Microfluidic devices are also well suited to recreate and study homotypic cell-cell interactions in physiologically relevant time- and length-scales, similar to heterotypic cell-cell interactions discussed above. For example, Klauke et al.79 developed a microfluidic platform to form cardiac myocyte doublets and measure the transmission of calcium waves through the intercellular junctions formed by these cells. Yin et al.80 described a microfluidic device based on dielectrophoretic forces to recreate interactions between a cell pair of endothelial cells and demonstrated that secreted matrix proteins and growth factors can have considerable guidance effects on cell motility.
Tissue specimen culture in microfluidic devices
The examples discussed above have amply demonstrated the utility of microfluidic systems for studying cocultures of dissociated cells to obtain valuable information about the cell-cell interactions. Although the cell-ECM interactions also play a critical role in maintaining physiological homeostasis (or in tumor progression), understanding these interactions in great detail has not been possible due to difficulties associated with culturing tissue slices. ECM regulates cell differentiation, migration, and proliferation, serves as a reservoir for growth factors, and provides a substrate for cell attachment and spreading.81 For example, ECM components of brain tissue (collagens, laminin, proteoglycans) play a major role in nerve cell guidance, axonal outgrowth, and synapse formations.82 Similarly, different subregions of the brain (e.g., prefrontal cortex, hippocampus, hypothalamus) communicate via localized neuronal pathways, thereby maintaining normal brain functionality. Elucidating these processes that guide cell-tissue and tissue-tissue interactions not only provides insight into the pathophysiology of diseases and disorders, but also unlocks the signaling pathways involved during embryogenesis. While in vivo experiments designed to understand the interactions between various brain tissue types (e.g., thalamocortical, cortex-hippocampal) have yielded valuable information,83, 84 they are limited by (i) global delivery of experimental conditions, usually through biomolecule injection in the blood circulation,85 (ii) lack of controlled manipulation of selective regions of tissues,86 and (iii) alterations in cellular properties such as growth rate, morphology, and metabolic activities87 due to experimental manipulation, which affects native signaling cascade between cells and surrounding tissues in that region. To overcome these limitations, there is a critical need to establish in vitro cell-tissue and tissue-tissue coculture systems, which closely mimic the complex in vivo microenvironment.88, 89
There are very few reports in the literature that detail the applications of microfluidic systems for culturing tissue sections in vitro. Queval et al.90 reported on the fabrication of a microfluidic system consisting of a perfusion chamber and a microfluidic probe for localized microperfusion of a small number of cells within a 400 μm slice of a mice hippocampal tissue. Advantages of this system over conventional perfusion chambers91 include its ease of assembly, adaptability to existing perfusion chambers, long-term low-shear stress on brain tissue slice and cells within, and simultaneous high-resolution fluorescence imaging. This system might permit local delivery of ions and drugs for selective manipulation of slice regions, and for studying neuronal interactions and remodeling within tissue using high-resolution imaging. Toward developing a high-throughput assay for personalized cancer therapy, Kim et al.92 developed and tested a microfluidic system for culturing thin sections of human breast cancer tissue or breast carcinoma cells. The device consisted of reservoirs for reagents, biomarker reservoirs, individual microvalves for each reservoir, and reaction channels to fit the breast tumor biopsy tissues. By employing quantitative histopathological techniques93 on these tissue sections, they showed that several cancer-related proteins (estrogen receptor, progesterone receptor, Ki-67, etc.) can be identified on a single tumor tissue slice, unlike conventional whole-section analysis. This device not only minimized the time and cost of analysis, reduced biopsy tissue size and reagents consumption, but also enhanced the sensitivity and repeatability of the assay.
Angiogenesis, the process of new blood vessel formation from existing blood vessels, plays an essential role in wound healing, fetal development, and tumor progression by supplying oxygen and vital nutrients.94 Previous in vitro microfluidic studies utilized monolayers of endothelial cells on a 3D gel and investigated the effect of angiogenic growth factors and matrix density on sprouting.22, 45, 95 Barkefors et al.96 utilized the recent development in microfluidic technology of creating stable growth-factor gradients, with the goal of investigating directional angiogenesis in 3D cultures of tissues and cells. The device design permits 3D culturing of differentiating stem cells or embryonic mouse kidney tissue section in the central chamber, with two fluidic channels on the sides across which a continuous stable growth-factor gradient can be established within 9 h. Results invariably showed that gradients of growth factors (VEGFA, FGF, VEGFC) promoted angiogenesis and invasive sprouting into matrix on the side of kidney tissue facing high levels of growth factor. Similarly, embryoid bodies of mouse embryonic stem cells exhibited significantly higher directional sprouting toward increasing concentration of VEGFA gradient in these devices. Thus, development of microfluidic devices such as the one described here offers a great advantage not only in developmental biology and molecular mechanism studies, but also in preclinical testing toward personalized medicine.
Recently, Gunther et al.97 developed a microfluidic device for loading a small artery tissue segment on-chip to probe its structure and function. This platform enabled long-term culture of the explants, on-chip imaging, immobilization, and spatiotemporal controlled delivery of drugs. The authors validated their method by demonstrating that the phenylephrine dose-response curves for mouse mesenteric arteries agreed very well with conventional pressure myograph measurements. To overcome current limitations of organotypic brain slice cultures,98 Berdichevsky et al.99 recently developed a microfluidic device to culture slices of rodent hippocampus over several weeks for electrophysiological studies. The device consisted of a designated miniwell for holding the tissue slice (∼350 μm thick), patterned microchannels for axonal guidance, and a large channel for medium circulation. When the tissue slice was placed on a glass cover slip coated with polylysine, firm tissue attachment and neurite outgrowth were observed within three days of culture, compared to uncoated surface cultures. Similarly, extensive microchannel-guided neurite extension (>500 μm) from hippocampal slices was observed after seven days of culture, which suggests broad applications in tissue engineering approaches designed for regenerating lost axonal pathways due to injury or disease. In a later study,100 the same group extended their previous platform to accommodate two designated chambers for holding brain slice tissues, and interconnected microchannels for facilitating axonal outgrowth. Results showed that cocultures of hippocampal slices or entorhinal cortex and hippocampal slices not only survived in these devices for up to four weeks, but also formed functional axon connectivity between them within 10–14 days of culture. Axonal outgrowth from these tissues was observed within three days, with the microchannels effectively isolating the tissue slices pharmacologically during two-channel perfusion with experimental media. The bidirectional axonal connectivity between tissue slices started synchronizing the two-slice network in the presence of GABA receptor antagonists after 16 days of culture in vitro, as evidenced from the recordings of individual external electrodes placed in these slices, thus signifying the applicability of this assay to study synapse formation in vitro under various pharmacological and physiological conditions.101
CONCLUSIONS AND FUTURE OUTLOOK
Considerable progress has been made in microfluidic cell culture just in the past few years, which has enabled the development of novel assays under well-defined biochemical and biophysical conditions. Studying homogeneous and heterogeneous cell populations in vitro has demonstrated the critical importance of cell-cell communication for basic cellular functions, such as survival, proliferation, migration, and differentiation under both normal and diseased conditions. By integrating heterotypic culture, microenvironmental control, and high-resolution real-time imaging, microfluidic technology is suited for creating new tissue engineering tools and novel cellular assays. The microfluidic assays described here have enabled the study of previously unexplored cellular interactions, and are already providing new insights into how biochemical and biophysical factors can regulate interactions between different cell populations. Progress in this area will have significant implications in drug discovery and artificial organ systems in vitro, and offers time and cost benefits for high-throughput drug screening.
Despite significant progress in the field of microfluidic cell culture, numerous challenges remain that need to be addressed in future studies. A vast majority of microfluidic platforms are PDMS-based, which has the advantage of rapid prototyping capabilities, but poses problems for surface chemistry modifications and mass production. In order for these assays to be compatible with standard pharmaceutical drug screening, hard plastic (polycarbonates, polystyrene) fabrication processes have to be adopted in the near future. Most of the current studies in microfluidic coculture platforms have focused on large cell populations (103–106 cells), and have only investigated a maximum of two different cell types. Integrating more than two cell types with biologically degradable 3D gels and controlled biochemical and biophysical conditions will allow for improved organotypic culture models, while also providing new insights into cellular interactions with heterotypic cells, soluble and insoluble factors. Scaling these platforms down to smaller cell populations will facilitate drug screening on patient samples and prove a valuable tool for personalized medicine. Finally, the development of microfluidic platforms incorporating real-time control over cell seeding and measurement of paracrine signals secretion will be important for understanding the dynamics of cellular interactions, an area which remains unexplored since most microfluidic studies have largely focused on measuring end-point cellular responses.
ACKNOWLEDGMENTS
The authors would like to acknowledge Yoojin Shin for his help with figure preparation, and Dr. Juliana Chan and Levi Wood for proofreading the manuscript. The research conducted by our group and presented in this review is supported by the National Science Foundation (Grant No. EFRI-0735997), IR&D Project No. DL-H-550151, Draper Laboratories, Inc., Singapore-MIT Alliance for Research and Technology (SMART), NIBIB (Grant No. EB003805), and Anonymous Foundations. S.C. is supported by a Korea University Grant and R.S. by KAKENHI (Grant No. 22680037).
References
- Hancock J. T., Cell Signalling, 2nd ed. (Oxford, New York, 2005). [Google Scholar]
- Nelson C. M. and Chen C. S., FEBS Lett. 514, 238 (2002). 10.1016/S0014-5793(02)02370-0 [DOI] [PubMed] [Google Scholar]
- Gnecchi M., Zhang Z., Ni A., and Dzau V. J., Circ. Res. 103, 1204 (2008). 10.1161/CIRCRESAHA.108.176826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. V. and Zukin R. S., Neuron 41, 495 (2004). 10.1016/S0896-6273(04)00043-1 [DOI] [PubMed] [Google Scholar]
- Chao D. L., Ma L., and Shen K., Nat. Rev. Neurosci. 10, 262 (2009). 10.1038/nrn2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Dooling L. J., and Asthagiri A. R., J. R. Soc., Interface 7, S341 (2010). 10.1098/rsif.2010.0066.focus [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond K., Deugnier M. A., Faraldo M. M., and Glukhova M. A., Curr. Opin. Cell Biol. 21, 623 (2009). 10.1016/j.ceb.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Poulton J. S. and Deng W. M., Dev. Biol. 311, 1 (2007). 10.1016/j.ydbio.2007.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva H., Polymenidou M., and Cleveland D. W., J. Cell Biol. 187, 761 (2009). 10.1083/jcb.200908164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. and Jain R. K., Nature (London) 407, 249 (2000). 10.1038/35025220 [DOI] [PubMed] [Google Scholar]
- Steeg P. S., Nat. Med. 12, 895 (2006). 10.1038/nm1469 [DOI] [PubMed] [Google Scholar]
- Joyce J. A. and Pollard J. W., Nat. Rev. Cancer 9, 239 (2009). 10.1038/nrc2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff J. B., Jones J. G., Condeelis J. S., and Segall J. E., Cancer Res. 60, 2504 (2000). [PubMed] [Google Scholar]
- Chung S., Sudo R., Vickerman V., Zervantonakis I. K., and Kamm R. D., Ann. Biomed. Eng. 38, 1164 (2010). 10.1007/s10439-010-9899-3 [DOI] [PubMed] [Google Scholar]
- Salieb-Beugelaar G. B., Simone G., Arora A., Philippi A., and Manz A., Anal. Chem. 82, 4848 (2010). 10.1021/ac1009707 [DOI] [PubMed] [Google Scholar]
- Young E. W. and Beebe D. J., Chem. Soc. Rev. 39, 1036 (2010). 10.1039/b909900j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paguirigan A. L. and Beebe D. J., BioEssays 30, 811 (2008). 10.1002/bies.20804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B. G. and Choo J., Electrophoresis 31, 3013 (2010). 10.1002/elps.201000137 [DOI] [PubMed] [Google Scholar]
- Gu W., Zhu X., Futai N., Cho B. S., and Takayama S., Proc. Natl. Acad. Sci. U.S.A. 101, 15861 (2004). 10.1073/pnas.0404353101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. W., Cavnar S. P., Walker A. C., Luker K. E., Gupta M., Tung Y. C., Luker G. D., and Takayama S., PLoS ONE 4, e5756 (2009). 10.1371/journal.pone.0005756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S., Sudo R., Mack P. J., Wan C. R., Vickerman V., and Kamm R. D., Lab Chip 9, 269 (2009). 10.1039/b807585a [DOI] [PubMed] [Google Scholar]
- Chung S., Sudo R., Zervantonakis I. K., Rimchala T., and Kamm R. D., Adv. Mater. (Weinheim, Ger.) 21, 4863 (2009). 10.1002/adma.200901727 [DOI] [PubMed] [Google Scholar]
- Kaji H., Yokoi T., Kawashima T., and Nishizawa M., Lab Chip 10, 2374 (2010). 10.1039/c004583g [DOI] [PubMed] [Google Scholar]
- Sudo R., Chung S., Zervantonakis I. K., Vickerman V., Toshimitsu Y., Griffith L. G., and Kamm R. D., FASEB J. 23, 2155 (2009). 10.1096/fj.08-122820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pattrick N., Zhang X., Klauke N., Cordingley H. C., Haswell S. J., and Cooper J. M., Anal. Chem. 80, 179 (2008). 10.1021/ac701958z [DOI] [PubMed] [Google Scholar]
- Young E. W. and Simmons C. A., Lab Chip 10, 143 (2010). 10.1039/b913390a [DOI] [PubMed] [Google Scholar]
- Kang L., Chung B. G., Langer R., and Khademhosseini A., Drug Discovery Today 13, 1 (2008). 10.1016/j.drudis.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M. H., Huang S. B., and Lee G. B., Lab Chip 10, 939 (2010). 10.1039/b921695b [DOI] [PubMed] [Google Scholar]
- Colgan S. P., Cell-Cell Interactions: Methods and Protocols (Humana, New Jersey, 2006), Vol. 341. [Google Scholar]
- Walsh E., Ueda Y., Nakanishi H., and Yoshida K., Neurosci. Lett. 138, 103 (1992). 10.1016/0304-3940(92)90482-M [DOI] [PubMed] [Google Scholar]
- Khodarev N. N., Yu J., Labay E., Darga T., Brown C. K., Mauceri H. J., Yassari R., Gupta N., and Weichselbaum R. R., J. Cell. Sci. 116, 1013 (2003). 10.1242/jcs.00281 [DOI] [PubMed] [Google Scholar]
- Krause S., Maffini M. V., Soto A. M., and Sonnenschein C., Tissue Engineering Part C: Methods 14, 261 (2008). 10.1089/ten.tec.2008.0030 [DOI] [PubMed] [Google Scholar]
- Stuelten C. H., DaCosta Byfield S., Arany P. R., Karpova T. S., Stetler-Stevenson W. G., and Roberts A. B., J. Cell. Sci. 118, 2143 (2005). 10.1242/jcs.02334 [DOI] [PubMed] [Google Scholar]
- Wang X., Sun L., Maffini M. V., Soto A., Sonnenschein C., and Kaplan D. L., Biomaterials 31, 3920 (2010). 10.1016/j.biomaterials.2010.01.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J. F., Harrington K., and Sahai E., Nat. Cell Biol. 9, 1392 (2007). 10.1038/ncb1658 [DOI] [PubMed] [Google Scholar]
- Kasuya J., Sudo R., Mitaka T., Ikeda M., and Tanishita K., Tissue Engineering Part A: Methods 17, 361 (2010). 10.1089/ten.tea.2010.0033 [DOI] [PubMed] [Google Scholar]
- Levenberg S., Rouwkema J., Macdonald M., Garfein E. S., Kohane D. S., Darland D. C., Marini R., van Blitterswijk C. A., Mulligan R. C., D’Amore P. A., and Langer R., Nat. Biotechnol. 23, 879 (2005). 10.1038/nbt1109 [DOI] [PubMed] [Google Scholar]
- Bhatia S. N., Balis U. J., Yarmush M. L., and Toner M., FASEB J. 13, 1883 (1999). [DOI] [PubMed] [Google Scholar]
- Kaji H., Camci-Unal G., Langer R., and Khademhosseini A., Biochim. Biophys. Acta 1810, 239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisawa Y.-s., Mosadegh B., Cavnar S. P., Ho M., and Takayama S., Tissue Engineering Part C: Methods, 17, 61 (2011). 10.1089/ten.tec.2010.0305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis K. and Palsson B. O., Proc. Natl. Acad. Sci. U.S.A. 94, 12258 (1997). 10.1073/pnas.94.23.12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi W., Rhee S. W., Lin F., Vahidi B., Chung B. G., and Jeon N. L., Biomed. Microdevices 9, 627 (2007). 10.1007/s10544-007-9051-9 [DOI] [PubMed] [Google Scholar]
- Wang S. J., Saadi W., Lin F., Minh-Canh Nguyen C., and Li Jeon N., Exp. Cell Res. 300, 180 (2004). 10.1016/j.yexcr.2004.06.030 [DOI] [PubMed] [Google Scholar]
- Haessler U., Kalinin Y., Swartz M. A., and Wu M., Biomed. Microdevices 11, 827 (2009). 10.1007/s10544-009-9299-3 [DOI] [PubMed] [Google Scholar]
- Vickerman V., Blundo J., Chung S., and Kamm R., Lab Chip 8, 1468 (2008). 10.1039/b802395f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zervantonakis I. K., Chung S., Sudo R., Zhang M., Charest J. L., and Kamm R. D., International Journal of Micro-Nano Scale Transport 1, 27 (2010). 10.1260/1759-3093.1.1.27 [DOI] [Google Scholar]
- Crane M. M., Chung K., Stirman J., and Lu H., Lab Chip 10, 1509 (2010). 10.1039/b927258e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regehr K. J., Domenech M., Koepsel J. T., Carver K. C., Ellison-Zelski S. J., Murphy W. L., Schuler L. A., Alarid E. T., and Beebe D. J., Lab Chip 9, 2132 (2009). 10.1039/b903043c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domachuk P., Tsioris K., Omenetto F. G., and Kaplan D. L., Adv. Mater. (Weinheim, Ger.) 22, 249 (2010). 10.1002/adma.200900821 [DOI] [PubMed] [Google Scholar]
- Paguirigan A. L. and Beebe D. J., Integr Biol (Camb) 1, 182 (2009). 10.1039/b814565b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Alexander C. M., and Beebe D. J., Lab Chip 7, 726 (2007). 10.1039/b618793e [DOI] [PubMed] [Google Scholar]
- Yin H., Zhang X., Pattrick N., Klauke N., Cordingley H. C., Haswell S. J., and Cooper J. M., Anal. Chem. 79, 7139 (2007). 10.1021/ac071146k [DOI] [PubMed] [Google Scholar]
- Carmeliet P., Nat. Med. 9, 653 (2003). 10.1038/nm0603-653 [DOI] [PubMed] [Google Scholar]
- Hirschi K. K., Rohovsky S. A., Beck L. H., Smith S. R., and D’Amore P. A., Circ. Res. 84, 298 (1999). [DOI] [PubMed] [Google Scholar]
- Korff T., Kimmina S., Martiny-Baron G., and Augustin H. G., FASEB J. 15, 447 (2001). 10.1096/fj.00-0139com [DOI] [PubMed] [Google Scholar]
- Jain R. K., Au P., Tam J., Duda D. G., and Fukumura D., Nat. Biotechnol. 23, 821 (2005). 10.1038/nbt0705-821 [DOI] [PubMed] [Google Scholar]
- Hwa A. J., Fry R. C., Sivaraman A., So P. T., Samson L. D., Stolz D. B., and Griffith L. G., FASEB J. 21, 2564 (2007). 10.1096/fj.06-7473com [DOI] [PubMed] [Google Scholar]
- Nahmias Y., Berthiaume F., and Yarmush M. L., Adv. Biochem. Eng./Biotechnol. 103, 309 (2007). 10.1007/10_029 [DOI] [PubMed] [Google Scholar]
- Sudo R., Mitaka T., Ikeda M., and Tanishita K., FASEB J. 19, 1695 (2005). [DOI] [PubMed] [Google Scholar]
- Toh Y. C., Zhang C., Zhang J., Khong Y. M., Chang S., Samper V. D., van Noort D., Hutmacher D. W., and Yu H., Lab Chip 7, 302 (2007). 10.1039/b614872g [DOI] [PubMed] [Google Scholar]
- Mareel M. and Leroy A., Physiol. Rev. 83, 337 (2003). [DOI] [PubMed] [Google Scholar]
- Condeelis J., Singer R. H., and Segall J. E., Annu. Rev. Cell Dev. Biol. 21, 695 (2005). 10.1146/annurev.cellbio.21.122303.120306 [DOI] [PubMed] [Google Scholar]
- Khuon S., Liang L., Dettman R. W., Sporn P. H., Wysolmerski R. B., and Chew T. L., J. Cell. Sci. 123, 431 (2010). 10.1242/jcs.053793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. H. and Dong C., Cell Mol Bioeng 2, 375 (2009). 10.1007/s12195-009-0067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji H., Yokoi T., Kawashima T., and Nishizawa M., Lab Chip 9, 427 (2009). 10.1039/b812510d [DOI] [PubMed] [Google Scholar]
- Friedl P. and Wolf K., Nat. Rev. Cancer 3, 362 (2003). 10.1038/nrc1075 [DOI] [PubMed] [Google Scholar]
- Bauer M., Su G., Beebe D. J., and Friedl A., Integr Biol (Camb) 2, 371 (2010). 10.1039/c0ib00001a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Lin B., and Qin J., Lab Chip 10, 1671 (2010). 10.1039/c000022a [DOI] [PubMed] [Google Scholar]
- Huang C. P., Lu J., Seon H., Lee A. P., Flanagan L. A., Kim H. Y., Putnam A. J., and Jeon N. L., Lab Chip 9, 1740 (2009). 10.1039/b818401a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S., Sahai E., Wyckoff J. B., Cammer M., Cox D., Pixley F. J., Stanley E. R., Segall J. E., and Condeelis J. S., Cancer Res. 65, 5278 (2005). 10.1158/0008-5472.CAN-04-1853 [DOI] [PubMed] [Google Scholar]
- Wyckoff J., Wang W., Lin E. Y., Wang Y., Pixley F., Stanley E. R., Graf T., Pollard J. W., Segall J., and Condeelis J., Cancer Res. 64, 7022 (2004). 10.1158/0008-5472.CAN-04-1449 [DOI] [PubMed] [Google Scholar]
- Dertinger S. K., Jiang X., Li Z., Murthy V. N., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. 99, 12542 (2002). 10.1073/pnas.192457199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujic Z., Giacomantonio C., Unni D., Rosoff W., and Goodhill G., J. Neurosci. Methods 170, 220 (2008). 10.1016/j.jneumeth.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Kothapalli C. R., Van Veen J. E., de Valence S., Chung S., Zervantonakis I. K., Gertler F. B., and Kamm R. D., Lab Chip 11, 497 (2010). 10.1039/c0lc00240b [DOI] [PubMed] [Google Scholar]
- Lavigne P., Shi Q., Jolicoeur F. C., Pelletier J. P., Martel-Pelletier J., and Fernandes J. C., Osteoarthritis Cartilage 10, 898 (2002). 10.1053/joca.2002.0846 [DOI] [PubMed] [Google Scholar]
- Schmidt C., Steinbach G., Decking R., Claes L. E., and Ignatius A. A., Biomaterials 24, 4191 (2003). 10.1016/S0142-9612(03)00317-X [DOI] [PubMed] [Google Scholar]
- Wei C.-W., Cheng J.-Y., and Young T.-H., Biomed. Microdevices 8, 65 (2006). 10.1007/s10544-006-6384-8 [DOI] [PubMed] [Google Scholar]
- Horowitz S. M., Rapuano B. P., Lane J. M., and Burstein A. H., Calcif. Tissue Int. 54, 320 (1994). 10.1007/BF00295957 [DOI] [PubMed] [Google Scholar]
- Klauke N., Smith G., and Cooper J. M., Lab Chip 7, 731 (2007). 10.1039/b706175g [DOI] [PubMed] [Google Scholar]
- Yin Z., Noren D., Wang C. J., Hang R., and Levchenko A., Mol. Syst. Biol. 4, 232 (2008). 10.1038/msb.2008.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R., and Yamada K. M., Nat. Rev. Mol. Cell Biol. 2, 793 (2001). 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- Rutka J. T., Apodaca G., Stern R., and Rosenblum N., J. Neurosurg. 69, 155 (1988). 10.3171/jns.1988.69.2.0155 [DOI] [PubMed] [Google Scholar]
- Bolz J., Novak N., Götz M., and Bonhoeffer T., Nature (London) 346, 359 (1990). 10.1038/346359a0 [DOI] [PubMed] [Google Scholar]
- Yamamoto N., Kurotani T., and Toyama K., Science 245, 192 (1989). 10.1126/science.2749258 [DOI] [PubMed] [Google Scholar]
- Uesaka N., Hirai S., Maruyama T., Ruthazer E. S., and Yamamoto N., J. Neurosci. 25, 1 (2005). 10.1523/JNEUROSCI.3855-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Turco D. and Deller T., Methods Mol. Biol. 399, 55 (2007). 10.1007/978-1-59745-504-6_5 [DOI] [PubMed] [Google Scholar]
- Weibel D. B., Garstecki P. G., and Whitesides G. M., Curr. Opin. Neurobiol. 15, 560 (2005). 10.1016/j.conb.2005.08.013 [DOI] [PubMed] [Google Scholar]
- Griffith L. G. and Swartz M. A., Nat. Rev. Mol. Cell Biol. 7, 211 (2006). 10.1038/nrm1858 [DOI] [PubMed] [Google Scholar]
- Zhang S., Nat. Biotechnol. 22, 151 (2004). 10.1038/nbt0204-151 [DOI] [PubMed] [Google Scholar]
- Queval A., Ghattamaneni N. R., Perrault C. M., Gill R., Mirzaei M., McKinney R. A., and Juncker D., Lab Chip 10, 326 (2010). 10.1039/b916669f [DOI] [PubMed] [Google Scholar]
- Ho C. L., Mou T. Y., Chiang P. S., Weng C. L., and Chow N. H., BioTechniques 38, 267 (2005). 10.2144/05382RR03 [DOI] [PubMed] [Google Scholar]
- Kim M. S., Kim T., Kong S.-Y., Kwon S., Bae C. Y., Choi J., Kim C. H., Lee E. S., and Park J.-K., PLoS ONE 5, e10441 (2010). 10.1371/journal.pone.0010441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M., Sørlie T., Eisen M. B., van de Rijn M., and Jeffrey S. S., Nature (London) 406, 747 (2000). 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- Folkman J. and Shing Y., J. Biol. Chem. 267, 10931 (1992). [PubMed] [Google Scholar]
- Shamloo A. and Heilshorn S. C., Lab Chip 10, 3061 (2010). 10.1039/c005069e [DOI] [PubMed] [Google Scholar]
- Barkefors I., Thorslund S., Nikolajeff F., and Kreuger J., Lab Chip 9, 529 (2009). 10.1039/b814691h [DOI] [PubMed] [Google Scholar]
- Günther A., Yasotharan S., Vagaon A., Lochovsky C., Pinto S., Yang J., Lau C., Voigtlaender-Bolz J., and Bolz S. S., Lab Chip 10, 2341 (2010). 10.1039/c004675b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahwiler B. H., J. Neurosci. Methods 4, 329 (1981). 10.1016/0165-0270(81)90003-0 [DOI] [PubMed] [Google Scholar]
- Berdichevsky Y., Sabolek H., Levine J. B., Staley K. J., and Yarmush M. L., J. Neurosci. Methods 178, 59 (2009). 10.1016/j.jneumeth.2008.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdichevsky Y., Staley K. J., and Yarmush M. L., Lab Chip 10, 999 (2010). 10.1039/b922365g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S. B. and Turrigiano G. G., Neuron 60, 477 (2008). 10.1016/j.neuron.2008.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]