Abstract
Susceptibility to scrapie is mainly controlled by point mutations at the PRNP locus. However, additional quantitative trait loci (QTL) have been identified across the genome including a region in OAR18. The gene which encodes the inducible form of the cytoplasmic Hsp90 chaperone (HSP90AA1) maps within this region and seems to be associated with the resistance/susceptibility to scrapie in sheep. Here, we have analyzed several polymorphisms which were previously described in the ovine HSP90AA1 5′ flanking region and in intron 10 in two naturally scrapie infected Romanov sheep populations. First, we have studied 58 ARQ/VRQ animals pertaining to the sire family where the QTL influencing scrapie incubation period in OAR18 was detected. We have found a significant association between polymorphisms localized at −660 and −528 in the HSP90AA1 5′ flanking region and the scrapie incubation period. These two polymorphisms have also been studied in a second sample constituted by 62 VRQ/VRQ sheep showing an extreme incubation period. Results are concordant with the first dataset. Finally, we have studied the HSP90AA1 expression in scrapie and control animals (N = 41) with different HSP90AA1 genotypes by real time PCR on blood samples. The HSP90AA1 expression rate was equivalent in CC−600AA−528 and CG−600AG−528 scrapie resistant animals (ARR/ARR) and was higher in their CC−600AA−528 than in their CG−600AG−528 scrapie susceptible counterparts (VRQ/VRQ). Our results support the hypothesis that the ovine HSP90AA1 gene acts as a modulator of scrapie susceptibility, contributing to the observed differences in the incubation period of scrapie infected animals with the same PRNP genotype.
Keywords: HSP90AA1, Scrapie, Sheep, QTL, Neurodegenerative disease
Introduction
Scrapie (SC) is a transmissible spongiform encephalopathy (TSE) of sheep and goat and has been endemic in sheep flocks for more than 200 years. TSEs occur as hereditary, sporadic, infectious, and iatrogenic diseases in various mammals including man (Prusiner 1998). Their etiology is still in dispute (DeArmond and Prusiner 1995; Deleault et al. 2003; Dickinson and Outram 1988; Lasmezas 2003; Weissmann 1991; Zeiler et al. 2003), but the prevailing theory assumes that a phosphatidilinositol-glycan anchored membrane protein called cellular prion protein (PrPC), a normal constituent of mammalian cells, plays a role in the pathogenesis of these fatal neurodegenerative diseases (Basler et al. 1986; Oesch et al. 1985). Through unknown posttranslational mechanisms, PrPC is converted into PrPSc, a protease resistant and less soluble isoform believed to be the causative principle (Prusiner 1982). However, the mechanism by which PrPSc causes neurodegeneration remains unclear.
Polymorphisms in the coding region of the prion protein gene (PRNP) are known to be strongly associated with different incubation times in humans, mice, and sheep. Susceptibility to SC is associated with polymorphisms in the amino acid sequence of the PrP protein. This PrP protein of 210 amino acids exists in at least 55 unique variants (Goldmann 2008). In particular, amino acid changes at positions V136A, R154H, and QH171R in the PRNP are strongly associated with the incidence of SC (Baylis et al. 2004). The influence of this polymorphism is well described in sheep with the identification of alleles associated with a higher (ARR/ARR) and lower (VRQ/VRQ) resistance. However, several quantitative trait loci (QTL) studies showed significant differences in the incubation period for this neurodegenerative disease in mice and sheep with the same PRNP genotype (Carlson et al. 1988; Diaz et al. 2005; Dickinson 1975; Westaway et al. 1987). Additionally, the existence of a polygenic variance involved in the modulation of the response to SC has been demonstrated (Diaz et al. 2005). For example, in mice, significant or suggestive evidence for QTLs on different chromosomes has been revealed (Lander and Green 1987; Lloyd et al. 2001, 2002; Manolakou et al. 2001; Moreno et al. 2003a, b, 2008; Stephenson et al. 2000). Within these regions, putative candidate genes have been proposed, and in a recent study, the influence of 20 candidate genes have been tested in vivo (Tamgüney et al. 2008).
Additionally, a QTL region has been identified in OAR 18 in sheep (Moreno et al. 2003a, 2008). Within this region, previous results indicate that the HSP90AA1 gene, which encodes the inducible form of the cytoplasmic Hsp90α chaperone, is a good functional and positional candidate gene acting as a modulator of the response to SC in sheep (Marcos-Carcavilla et al. 2008).
Studies in Saccharomyces cerevisiae have revealed that prions can exploit the molecular chaperone machinery of the cell in order to ensure stable propagation of the infectious, aggregation prone form. The disaggregation of yeast prion aggregates by molecular chaperones generates forms of the prion protein that can seed the protein polymerization that underlies the prion replication cycle (Jones and Tuite 2005).
We previously hypothesized that after SC infection, PrPSc starts to accumulate because its degradation is impaired (Marcos-Carcavilla et al. 2008). Therefore, Hsp90 could contribute to the correct folding of PrPSc instead of participating in other functions as the repression of heat shock factor 1 (HSF1) or the maintenance of the 26S proteasome and, thus, result in an increased expression of other heat shock proteins and in a minor proteasomal function. Additionally, as a consequence of the reduced proteasomal activity, the PrPC, which in normal circumstances is efficiently degraded by this machinery, would accumulate in the cytoplasm, increasing the amount of substrate susceptible to being transformed to the PrPSc isoform.
Based on previous results developed in a sample of 80 ARQ/ARQ Rasa Aragonesa breed sheep pertaining to six different flocks affected by natural SC, we proposed that mutations at −660 and −528 positions in the HSP90AA1 5′ flanking region and six linked SNPs at intron 10 (positions 40, 165, 178, 205, 220, and 239) could affect HSP90AA1 expression by altering possible transcription and splicing factors binding sites, respectively. Thus, these polymorphisms might affect the development of the disease (Marcos-Carcavilla et al. 2008).
The present work aims to corroborate the above mentioned results in a sample of Romanov sheep pertaining to the same naturally SC-infected flock and showing different incubation periods. Real-time PCR studies were also carried out to test the possible effect of the mutations at 5′ flaking region on the HSP90AA1 gene expression rate.
Materials and methods
Animal samples
The 161 animal samples analyzed in the present work have been maintained at the Langlade experimental INRA farm and belong to a flock of Romanov breed which has been naturally infected by SC in 1993 with an incidence close to 30% (Elsen et al. 1999). Details about general population and measured phenotype are shown by Moreno and coworkers (Moreno et al. 2008). Three datasets have been analyzed in the present work. Dataset 1 comprised a sire family (58 ARQ/VRQ animals) where a QTL affecting SC incubation period in OAR 18 (Ovis aries18) was detected (Moreno et al. 2008). Dataset 2 comprised 62 VRQ/VRQ sheep selected from the total Langlade farm population. Extreme values of SC incubation period corrected for litter size, type of rearing, PRNP genotype of the animal, PRNP genotype of the dam, and lambing season by years have been considered. These effects were selected as in Moreno and coworkers (submitted). Dataset 3 comprised of 41 live sheep (31 VRQ/VRQ 1.5 years old and 10 ARR/ARR 2.5 years old) which have been selected to study HSP90AA1 expression. No incubation period was available for these samples because the animals were still living at the moment of the sample collection. However, the VRQ/VRQ sheep showed the first clinical signs of SC. All animals used in these experiments were treated according to EEC recommendations for animal welfare and the supervision of the local INRA Ethics Committee.
Polymorphism genotyping
Previous work comparing SC versus control ARQ/ARQ Rasa Aragonesa sheep identified a total of 34 polymorphisms in the HSP90AA1 locus (Marcos-Carcavilla et al. 2008). Besides, the possible implication of several mutations at both HSP90AA1 5′ flanking region and intron 10 in Hsp90α synthesis have been proposed (Marcos-Carcavilla et al. 2008). Here, we have analyzed 22 polymorphisms located in these two regions. With this purpose, 60 to 120 ng of genomic DNA were amplified in a final volume of 50 μl containing 1 μM of each primer, 200 μM of dNTPs, 1.5 mM MgCl2, 10 μl of 5× buffer MgCl2 free (promega Go Taq Flexi), 2.5 U Taq polymerase (promega Go Taq Flexi) and 10% DMSO. The following PCR conditions were used for the amplification of the 5′ flanking region: denaturation at 94°C for 5 min, 40 amplification cycles of denaturation at 94°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min followed by a final 5 min extension at 72°C. In the case of intron 10, no DMSO was used. The following PCR conditions were used for its amplification: denaturation at 94°C for 5 min, 30 amplification cycles of denaturation at 94°C for 45 s, annealing at 58°C for 30 s, and extension at 72°C for 1 min followed by a final 5 min extension at 72°C. The resulting PCR fragments were purified with MultiScreen HTS™ PCR (Millipore) and sequenced with the primers indicated in Table 1. For dataset 1, the HSP90AA1 5′ flanking region and intron 10 have been sequenced. For datasets 2 and 3, only the HSP90AA1 5′ flanking region was sequenced.
Table 1.
List of primers used to genotype the polymorphisms studied in the ovine HSP90AA1 gene
| 5′ flanking region | Intron 10 | |
|---|---|---|
| Forward (5′–3′) | GCGCCCTAGACCCTCTAATC | ACAGGATCTACAGGATGATa |
| Reverse (5′–3′) | GAACCTTCCGGAAGAACCCa | ACTAGCTCTGCTTTAGTGC |
| Amplicon size | 520 bp | 700 bp |
aPrimers used for the amplicon sequencing
Association analysis
Dataset 1: The QTL family The genotyping of the ARQ/VRQ animals of the QTL family was carried out in order to analyze if any of the 22 previously described polymorphisms in the Spanish Rasa Aragonesa breed affecting both the HSP90AA1 5′ flanking region and intron 10 (Marcos-Carcavilla et al. 2008) were associated with SC incubation period. The HSP90AA1 genotype effects of each detected polymorphism were tested using the SAS GLM procedure (SAS 1990).
Dataset 2: Extreme animals for the incubation period The C/G and A/G mutations located at −660 and −528 positions in the 5′ flanking region, respectively, were analyzed. The HSP90AA1 genotype effects of each polymorphism were tested using the SAS GLM procedure in each PRNP genotype group (SAS 1990).
Expression analysis
Total RNA isolation and cDNA synthesis To determine the effect of SC infection on HSP90AA1 expression and the possible differences associated with the mutations at −660 and −528 locations, real-time PCR was performed on dataset 3. HSP90AA1 expression was compared among 5 ARR/ARR CC−660AA−528, 5 ARR/ARR CG−660AG−528, 15 VRQ/VRQ CC−660AA−528, and 16 VRQ/VRQ CG−660AG−528 animals.Total RNA was isolated from blood samples using the LeukoLock kit (Ambion) following manufacturer’s instructions. In order to avoid contamination with genomic DNA, samples were treated with 3 μl of RQ1 RNase-Free DNase (Promega) at 37°C for 30 min. The quality of the RNA was assessed based on the demonstration of distinct intact 28S and 18S ribosomal RNA bands. RNA concentration was determined with the nanodrop spectrophotometer. cDNA was synthesized from 0.5 μg of each RNA sample using random hexamers (1 μl) and oligo(dT) (1 μl) primers with the ImProm-II Reverse Transcriptase (Promega).
Real-time PCR Real-time PCRs were carried out on a Mastercycler ep realplex (Eppendorf) in a final volume of 25 μl containing 12.5 μl of Absolute Blue QPCR Green ROX Mix (ABgene), 2 μl of cDNA diluted 1/20, and 300 nM of each primer. After preheating the mix at 95°C for 15 min, 40 cycles of 95°C for 15 s and 60°C for 1 min were carried out. A melting curve consisting of 95°C for 15 s; 60°C for 15 s; and a ramp of 60°C to 95°C during 20 min was performed afterwards to confirm reaction specificity. All the reactions were run in triplicate. As plate controls, the same two animals were run in each plate. Primers used for gene expression analysis, their concentrations, and amplicon sizes are shown in Table 2.
Table 2.
List of primers and probes used to perform the real-time RT-PCR
Statistical analyses First, raw Ct data were pre-corrected by considering the average differences in Ct among plates for the two animals used as plate controls. Thus, pre-corrected Ct values, corresponding to the three replicates of each animal, were averaged in a single value (Ct med). Results were normalized to the GAPDH housekeeping gene using the delta–delta–Ct method (Livak and Schmittgen 2001). A general linear model analysis including PRNP genotypes (ARR/ARR or VRQ/VRQ) nested to HSP90AA1 genotypes (CC−660AA−528 or CG−660AG−528) as fixed effects was developed to assess the association (F ratio test) between HSP90AA1 genotypes and gene expression level. We have considered that the ARR/ARR animals were not infected in the Langlade farm even if they were in a contaminated environment. Indeed, we have not detected SC infection among the 90 ARR/ARR animals tested for SC status by histopathology and immunochemistry of neural and lymphoid tissues during 15 years of observation. Nevertheless, all the VRQ/VRQ adult sheep have surrendered to the infection. Thus, we have considered the ARR/ARR group as the control to compare the expression of the VRQ/VRQ SC-infected group.
Results
Association analysis
Dataset 1: In the QTL families Twenty-two SNPs previously described in the HSP90AA1 5′ flanking region and intron 10 in a Spanish Rasa Aragonesa breed (Marcos-Carcavilla et al. 2008) have been studied in the 58 sequenced ARQ/VRQ animals pertaining to the family where the QTL associated with the incubation period to SC was previously described in OAR18 (Marcos-Carcavilla et al. 2008; Moreno et al. 2003a). Among these 22 SNPs, 16 were polymorphic, but only six were represented in more than five animals for the smallest genotype frequency. These SNPs are located at −660, −528, −524, −468, −295, and 84 positions from the HSP90AA1 transcription start point. Among the six observed variations, only the two substitutions located at −660 and −528 positions in the 5′ flanking region, which appear generally linked, showed a significant association (p < 0.05) with the SC incubation period. Thus, the incubation period of animals bearing the CC−660AA−528 and CG−660AG−528 genotypes were 945 and 781 days, respectively (Table 3). The GG−660GG−528 genotype was not found within the sample used.
Table 3.
Significant associations between the polymorphisms located at −660 and −528 positions in the ovine HSP90AA1 5′ flankig region and SC resistance
| Trait | Population | HSP90AA1 haplotypea | Parameter (N) | Adjusted means (in days) | Estimated effect (SD) | P valueb |
|---|---|---|---|---|---|---|
| Incubation period | Data set-1: QTL ARQ/VRQ family (Moreno et al. 2008) | CC−660AA−528 | 23 | 945 | 0.57 | 0.04 |
| CG−660AG−528 | 35 | 781 | ||||
| Residuec of incubation period | Data set-2: VRQ/VRQ population | CC−660AA−528 | 38 | 705 | 0.48 | 0.06 |
| CG−660AG−528 | 24 | 695 |
aOnly haplotypes with more than five genotyped individuals were considered in the analyses
bLevel of significance of the HSP90AA1 haplotype
cEstimation of incubation time corrected for fixed effects (when the total SC Langlade population is considered (Moreno et al. unpublished))
SD standard deviation of the scrapie incubation time
Dataset 2: Extreme VRQ/VRQ animals for the SC incubation period The polymorphisms at −660 and −528 locations had a borderline significant effect (p = 0.06) in this analysis. These results are concordant with those obtained from dataset 1. Thus, the incubation period in animals bearing the CC−660AA−528 genotype was slightly borderline higher than in those bearing the CG−660AG−528 genotype (Table 3). The effect of HSP90AA1 polymorphisms in the VRQ/VRQ population is close to the effect observed for ARQ/VRQ. This effect expressed in days is not so high because the VRQ/VRQ population has a smaller standard deviation of incubation period than the ARQ/VRQ population (Table 3).
Expression analysis
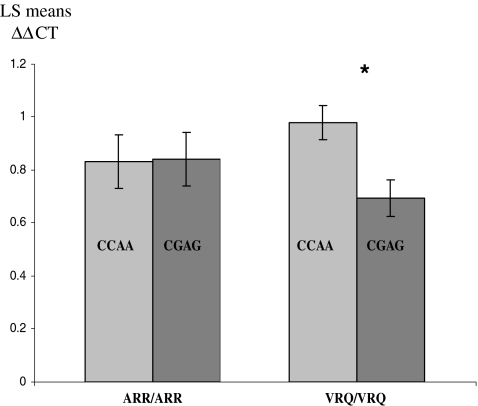
All the animals constituting the sample used in the expression study developed here were chosen from a permanent naturally SC-infected flock. Due to the sample availability, and considering that association analysis can be influenced by many different physiological factors, the possible effect of the mutations at −660 and −528 on HSP90AA1 expression was analyzed in ARR/ARR and VRQ/VRQ animals. ARR/ARR animals have been considered as SC-uninfected animals and VRQ/VRQ animals as SC-infected animals. Results from the GLM analysis showed a significant effect of the HSP90AA1 genotype within the different PRNP groups (p < 0.05). The smallest square means for the number of HSP90AA1 transcripts depending on both HSP90AA1 and PRNP genotypes were 0.83, 0.84, 0.98, and 0.69 in ARR/ARR CC−660AA−528, ARR/ARR CG−660AG−528, VRQ/VRQ CC−660AA−528, and VRQ/VRQ CG−660AG−528 animals, respectively. As it is shown in Fig. 1, comparison between genotypes showed no statistical differences in transcription rate between ARR/ARR animals with different HSP90AA1 genotypes (ARR/ARR CC−660AA−528 and ARR/ARR CG−660AG−528). Conversely, significant differences (p = 0.0236) were observed between VRQ/VRQ CC−660AA−528 and VRQ/VRQ CG−660AG−528 animals (p < 0.05). If HSP90AA1 expression in ARR/ARR animals is considered as the basal expression, VRQ/VRQ CC−660AA−528 animals presented an increase of 16% while VRQ/VRQ CG−660AG−528 animals presented a decrease of 26% in the amount of HSP90AA1 transcripts (Fig. 1). No differences between ARR/ARR and VRQ/VRQ samples were detected in our study if the HSP90AA1 genotype was neglected.
Fig. 1.
Expression results determined by real time PCR. The square means shown represent the relative amount of mRNA quantified in five ARR/ARR CC−660AA−528, five ARR/ARR CG−660AG−528, 15 VRQ/VRQ CC−660AA−528, and 16 VRQ/VRQ CG−660AG−528 animals maintained at permanent SC infected flock. *p < 0.05, statistically significant between VRQ/VRQ CC−660AA−528 and VRQ/VRQ CG−660AG−528 animals
Discussion
In the present work, several polymorphisms which were suggested to be involved in the modulation of the resistance/susceptibility to SC by affecting different putative regulatory elements in the HSP90AA1 gene were studied in order to test their possible implication in SC incubation period. We have found a significant association with the substitutions at positions −660 and −528 in the 5′ flanking region and the SC incubation period in our dataset 1, and these results are concordant with dataset 2.
The difference (in day unit) of effect size between VRQ/VRQ and ARQ/VRQ populations is due to a very different standard deviation of SC incubation time trait in the populations. Using the same scrapie resistance trait, Moreno et al. (submitted) observed also a difference of standard deviation between VRQ/VRQ and ARQ/VRQ populations of Langlade Farm.
Based on these data, we can conclude that polymorphisms at HSP90AA1 locus have a significant effect, with an ARQ/VRQ or VRQ/VRQ PRNP genotype. They appeared to be significant at 10% level in two independent populations having different susceptible PRNP genotypes. Consequently, the effect HSP90AA1 polymorphisms could explain part of the heritability for the SC incubation period not associated with the PRNP genotype (Diaz et al. 2005; Moreno et al. submitted). In addition, a significant effect of HSP90AA1 was previously found in a SC case control study where selected animals had ARQ/ARQ genotype and came from Rasa Aragonesa breed (Marcos-Carcavilla et al. 2008). CC−660AA−528 genotype was also the most resistant genotype. Moreover, the HSP90AA1 gene was located in the confidential interval of QTL of chromosome 18 found in different subset of Langlade Farm sheep: a ARQ/VRQ sire family, the global VRQ/VRQ population, and the global ARQ/ (VRQ or ARQ) population.
On the other hand, expression results revealed no differences in the HSP90AA1 transcription rate between CC−600AA−528 and CG−600AG−528 animals presenting the SC-resistant PRNP genotype (ARR/ARR). Conversely, HSP90AA1 expression was higher in VRQ/VRQ CC−600AA−528 and lower in VRQ/VRQ CG−600AG−528 SC animals (Fig. 1).
Interestingly, in a parallel study (Marcos-Carcavilla et al. 2009) performed on ARR/ARR Manchega sheep breed, it has been observed that after several days of high temperatures, HSP90AA1 expression increased significantly (p < 0.05) in animals bearing the CC−660AA−528 genotype. Conversely, no significant differences were detected in their CG−660AG−528 counterparts. No changes were observed in any of these genotypes in the same group of animals when the samples were collected in spring, when daily maximum temperatures were still moderate. These data support the hypothesis that mutations at −660 and −528 affect HSP90AA1 inducible expression under stress conditions. Thus, animals induce expression of this chaperone when they are exposed to a stressor. In the case of ARR/ARR animals, high temperatures were enough to trigger a response while no changes were observed as consequence of prion infection. This fact could indicate that prion infection, or at least the SC strain of Langlade, is not a source of stress in ARR/ARR animals. These results, together with the data presented here, lead to the hypothesis that the polymorphisms at −660 and −528 in the HSP90AA1 5′ flanking region modify the inducible expression of this gene.
It is necessary to consider that most changes in transcript abundance, whether they reflect effects of mRNA and protein stability or adaptive alterations in protein concentrations, are likely to be important. In this regard, stressful stimuli, as prion deposition or temperature changes, might trigger different signaling pathways which will lead to the activation of different transcription factors which will exert different effects depending on both the target gene and additional transcription factors they interact with. In the context of the present study, the SNPs at −660 and −528 positions could be altering binding sites for zinc-finger protein (ZBP-89) and Sis-inducible factor (SIF) transcription factors, respectively. ZBP-89 is ubiquitously expressed and possesses multiple functions, including transcriptional regulation of a variety of genes (Bai et al. 2002; Bai and Merchant 2003), cell growth (Bai and Merchant 2003), cell growth arrest (Remington et al. 1997), and cell death (Bai and Merchant 2001). The (C)(C)CCCCCCC/gA sequence, located between −667 and −660 positions, is identical with a ZBP-89 binding site described in the 5′ region of the vimentin gene (GGACCCCCCCC) by Zhang and coworkers (2003). ZBP-89 acts both as transcriptional activator and repressor (Yamada et al. 2001).
On the other hand, the SNP at −528 affects a putative SIF binding site (CCCG/aTM). The sequence of this putative regulatory element is very similar to the core sequence of the SIF binding site described by Wagner and coworkers (1990) in the c-fos promoter (CCCGTC). SIF complexes are dimerized forms of signal transducers and activators of transcription factors which, after being activated by different ligands, translocate into the nucleus to direct the transcription of specific target genes (Wang et al. 2006).
Different HSP90AA1 transcription efficiency might have important effects on SC incubation period. Prion accumulation might sequester cytoplasmic Hsp90 (Hsp90 α and Hsp90 β) leading to the activation of the HSF1 and, hence, the activation of the stress response and compromising the proteasomal activity. Several transcription factors will participate in the restoration of Hsp90 normal levels. Thus, mutations at these sites might yield different rates of ovine HSP90AA1 transcription due to positive or negative interactions between these factors. This fact might have consequences in SC development by affecting prion aggregation and/or degradation.
Results obtained in the present work support the hypothesis that the ovine HSP90AA1 may act as a modulator of scrapie susceptibility. We have demonstrated that the polymorphisms at −660 and −528 in the HSP90AA1 5′ flanking region modify the inducible expression of this gene. The G−660G−528 allele is associated with higher susceptibility to SC, possibly due to a reduced efficiency in HSP90AA1 transcription. Although additional experiments need to be carried out, it is important to specify that our results have been obtained from two independent populations with different PRNP genotypes.
Acknowledgments
We thank Langlade farm for kindly providing animal samples. This work was supported by the RTA2006-00104 INIA project.
References
- Bai L, Logsdon C, Merchant JL. Regulation of epithelial cell growth by ZBP-89: potential relevance in pancreatic cancer. International Journal of Gastrointestinal Cancer. 2002;31:79–88. doi: 10.1385/IJGC:31:1-3:79. [DOI] [PubMed] [Google Scholar]
- Bai L, Merchant JL. ZBP-89 promotes growth arrest through stabilization of p53. Mol Cell Biol. 2001;21:4670–4683. doi: 10.1128/MCB.21.14.4670-4683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Merchant JL. Transcription factor ZBP-89 is required for STAT1 constitutive expression. Nucl Acids Res. 2003;31:7264–7270. doi: 10.1093/nar/gkg929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler K, Oesch B, Scott M, Westaway D, Walchli M, Groth DF, McKinley MP, Prusiner SB, Weissmann C. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell. 1986;46:417–428. doi: 10.1016/0092-8674(86)90662-8. [DOI] [PubMed] [Google Scholar]
- Baylis M, Chihota C, Stevenson E, Goldmann W, Smith A, Sivam K, Tongue S, Gravenor MB. Risk of scrapie in British sheep of different prion protein genotype. J Gen Virol. 2004;85:2735–2740. doi: 10.1099/vir.0.79876-0. [DOI] [PubMed] [Google Scholar]
- Carlson GA, Goodman PA, Lovett M, Taylor BA, Marshall ST, Peterson-Torchia M, Westaway D, Prusiner SB. Genetics and polymorphism of the mouse prion gene complex: control of scrapie incubation time. Mol Cell Biol. 1988;8:5528–5540. doi: 10.1128/mcb.8.12.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeArmond SJ, Prusiner SB. Etiology and pathogenesis of prion diseases. Am J Pathol. 1995;146:785–811. [PMC free article] [PubMed] [Google Scholar]
- Deleault NR, Lucassen RW, Supattapone S. RNA molecules stimulate prion protein conversion. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- Diaz C, Vitezica ZG, Rupp R, Andreoletti O, Elsen JM. Polygenic variation and transmission factors involved in the resistance/susceptibility to scrapie in a Romanov flock. J Gen Virol. 2005;86:849–857. doi: 10.1099/vir.0.80412-0. [DOI] [PubMed] [Google Scholar]
- Dickinson AG. Host-pathogen interactions in scrapie. Genetics. 1975;79(Suppl):387–395. [PubMed] [Google Scholar]
- Dickinson AG, Outram GW. Genetic aspects of unconventional virus infections: the basis of the virino hypothesis. Ciba Found Symp. 1988;135:63–83. doi: 10.1002/9780470513613.ch5. [DOI] [PubMed] [Google Scholar]
- Elsen JM, Amigues Y, Schelcher F, Ducrocq V, Andreoletti O, Eychenne F, Khang JV, Poivey JP, Lantier F, Laplanche JL. Genetic susceptibility and transmission factors in scrapie: detailed analysis of an epidemic in a closed flock of Romanov. Arch Virol. 1999;144:431–445. doi: 10.1007/s007050050516. [DOI] [PubMed] [Google Scholar]
- Goldmann W. PrP genetics in ruminant transmissible spongiform encephalopathies. Vet Res. 2008;39:30. doi: 10.1051/vetres:2008010. [DOI] [PubMed] [Google Scholar]
- Jones GW, Tuite MF. Chaperoning prions: the cellular machinery for propagating an infectious protein? Bioessays. 2005;27:823–832. doi: 10.1002/bies.20267. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. PNAS. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasmezas CI. The transmissible spongiform encephalopathies. Rev Sci Tech Off Int Epizoot. 2003;22:23–36. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lyahyai J, Bolea R, Serrano C, Monleon E, Moreno C, Osta R, Zaragoza P, Badiola JJ, Martin-Burriel I. Correlation between Bax overexpression and prion deposition in medulla oblongata from natural scrapie without evidence of apoptosis. Acta Neuropathol. 2006;112:451–460. doi: 10.1007/s00401-006-0094-4. [DOI] [PubMed] [Google Scholar]
- Lloyd SE, Onwuazor ON, Beck JA, Mallinson G, Farrall M, Targonski P, Collinge J, Fisher EM. Identification of multiple quantitative trait loci linked to prion disease incubation period in mice. Proc Natl Acad Sci USA. 2001;98:6279–6283. doi: 10.1073/pnas.101130398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SE, Uphill JB, Targonski PV, Fisher EM, Collinge J. Identification of genetic loci affecting mouse-adapted bovine spongiform encephalopathy incubation time in mice. Neurogenetics. 2002;4:77–81. doi: 10.1007/s10048-002-0133-9. [DOI] [PubMed] [Google Scholar]
- Manolakou K, Beaton J, McConnell I, Farquar C, Manson J, Hastie ND, Bruce M, Jackson IJ. Genetic and environmental factors modify bovine spongiform encephalopathy incubation period in mice. PNAS. 2001;98:7402–7407. doi: 10.1073/pnas.121172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, Calvo JH, González C, Moazami-Goudarzi K, Laurent P, Bertaud M, Hayes H, Beattie AE, Serrano C, Lyahyai J, Martin-Burriel I, Serrano M. Structural and functional analysis of the HSP90AA1 gene: distribution of polymorphisms among sheep with different responses to scrapie. Cell Stress Chaperones. 2008;13:19–29. doi: 10.1007/s12192-007-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos-Carcavilla A, Mutikainen M, González C, Calvo JH, Kantanen J, Sanz A, Marzanov NS, Pérez-Guzmán MD, Serrano M (2009) A SNP in the HSP90AA1 gene 5′ flanking region is associated with the adaptation to differential thermal conditions in the ovine species. Cell Stress & Chaperones, doi:10.1007/s12192-009-0123-z [DOI] [PMC free article] [PubMed]
- Moreno CR, Cosseddu GM, Andreoletti IO, Schibler L, Roig A, Moazami-Goudarzi K, Echeynne F, Lajous D, Schelcher F, Cribiu EP, Vaiman D, Elsen JM (2003a) Identification of quantitative trait loci (QTL) modulating prion incubation period in sheep. (Identification de QTL affectant la durée d'incubation de la tremblante chez les ovins.). Toulouse: Proceedings of the International Workshop on Major Genes and QTL in Sheep and Goat
- Moreno CR, Lantier F, Lantier I, Sarradin P, Elsen JM. Detection of new quantitative trait Loci for susceptibility to transmissible spongiform encephalopathies in mice. Genetics. 2003;165:2085–2091. doi: 10.1093/genetics/165.4.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno CR, Cosseddu GM, Schibler L, Roig A, Moazami-Goudarzi K, Andreoletti O, Eychenne F, Lajous D, Schelcher F, Cribiu EP, Laurent P, Vaiman D, Elsen JM. Identification of new quantitative trait Loci (other than the PRNP gene) modulating the scrapie incubation period in sheep. Genetics. 2008;179:723–726. doi: 10.1534/genetics.108.088146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesch B, Westaway D, Walchli M, McKinley MP, Kent SB, Aebersold R, Barry RA, Tempst P, Teplow DB, Hood LE, Prusiner SB, Weissmann C. A cellular gene encodes scrapie PrP 27–30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. PNAS. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington MC, Tarle SA, Simon B, Merchant JL. ZBP-89, a Kruppel-type zinc finger protein, inhibits cell proliferation. Biochem Biophys Res Commun. 1997;237:230–234. doi: 10.1006/bbrc.1997.7119. [DOI] [PubMed] [Google Scholar]
- SAS/STAT user’s guideline. Cary: SAS; 1990. [Google Scholar]
- Stephenson DA, Chiotti K, Ebeling C, Groth D, DeArmond SJ, Prusiner SB, Carlson GA. Quantitative trait loci affecting prion incubation time in mice. Genomics. 2000;69:47–53. doi: 10.1006/geno.2000.6320. [DOI] [PubMed] [Google Scholar]
- Tamgüney G, Giles K, Glidden DV, Lessard P, Wille H, Tremblay P, Groth DF, Yehiely F, Korth C, Moore RC, Tatzelt J, Rubinstein E, Boucheix C, Yang X, Stanley P, Lisanti MP, Dwek RA, Rudd PM, Moskovitz J, Epstein CJ, Cruz TD, Kuziel WA, Maeda N, Sap J, Ashe KH, Carlson GA, Tesseur I, Wyss-Coray T, Mucke L, Weisgraber KH, Mahley RW, Cohen FE, Prusiner SB. Genes contributing to prion pathogenesis. J Gen Virol. 2008;89:1777–1788. doi: 10.1099/vir.0.2008/001255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner BJ, Hayes TE, Hoban CJ, Cochran BH. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. Embo J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XD, Chen XM, Wang JZ, Hong Q, Feng Z, Fu B, Zhou F, Wang FY, Fan DM. Signal transducers and activators of transcription 3 mediates up-regulation of angiotensin II-induced tissue inhibitor of metalloproteinase-1 expression in cultured human senescent fibroblasts. Chin Med J. 2006;119:1094–1102. [PubMed] [Google Scholar]
- Weissmann C. A ‘unified theory’ of prion propagation. Nature. 1991;352:679–683. doi: 10.1038/352679a0. [DOI] [PubMed] [Google Scholar]
- Westaway D, Goodman PA, Mirenda CA, McKinley MP, Carlson GA, Prusiner SB. Distinct prion proteins in short and long scrapie incubation period mice. Cell. 1987;51:651–662. doi: 10.1016/0092-8674(87)90134-6. [DOI] [PubMed] [Google Scholar]
- Yamada A, Takaki S, Hayashi F, Georgopoulos K, Perlmutter RM, Takatsu K. Identification and characterization of a transcriptional regulator for the lck proximal promoter. J Biol Chem. 2001;276:18082–18089. doi: 10.1074/jbc.M008387200. [DOI] [PubMed] [Google Scholar]
- Zeiler B, Adler V, Kryukov V, Grossman A. Concentration and removal of prion proteins from biological solutions. Biotechnol Appl Biochem. 2003;37:173–182. doi: 10.1042/BA20020087. [DOI] [PubMed] [Google Scholar]
- Zhang X, Diab IH, Zehner ZE. ZBP-89 represses vimentin gene transcription by interacting with the transcriptional activator, Sp1. Nucl Acids Res. 2003;31:2900–2914. doi: 10.1093/nar/gkg380. [DOI] [PMC free article] [PubMed] [Google Scholar]