Abstract
DNA repair is required to maintain genome stability in stem cells and early embryos. At critical junctures, oxidative damage to DNA requires the base excision repair (BER) pathway. Since early zebrafish embryos lack the major polymerase in BER, DNA polymerase ß, repair proceeds via replicative polymerases, even though there is ample polb mRNA. Here, we report that Polb protein fails to appear at the appropriate time in development when AP endonuclease 1 (Apex), the upstream protein in BER, is knocked down. Because polb contains a Creb1 binding site, we examined whether knockdown of Apex affects creb1. Apex knockdown results in loss of Creb1 and Creb complex members but not Creb1 phosphorylation. This effect is independent of p53. Although both apex and creb1 mRNA rescue Creb1 and Polb after Apex knockdown, Apex is not a co-activator of creb1 transcription. This observation has broad significance, as similar results occur when Apex is inhibited in B cells from apex+/− mice. These results describe a novel regulatory circuit involving Apex, Creb1 and Polb and provide a mechanism for lethality of Apex loss in higher eukaryotes.
INTRODUCTION
Oxidative damage to DNA requiring the base excision repair (BER) pathway is important in normal embryological development and cell differentiation (1–4). Two critical enzymes in the pathway are AP endonuclease 1 (Apex) and DNA polymerase ß (Polb). The former makes a single nick 5′ to an abasic site in double-stranded DNA (5), while in the next step the latter inserts the replacement nucleotide(s) (5). Although the two proteins do not form a stable complex, they associate with one another during the repair process (6,7). Mouse embryos die shortly after gastrulation when Apex is knocked out (8), while zebrafish embryos develop with brain, heart and neuron abnormalities that result in termination of development later, after the primordia for all organs have formed (9). To date, no viable cultured line has been recovered from knockout mouse embryos. Furthermore, inhibition of Apex in primary cultures of Apex deficient murine B cells results in failure of class switching during lymphocyte maturation (10).
Because BER is so important in embryogenesis, we examined the major polymerase involved in the repair pathway. Although mRNA for Polb is present throughout embryogenesis, zebrafish embryos lack Polb protein before gastrulation (11). This suggests that there may be an important mechanism regulating transcription and/or translation. We asked here how this process might occur. Our results show that Apex regulates transcription of Creb1 and its binding partners, which then regulates Polb. These studies are the first to indicate a role for Apex in regulating downstream participants in BER and the first to identify a regulator for protein levels of the important transcription factor Creb1 and participants in the Creb complex.
MATERIALS AND METHODS
Mouse and fish husbandry and breeding
Wild-type zebrafish, purchased from Aquatic Tropicals (Plant City, FL), were maintained and bred as described (9,11). Homozygous p53 mutant zebrafish adults (p53M214K/ M214K) were the kind gift of Dr Thomas Look (12). Wild type C57BL/6 mice were housed in the IACUC-approved specific pathogen-free facility at the University of Massachusetts Medical School. The mice were bred and used according to the guidelines from the University of Massachusetts Animal Care and Use Committee.
Cell culture, total RNA isolation, electrophoresis, transfer and northern blotting
Total RNA was extracted using TRIzol® reagent (Invitrogen, Carlsbad, CA, USA). After adult fish were ground in liquid nitrogen, 50 mg homogenate was processed in 1 ml TRIzol reagent. Embryos were homogenized in TRIzol reagent by passing the embryos through a 22.5 gage needle. Total RNA was extracted as described in the manufacturer’s instructions, and then subjected to phenol/chloroform purification to improve quality. For mouse B-cell cultures, spleens of WT C57BL/6 mice were dispersed and depleted of T cells, and B cells were cultured at 4 × 105/ml for 3 days with lipopolysaccharide (50 µg/ml, Sigma-Aldrich, St. Louis, MO, USA), anti-IgD-dextran (0.3 ng/ml) in the absence of BLyS with DMSO or 300 µM CRT0044876 (Maybridge Trevillet) added as previously described (10).
For northern blots, ∼30 µg total RNA was resolved in a 1.2% denaturing agarose gel containing 18% deionized formaldehyde at 70 V for 3 h. After electrophoresis, RNA was transferred to nitrocellulose membranes as described in Supplementary Data for Southern blotting and stored at 4°C. To probe for polb mRNA, the same DNA probe used in Southern blot analysis was prepared for northern blots. The membrane was first prehybridized at 68°C by immersing the membrane in ExpressHyb™ solution for 1.5 h. Hybridization at 68°C for 3 h was followed by one wash at room temperature and a second at 55°C for 1 h each. The distribution of isotopically labeled probe was determined by phosphorImager analysis and quantitation using ImageQuant software (11).
Quantitative real-time polymerase chain reaction
Total RNA, extracted from different stage embryos and digested with RNase-free DNase, was reverse transcribed using High-capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time polymerase chain reaction (qRT–PCR) was performed as described (11) using the following thermal cycle parameters: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 s at 95°C and 1 min at 60°C. The mean value of triplicate determinations was normalized to transcript levels of B-actin that served as the internal control.
Protein extraction, gel electrophoresis, transfer and western blotting
Protein extraction and western blotting were performed as described (9,11). Anti-zfApex1 antibody was prepared against zebrafish residues 140–155 by Sigma-Genosys (The Woodlands, TX, USA) (9). Unless indicated otherwise, all western blots detecting Apex were performed using this antibody. For antibody directed against human AP endonuclease 1 (hApex), we used antibody purchased from Novus Biologicals (Littleton, CO, USA). For antibody directed against Polb, we used either a mouse monoclonal anti-rat Polb antibody (Thermo scientific, Fremont, CA, USA) or a rabbit polyclonal custom antibody (21 Century Biochemicals, Marlboro, MA, USA) prepared against zebrafish Polb residues 324–339 (11). Polyclonal rabbit antibodies to detect Creb1 and Creb1 complex peptides conserved in zebrafish were obtained from Abcam Inc. (Cambridge, MA, USA) for Creb1 and p133Creb1, or from Cell Signaling (Santa Cruz Biotech Inc., Santa Cruz, CA, USA) for Crtc1, Cbp and Crem. To our knowledge there is no commercially available antiserum for Crtc3 at this time. In all cases bands of the molecular weight expected based on the sequence of the appropriate zebrafish protein were detected. Images were quantified using ImageJ software (http://rsbweb.nih.gov/ij/) and normalized to intensities of B-actin obtained with antibody purchased from GeneTex Inc. (Irvine, CA, USA).
Knockdown of selected genes by morpholino microinjection
All MOs, synthesized by GeneTools, LLC (Philomath, OR), are listed in Supplementary Table S1. Two nanoliter MO at 3 ng/nl was injected into 1–2 cell stage embryos, using phenol red as an injection indicator. It is necessary to microinject prior to the 8-cell stage so that the MO will distribute equally to all cells in the embryo. Injection volume was determined by calibration performed on a 1× 0.01 mm stage micrometer (Thermo scientific, Fremont, CA, USA). Injected embryos were raised at 28.6°C to the desired developmental stages. Phenotypes were examined daily using a Leica stereomicroscope (Bannockburn, IL, USA) and photographed or harvested for biochemistry.
Plasmid construction and capped RNA synthesis
Supplementary Table S1 lists all primers used in this study. To construct pCS2+-GFP-Polb, enhanced GFP gene (eGFP) was amplified from p3E-eGFPpA vector with primer set eGFP-BamHI-For and eGFP-EcoRI-Rev and cloned into the pCS2+ expression vector between the BamHI and EcoRI cloning sites. Zebrafish polb gene was amplified from first-strand cDNA using the primer set polb-EX-For/Rev. Zebrafish polb coding region was then cloned into pCS2+-eGFP plasmid downstream of the eGFP gene.
To construct the pCreb1-GFP plasmid, 3040 bp of the creb1 promoter preceding the ATG start codon was amplified using creb1 promoter primers For/Rev (CrebP-For/Rev). After digestion with XhoI and BamHI, the creb1 promoter sequence was inserted into peGFP-N3 vector between the XhoI and BamHI sites to displace the original cytomegalovirus promoter (13). To construct pCS2+-creb1, the 957 bp creb1 open reading frame (ORF) was cloned from first-strand cDNA with the creb1 ORF-For/Rev primer set. After digestion with EcoRI and SneBI, creb1 ORF was cloned into pCS2+ between the EcoRI and SneBI sites.
To obtain capped RNA for eGFP tagged polb or for creb1 RNA, in vitro transcription was performed using Ambion® mMESSAGE mMACHINE® SP6 Kit (Applied Biosystems, Austin, TX, USA), followed by purification with the MEGAclear™ Kit (Applied Biosystems). RNA (540 pg) was microinjected into 1–2 cell stage embryos. eGFP expression and embryonic development were examined daily by fluorescence microscopy. Western blot analysis was performed to confirm the overexpression of Polb protein with both zebrafish specific Polb antibody and anti-GFP antibody (Cell Signaling Technology Inc., Danvers, MA, USA).
Methyl methanesulfonate treatment
Water-injected (control), MO knockdown and polb mRNA rescued embryos were subjected to methyl methanesulfonate (MMS) treatment beginning at 4 hours post-fertilization (hpf). Rescue experiments were performed by co-injection of MO2 and eGFP-polb mRNA. Chronic treatment was performed by placing embryos in MMS diluted in fish water for a total of 5 days, with daily changes of medium. Embryos were examined daily for developmental progress. Death was evaluated as cessation of heartbeat and circulation on or before 5 dpf. Percent survival was recorded as surviving embryos at 5 dpf divided by total number of embryos subjected to the treatment.
RESULTS
Knocking down Polb protein results in increased sensitivity to alkylation agents
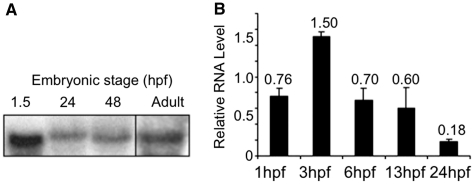
Although eggs and early zebrafish embryos lack Polb protein (11), they had ample polb mRNA, as shown by northern blots of RNA obtained from embryos at various stages and adult fish (Figure 1). The single band matched the 1594 bp cDNA expected from the sequence cloned from the single copy identified in zebrafish genomic DNA (Supplementary Figures S1 and S2). Expression of polb message peaked at the beginning of the midblastula (MBT) transition ∼3 hpf, after which transcript levels decreased gradually to approximately one-eighth of the amount present in 3 hpf embryos (Figure 1B). Since Polb protein was not detected before 13 hpf (11), protein synthesis for this protein did not correlate with mRNA expression. Overexpression of Polb had no obvious morphological effect in developing embryos.
Figure 1.
Messenger RNA for polb is plentiful in early stage zebrafish embryos. Developmental stages are indicated as hours post-fertilization (hpf). Values represent the average of 3 independent experiments ± SD of the mean. (A) Northern blot of total RNA extracted from different stage embryos and adults. Each lane was loaded with 30 µg total RNA and probed with α[32P] dCTP labeled polb cDNA. (B) qRT–PCR quantitation of polb mRNA expression within the first 24 h of development. Data were generated from two independent experiments.
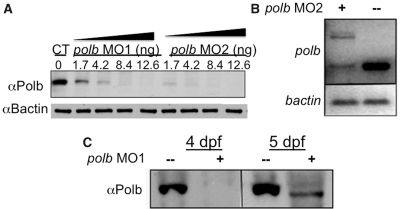
Knockdown by microinjection of morpholino oligonucleotides (MO) targeted to the translation start site (MO1) or to the junction of intron 1 and exon 2 (MO2) delayed appearance of Polb for up to 4 dpf as shown by both qRT-PCR and western blotting (Figure 2). Failure to process polb pre-mRNA confirmed successful knockdown (Figure 2B). After knockdown, Polb protein levels did not recover until 5 dpf. Consistent with results from cultured mammalian cells and murine embryos (11,12), there were no obvious morphological abnormalities in early knockdown embryos. Antibody staining with MF20, which recognizes sarcomeric myosin in both skeletal and cardiac muscle, Alcian blue cartilage staining and o-dianisidine red blood cell staining all showed no difference between Polb knockdown and control embryos (data not shown). Hence, Polb did not play an indispensable role during very early morphogenesis, which is consistent with the case in early murine embryogenesis (14,15). However, 84 and 61% larvae microinjected with MO1 and MO2, respectively, failed to inflate their swim bladders. Although failure to inflate the swimbladder is a common sign of stress in zebrafish, the swimbladder is the teleost equivalent of the mammalian lung, which is defective in polb−/− mouse knockouts, so that polb−/− pups die shortly after birth (15). Therefore the failure to inflate may be a specific response to loss of Polb.
Figure 2.
Both polb morpholinos MO1 and MO2 can effectively block Polb translation which does not resume until 5 dpf. (A) Western blot showing loss of Polb protein after microinjection of different concentrations of MO1 (directed against the translation start site of polb) or MO2 (directed against the intron1/exon2 splice site). Protein extracts were prepared from 1 dpf (days post fertilization) embryos. (B) qRT–PCR shows that MO2, a splicing blocker targeting the intron 1/exon 2 junction, produced a polb splice variant that retains intron1 (upper band), besides the normal transcript (lower band). B-actin, shown in the lower panel, served as control. (C) Western blotting confirms that after knockdown of polb, Polb protein does not appear until 5 dpf. Total protein from Polb knockdown embryos and controls was extracted at 4 and 5 dpf, and examined by western blot analysis for the presence of Polb protein.
Although there were no major morphological alterations in development due to loss or overexpression of Polb, cell lines from Polb knockdown embryos are hypersensitive to the alkylating agent, MMS, which causes alkylation damage to DNA requiring repair by BER (16). As predicted, zebrafish embryos in which polb had been knocked down were more sensitive to MMS administered as a chronic exposure beginning at 4 hpf. The knockdown embryos had significantly (P < 0.05) lower median lethal dosage (LC50) values (0.083 ± 0.003 mM) than those in untreated control embryos (0.113 ± 0.003 mM). Microinjection of ectopic eGFP tagged wild-type zebrafish polb mRNA along with MO2 into 1-cell stage embryos partially restored the resistance to MMS in knockdown embryos (0.096 ± 0.010 mM). MO2 did not interfere with the expression of ectopic eGFP-zfpolb mRNA, because MO2 blocked splicing but not transcription initiation (Figure 2B). Successful rescue was confirmed by western blot analysis using anti-GFP antibody. Therefore, loss of Polb protein in early embryogenesis resulted in blunting of DNA repair by BER.
Reduction of Apex results in loss of Polb protein via Creb1
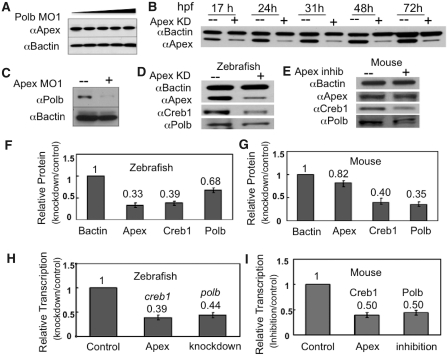
The protein upstream of Polb in the BER pathway is Apex (5), which is likely to interact directly with Polb during BER (6,7,17). We determined the effects of reducing the levels of the BER partners on each other when we altered the level of each protein. Western blot analysis showed that knocking down Polb did not alter levels of Apex (Figure 3A). In contrast and to our surprise, knocking down Apex, the effect of which lasted for up to 4 days (Figure 3B), resulted in a substantial reduction of both Polb protein (Figure 3C, D and F) and polb message (Figure 3H). Recovery of Polb levels corresponded to the reappearance of Apex at ∼5 dpf. These results suggest that Apex is an important regulator of its partner in the BER pathway by regulating expression of the enzyme immediately downstream from it.
Figure 3.
While knockdown of Polb does not affect Apex levels, knockdown of Apex has a profound effect on Polb and Creb1 in both zebrafish embryos and primary cultures of murine B cells. (A) Knockdown of Polb does not alter Apex protein levels. Extracts were prepared from 1 dpf embryos in which Polb had been knocked down by microinjection of increasing amounts of MO1 and probed by western blot analysis using rabbit polyclonal anti-Apex or anti-B actin (9). Embryos were microinjected with Polb MO1 or water; extracts were prepared at 1 dpf. (B) Knockdown of Apex by MO results in Apex loss for the duration of these experiments. Western blot analysis of protein extracts prepared on the indicated days was performed using anti-Apex and anti-B-actin. Loss of Apex, shown here for 3 days after knockdown, was usually detected for up to 5 dpf. (C) Knockdown of Apex results in failure of Polb protein to appear at the appropriate time in zebrafish embryos. Protein extracts were prepared from 3 dpf embryos. Apex hypomorphic embryos (+) were created by microinjecting 0.15 mM Apex MO at the 1–2 cell stage. (D and E) Western blotting analysis of Creb1 and Polb protein levels in Apex knockdown embryos at 24 hpf or Apex-inhibited mouse primary B lymphocytes. Note that in the latter, Apex activity is inhibited by CRT0044876 but that Apex protein is only slightly diminished. (F and G) Quantitation of western blot results in Apex knockdown embryos and Apex-inhibited mouse primary B lymphocytes. Data for (F) are the average of four independent experiments ± SD of the mean. Values for (G) are the average of three independent experiments ± SD of the mean. (H and I) Quantitative RT–PCR analysis of creb1 and polb mRNA levels in Apex knockdown zebrafish embryos and Apex-inhibited mouse lymphocytes. Loss of Apex in either mouse cells or zebrafish embryos results in diminution of both creb1 and polb mRNA. The values for H are the average of five independent experiments ± SD of the mean, while the values for I are the average of three independent experiments ± SD of the mean.
Previous work has shown that Creb1 is required for polb transcription (18,19). Since we had just determined that Apex was important for polb transcription, we examined whether there might be a connection between Apex, Creb1 and Polb. Indeed, microinjection of MO directed against the Apex translation start site resulted in loss not only of Apex protein and failure of Polb to appear in a timely fashion, but also of ∼50% Creb1 protein and message at 24 hpf (Figure 3D, F and H).
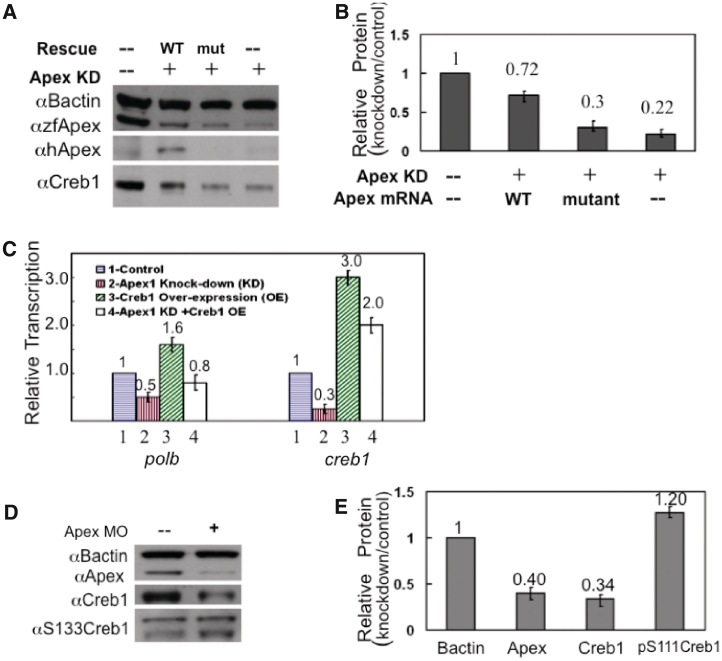
To confirm the relationship between Creb1, Apex and Polb, we first rescued Apex knockdown by co-injection of MO directed against the Apex translation start site with or without capped mRNA encoding WT human AP endonuclease 1 or the catalytically inactive site-directed mutant containing a triple mutation in the active site (Y171F-P173L-N174K) (Figure 4A). Although this mutant is enzymatically inactive, it retains conformation similar to the native enzyme (20). The gene for the human protein was chosen for rescue experiments, because the knockdown MO designed to target the ATG start site of the zebrafish message does not interfere with expression of the human message (9). Extracts prepared at 24 hpf were examined for the presence of apex, creb1 and polb message and protein (Figure 4). Results were entirely consistent with earlier findings (9), that rescue required the endonuclease function of the protein, since mRNA for WT human Apex but not the active site mutant lacking endonuclease activity resulted in rescue. We then overexpressed creb1 mRNA with or without knockdown of Apex. We observed increased creb1 mRNA in control embryos that had not been microinjected with MO and entirely restored loss of creb1 mRNA in embryos in which Apex had been knocked down (Figure 4C). Furthermore, overexpression of creb1 message resulted in enhanced levels of polb message when Apex had not been knocked down and restored polb message in Apex knockdowns.
Figure 4.
Rescue experiments confirm the role of Apex and Creb in maintaining Polb; Apex knockdown does not alter Creb1 phosphorylation at 24 hpf. (A) Microinjection of capped mRNA encoding WT human AP endonuclease 1 (hApex) rescued both Apex and Creb1 levels in Apex knockdown embryos. Note that antiserum directed against peptide 140–155 of the zebrafish protein (αzfApex) was able to recognize both WT and mutant hApex proteins, while antibody against hApex (αhApex) recognized only WT hApex protein. (B) Quantitation of relative Creb protein levels from (A). These data represent the mean ± SD of three independent experiments. (C) Microinjection of capped creb1 mRNA rescued creb1 and polb transcription in early zebrafish embryos. Apex was knocked down by microinjection of TS-MO in the presence or absence of creb1 capped RNA at the 1–2 cell stage. Total RNA was prepared from 24 hpf extracts. Messenger RNA levels for the indicated genes under the different conditions relative to b-actin transcription were determined by qRT–PCR. Data were then normalized to the mRNA level found in control extracts from embryos microinjected with water. This panel shows that co-injection of creb1 mRNA restores polb mRNA levels. Values represent the average of three independent experiments ± SD of the mean. (D) Knockdown of Apex did not alter Creb phosphorylation at 24 hpf. Western blot analysis of different forms of Creb1 protein in 24 hpf embryos. Note that despite the fact that Creb1 itself was diminished, p133Creb remained unchanged at 24 hpf. (E) Quantitation of western blot results shown in Figure 4D. The values are the average of three independent experiments ± SD of the mean.
In some systems, PKA-mediated phosphorylation of Creb1 promotes Creb1 nuclear localization and complex binding (21–23). To determine whether loss of Polb was mediated through PKA, we used western blot analysis to examine whether Creb1 phosphorylation was altered at Ser111 when Apex was knocked down. (Note that Ser133 is the corresponding residue in the human ortholog.) Phosphorylation at that residue was not altered at 24 hpf (Figure 4D and E). Consequently, the ratio of pSer111 relative to remaining Creb was increased an average of 2-fold at 24 hpf. The elevated level of phosphorylated Creb1 was not maintained, however, since by 48 hpf phosphorylation at Ser111 had decreased to 60% of control levels. Creb1 can also be phosphorylated at two additional serine residues, Ser107 and Ser120, which are equivalent to Ser129 and Ser142 in the human protein. Phosphorylation at these two residues decreased commensurately with the loss in Creb1 (data not shown). Thus, any change in PKA activity was secondary to the initial loss of Creb1 protein.
Apex regulates other members of the Creb complex
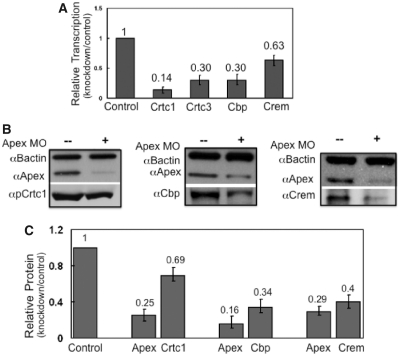
To examine whether participants in the Creb1 complex (24,25), such as Creb response element modulator (Crem), Creb binding protein (Cbp), and Creb regulated transcription coactivator (Crtc) 1 and 3, were affected by loss of Apex, we performed qRT–PCR and western blot analysis on extracts from 24 hpf control and Apex knockdown embryos. Loss of Apex resulted in substantial (40–85%) loss of crem, cbp and crtc 1 and 3 mRNA (Figure 5A). Corresponding protein levels were reduced up to 65% (Figure 5B and C). Therefore, Apex regulates not only Creb1 levels but also levels of Creb binding partners.
Figure 5.
Apex regulates proteins in the Creb-dependent transcription complex. (A) Transcripts of proteins that participate in the Creb complex were measured by qRT–PCR in control and Apex knockdown 24 hpf embryos. Messenger RNA for Crtc1, Crtc3, Cbp and Crem were examined using primers described in Supplementary Table S1. Data are presented as ratio of control normalized for b-actin message. Values are the average of three independent experiments ± SD of the mean. (B) Western blot analysis showed that proteins in the Creb complex (Creb1 itself, Crtc1, Cbp and Crem) are decreased when Apex is knocked down. We did not quantify Crtc3, as there is no commercially available antiserum. (C) Quantitation of western blot results of Creb complex proteins in 24 hpf control and Apex knockdown embryos. Values are the average of three independent experiments ± SD of the mean.
Apex regulation of Creb1 is independent of p53
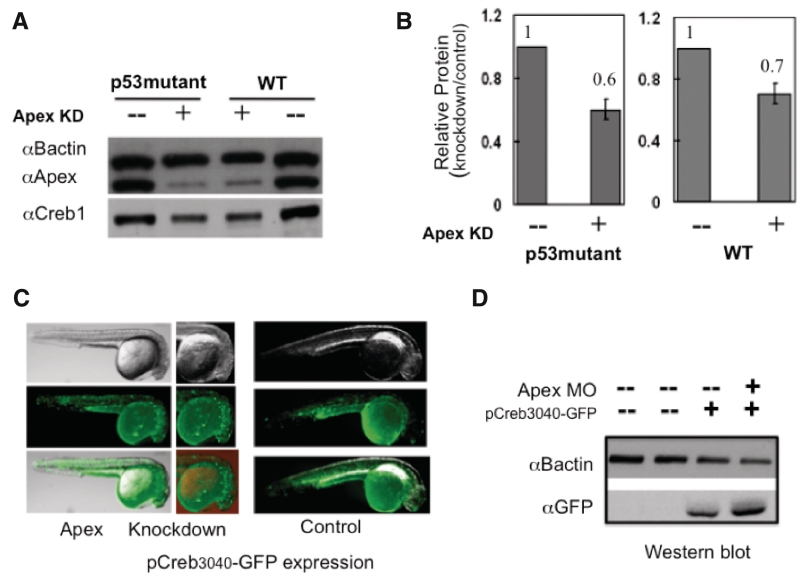
Because apoptosis is the most pronounced result of knocking down Apex in most cultured cells, we asked whether p53 might be involved in loss of Creb. We repeated the knockdown experiments in homozygous p53 (p53M214K/ M214K) mutant zebrafish. These mutant fish fail to upregulate p21, do not arrest at the G1/S checkpoint and fail to undergo radiation-induced apoptosis (12). Since knockdown of Apex resulted in diminution of Creb in the absence of p53 (Figure 6A and B), we conclude that Creb regulation by Apex is p53 independent. Our results may explain some of the p53 independent effects in mammalian cells exposed to MMS (26).
Figure 6.
Effects of Apex knockdown are independent of p53; Apex is not a co-activator of Creb1. (A) To examine whether loss of Creb was p53-dependent, we performed Apex knockdown in WT and p53M214K/ M214K fish. Cell extracts were prepared at 24 hpf and analyzed for Apex and Creb by western blotting. Apex knockdown resulted in similar loss of Creb in both WT and p53M214K/ M214K fish. (B) Quantitation of relative Creb protein levels from (A). The values are the average of 3 independent experiments ± SD of the mean. (C) To examine whether Apex is a co-activator of creb transcription we microinjected pCreb1P3040-eGFP with or without Apex knockdown and examined the embryos by fluorescence microscopy or western blot analysis. Distribution and intensity of GFP increased somewhat after Apex knockdown. Note the regions of greatest intensity in 24 hpf embryos in the head region, which remained unaltered by Apex knockdown. (D) Western blot, using rabbit antiGFP antibody, confirmed increased levels of GFP protein after knockdown of Apex.
Apex is not a coactivator of Creb1
Since the mRNA for many, if not all, members of the Creb1 complex and for Polb contains a Creb1 binding site, we asked whether Apex was a co-activator of Creb1. To that end, we inserted the 3040 bp sequence upstream of the creb1 gene in front of the coding sequence for green fluorescent protein (GFP) to obtain pCreb1P3040-eGFP (13). After microinjection of 50 pg pCreb1P3040-eGFP per embryo with or without the MO to knockdown Apex, we followed the appearance and localization of GFP (Figure 6C and D). GFP in control embryos appeared in selected regions of the head and along the trunk. After knockdown of Apex, the fluorescence was intensified, while the location remained unaltered. Western blot results confirmed knockdown of Apex and increased GFP. Therefore, we conclude that Apex is not a co-activator of the creb1 promoter, or at least not one acting within 3000 bp of the Creb binding site.
Loss of creb1 and polb message and protein due to reduction of Apex is not limited to teleosts
Murine primary B lymphocytes divide more slowly and fail to undergo class switching when Apex is inhibited with 7-nitro-1H-indole-2-carboxylic acid (CRT0044876) (10). To determine whether the results with zebrafish embryos were unique to embryos or to fish, we examined changes in creb1 and polb message and protein in primary cultures of murine B lymphocytes cultured in the presence or absence of 300 µM CRT0044876. We observed the loss of polb and creb1 message and protein after inhibition of Apex activity (Figure 3E, G and I), although the amount of Apex protein in B lymphocytes changed little and viability was not substantially affected. Therefore, Apex regulates Creb1 function in higher vertebrates including mouse B cells and zebrafish embryos. Creb1 is important at many stages of B cell development and for B cell survival (27). Furthermore, these data are consistent with the observation that Apex+/− mice have reduced levels of PolB (28).
DISCUSSION
We have provided conclusive evidence for a novel regulatory circuit in BER, one in which AP endonuclease 1 regulates the level of creb1 transcript and Creb1 protein in zebrafish early embryos and primary cultures of murine B cells. It also regulates levels of mRNA and protein levels of other proteins in the Creb1 complex, including Cbp, Crem and Crtc1 and 3 and thereby regulates the level of Polb, its partner in the BER pathway. Figure 7 is a cartoon summarizing our results. Since each of the proteins in the Creb complex is reduced by 60–80% when Apex function is reduced, the opportunity to form the Creb complex could be reduced by as much as 95%. Therefore, transcription of other genes that are positively regulated by Creb1 could be reduced to <5% control levels. Since reduction in the individual proteins is commensurate with reduction in mRNA levels, the primary effect is at the transcriptional or message stability level, rather than at the level of protein stability. These data provide the first evidence for a protein that regulates creb1 transcription and Creb1 protein levels. Since Creb1 regulates over 100 pathways (29–33), AP endonuclease 1, first known for its participation as an endonuclease in the base excision DNA repair pathway, serves as a driver gene for a master regulator of multiple major pathways in higher eukaryotic cells.
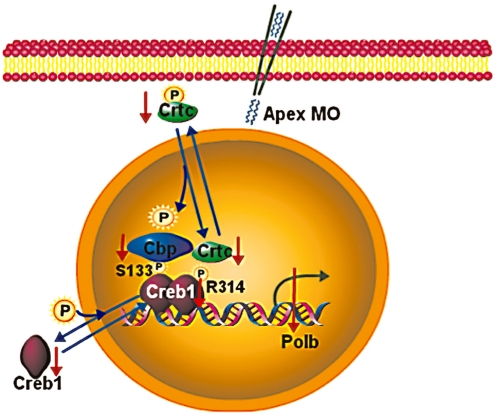
Figure 7.
Cartoon showing how Apex might regulate DNA polymerase β via the Creb complex. Note that the numbering of residues on Creb1 is that of the human homologue in order to avoid confusion in the literature. However, Ser133 in the human protein is Ser111 in the zebrafish protein. Similarly, Arg314 in the human protein, which binds Crtc1 when it is phosphorylated, is Arg291 in the zebrafish protein. These data indicate that Apex maintains levels of Creb1 and its partners without affecting PKA-mediated phosphorylation of Creb1. Consequently, in the absence of Apex levels of Creb1 complex diminish and Creb-mediated transcription is altered.
Loss of Apex also results in loss of polb mRNA, which corresponds to delayed appearance of Polb in development. Since Creb is required not only for transcription of polb (18,19) but also for transcription of sp1 and NFκB, two additional transcription factors required for polb transcription, failure of Polb to appear in a timely fashion is likely to be the result of diminution of Creb and the members of its complex. Because Apex is required for the synthesis of Polb, the next protein in the BER pathway, Apex can be designated a ‘feed forward activator’ (34–36). However, the presence of Apex is not sufficient to ensure the synthesis of Polb, since zebrafish eggs and early-stage embryos contain a surfeit of Apex protein and polb mRNA but lack detectable Polb protein. Other regulators must also be involved in preventing synthesis of Polb protein in early embryos.
Indeed, other regulators are likely to be involved in selecting which transcripts are up- or down-regulated after Apex reduction, since some Creb1-dependent transcripts are upregulated under knockdown conditions. These results are in keeping with studies that show that which Creb1-dependent genes are altered at any time depends on other members of the complex, such as Cbp, Torc and/or p300 (37–39).
Although loss of Apex is ultimately lethal for both mouse and zebrafish embryonic development, our findings reinforce the requirement for endonuclease activity rather than any redox activity for successful embryogenesis and cell survival. Not only does the WT zebrafish protein lack the requisite cysteine residue which renders it redox inactive in standard electrophoretic mobility shift assays (9,40) but also rescue in knockdown zebrafish embryos requires the endonuclease function. Furthermore, development of mouse embryos is not affected when the corresponding redox-relevant cysteine (Cys64) is converted to alanine (41). How might the endonuclease activity regulate creb transcription/stability? Since Apex does not make a Schiff base when it cleaves an AP site, the mechanism by which it regulates creb1 levels is different from that of Ku’s cleavage of AP sites in proximity to double strand breaks in DNA (42). Apex does have the ability to cleave RNA (43,44), which requires the same active site as cleavage of an AP site in DNA (43). Although the requirement for an enzymatically active site is consistent with our knockdown/rescue results, how loss of an RNase activity might account for reduction of creb1 mRNA is not clear. Regardless of the mechanism, that loss of Apex results in the loss of Creb1 and its complex partners, which regulate multiple interlocking pathways (45–47), can account for lethality in higher eukaryotic cells and embryos.
Because zebrafish embryos continue to develop through hatching without a functional heart, we were able to follow important pathways and cell types that depend on Apex. These studies could not have been done in mammalian embryos, since abnormal cardiac development in mouse embryos leads to resorption shortly after gastrulation. Nor could they could have been done in cultured cells, since most cultured cells undergo apoptosis when Apex levels are reduced to the levels obtained here. The finding that Apex regulates Creb1 levels in the absence of p53 implies that the effects of Apex reduction may be independent of p53-mediated apoptosis in cultured cells. Furthermore, since Creb1 regulates expression of the antiapoptotic genes mcl-1 and bcl-2 in vitro in B cells (48), these data suggest that Apex regulation of Creb may be a mechanism to protect against cell death, perhaps by preventing apoptosis to allow time for DNA to be repaired. This mechanism could be particularly critical for cephalic, neural and cardiac tissues that comprise the Apex knockdown phenotype and are critically sensitive to oxidative damage.
Besides serving as a driver for Creb, Apex has been proposed to participate in regulation of transcription as a trans-acting transcription factor (49–52). Apex also associates with HIF-1α, p300 and STAT3 in a complex to regulate VEGF transcription (38,53) and enhances binding of Egr-1 to its cognate sequence, which in turn activates PTEN expression (54,55). Although our data indicate that Apex is unlikely to serve as a co-activator of Creb1, both Egr1 and HIF1α contain Creb binding sites and the effect could be mediated through the Creb complex.
Creb1 anchors a transcription complex that regulates multiple pathways in cell physiology. Specificity is likely to be conferred by individual factor(s) completing the complex. Despite the intense focus on Creb1 over the years, how its protein level is regulated was not evident until our study. We have clearly demonstrated for the first time that AP endonuclease 1 regulates not only Creb1 protein levels but also the protein levels of other Creb complex members. In so doing, it controls the protein levels of DNA polymerase ß, its downstream partner in the BER pathway. This finding opens new horizons to further understand the importance of the multifunctional protein, Apex, in regulating cell physiology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The G. Harold and Leila Y. Mathers Charitable Foundation (to P.R.S.); Aid for Cancer Research and Northeastern University (to P.R.S); National Natural Science Foundation of China (Grant No. 30700607 to D.S.P.); National Institutes of Health (RO1 AI065639 to C.E.S.); Cancer Research Institute fellowship (to J.E.J.G.). Funding for open access charge: The G. Harold and Leila Y. Mathers Charitable Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Thomas Look for the p53 mutant zebrafish.
REFERENCES
- 1.Kawamura Y, Uchijima Y, Horike N, Tonami K, Nishiyama K, Amano T, Asano T, Kurihara Y, Kurihara H. Sirt3 protects in vitro-fertilized mouse preimplantation embryos against oxidative stress-induced p53-mediated developmental arrest. J. Clin. Invest. 2010;120:2817–2828. doi: 10.1172/JCI42020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menezo Y, Dale B, Cohen M. DNA damage and repair in human oocytes and embryos: a review. Zygote. 2010;18:357–365. doi: 10.1017/S0967199410000286. [DOI] [PubMed] [Google Scholar]
- 4.Wells PG, McCallum GP, Chen CS, Henderson JT, Lee CJ, Perstin J, Preston TJ, Wiley MJ, Wong AW. Oxidative stress in developmental origins of disease: teratogenesis, neurodevelopmental deficits, and cancer. Toxicol. Sci. 2009;108:4–18. doi: 10.1093/toxsci/kfn263. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2 edn. Washington, DC: ASM Press; 2006. [Google Scholar]
- 6.Bennett RA, Wilson DM, 3rd, Wong D, Demple B. Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway. Proc. Natl Acad. Sci. USA. 1997;94:7166–7169. doi: 10.1073/pnas.94.14.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abyzov A, Uzun A, Strauss PR, Ilyin VA. An AP endonuclease 1-DNA polymerase beta complex: theoretical prediction of interacting surfaces. PLoS Comput. Biol. 2008;4:e1000066. doi: 10.1371/journal.pcbi.1000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T. The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc. Natl Acad. Sci. USA. 1996;93:8919–8923. doi: 10.1073/pnas.93.17.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Shupenko CC, Melo LF, Strauss PR. DNA repair protein involved in heart and blood development. Mol. Cell. Biol. 2006;26:9083–9093. doi: 10.1128/MCB.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortier S, Yang X, Wang Y, Bennett RA, Strauss PR. Base excision repair in early zebrafish development: evidence for DNA polymerase switching and standby AP endonuclease activity. Biochemistry. 2009;48:5396–5404. doi: 10.1021/bi900253d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl Acad. Sci. USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei DS, Sun YH, Chen CH, Chen SP, Wang YP, Hu W, Zhu ZY. Identification and characterization of a novel gene differentially expressed in zebrafish cross-subfamily cloned embryos. BMC Dev. Biol. 2008;8:29. doi: 10.1186/1471-213X-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996;379:183–186. doi: 10.1038/379183a0. [DOI] [PubMed] [Google Scholar]
- 15.Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000;19:1397–1404. doi: 10.1093/emboj/19.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton JK, Joyce-Gray DF, Pachkowski BF, Swenberg JA, Wilson SH. Hypersensitivity of DNA polymerase beta null mouse fibroblasts reflects accumulation of cytotoxic repair intermediates from site-specific alkyl DNA lesions. DNA Repair. 2003;2:27–48. doi: 10.1016/s1568-7864(02)00184-2. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Prasad R, Beard WA, Kedar PS, Hou EW, Shock DD, Wilson SH. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J. Biol. Chem. 2007;282:13532–13541. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayan S, He F, Wilson SH. Activation of the human DNA polymerase beta promoter by a DNA-alkylating agent through induced phosphorylation of cAMP response element-binding protein-1. J. Biol. Chem. 1996;271:18508–18513. doi: 10.1074/jbc.271.31.18508. [DOI] [PubMed] [Google Scholar]
- 19.He F, Narayan S, Wilson SH. Purification and characterization of a DNA polymerase beta promoter initiator element-binding transcription factor from bovine testis. Biochemistry. 1996;35:1775–1782. doi: 10.1021/bi9525987. [DOI] [PubMed] [Google Scholar]
- 20.Kanazhevskaya LY, Koval VV, Zharkov DO, Strauss PR, Fedorova OS. Conformational transitions in human AP endonuclease 1 and its active site mutant during abasic site repair. Biochemistry. 2010;49:6451–6461. doi: 10.1021/bi100769k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 22.Mayr BM, Canettieri G, Montminy MR. Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc. Natl Acad. Sci. USA. 2001;98:10936–10941. doi: 10.1073/pnas.191152098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sands WA, Palmer TM. Regulating gene transcription in response to cyclic AMP elevation. Cell Signal. 2008;20:460–466. doi: 10.1016/j.cellsig.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc. Natl Acad. Sci. USA. 2003;100:12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol. Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J. Biol. Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]
- 27.Chen HC, Byrd JC, Muthusamy N. Differential role for cyclic AMP response element binding protein-1 in multiple stages of B cell development, differentiation, and survival. J. Immunol. 2006;176:2208–2218. doi: 10.4049/jimmunol.176.4.2208. [DOI] [PubMed] [Google Scholar]
- 28.Raffoul J, Cabelof D, Nakamura J, Meira L, Friedberg E, Heydari A. Apurinic/apyrimidinic endonuclease (APE/ref-1) haploinsufficient mice display tissue-specific differences in DNA polymerase beta-dependent base excision repair. J. Biol. Chem. 2004;279:18425–18433. doi: 10.1074/jbc.M313983200. [DOI] [PubMed] [Google Scholar]
- 29.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 30.Conkright MD, Guzman E, Flechner L, Su AI, Hogenesch JB, Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol. Cell. 2003;11:1101–1108. doi: 10.1016/s1097-2765(03)00134-5. [DOI] [PubMed] [Google Scholar]
- 31.Euskirchen G, Royce TE, Bertone P, Martone R, Rinn JL, Nelson FK, Sayward F, Luscombe NM, Miller P, Gerstein M, et al. CREB binds to multiple loci on human chromosome 22. Mol. Cell. Biol. 2004;24:3804–3814. doi: 10.1128/MCB.24.9.3804-3814.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl Acad. Sci. USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartzell DD, Trinklein ND, Mendez J, Murphy N, Aldred SF, Wood K, Urh M. A functional analysis of the CREB signaling pathway using HaloCHIP-chip and high throughput reporter assays. BMC Genomics. 2009;10:497. doi: 10.1186/1471-2164-10-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl Acad. Sci. USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacGillavry HD, Stam FJ, Sassen MM, Kegel L, Hendriks WT, Verhaagen J, Smit AB, van Kesteren RE. NFIL3 and cAMP response element-binding protein form a transcriptional feedforward loop that controls neuronal regeneration-associated gene expression. J. Neurosci. 2009;29:15542–15550. doi: 10.1523/JNEUROSCI.3938-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, Sauer S, Hadjur S, Leleu M, Naoe Y, Telfer JC, et al. Runx proteins regulate Foxp3 expression. J. Exp. Med. 2009;206:2329–2337. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR, 3rd, Montminy M. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. Embo J. 2007;26:2880–2889. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, Gallick GE. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–3120. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]
- 39.Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 40.Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat. Res. 2008;643:54–63. doi: 10.1016/j.mrfmmm.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ordway JM, Eberhart D, Curran T. Cysteine 64 of Ref-1 is not essential for redox regulation of AP-1 DNA binding. Mol. Cell Biol. 2003;23:4257–4266. doi: 10.1128/MCB.23.12.4257-4266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SE, Gorrell A, Rader SD, Lee CH. Endoribonuclease activity of human apurinic/apyrimidinic endonuclease 1 revealed by a real-time fluorometric assay. Anal. Biochem. 2010;398:69–75. doi: 10.1016/j.ab.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 45.Dworkin S, Heath JK, deJong-Curtain TA, Hogan BM, Lieschke GJ, Malaterre J, Ramsay RG, Mantamadiotis T. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Dev. Biol. 2007;307:127–141. doi: 10.1016/j.ydbio.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 46.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Sandoval S, Pigazzi M, Sakamoto KM. CREB: A Key Regulator of Normal and Neoplastic Hematopoiesis. Adv. Hematol. 2009;2009:634292. doi: 10.1155/2009/634292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang H, Wang J, Boxer LM. Role of the cyclic AMP response element in the bcl-2 promoter in the regulation of endogenous Bcl-2 expression and apoptosis in murine B cells. Mol. Cell. Biol. 2006;26:8599–8606. doi: 10.1128/MCB.01062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhakat K, Izumi T, Yang S, Hazra T, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. Embo J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid. Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okazaki T, Chung U, Nishishita T, Ebisu S, Usuda S, Mishiro S, Xanthoudakis S, Igarashi T, Ogata E. A redox factor protein, ref1, is involved in negative gene regulation by extracellular calcium. J. Biol. Chem. 1994;269:27855–27862. [PubMed] [Google Scholar]
- 52.Fuchs S, Philippe J, Corvol P, Pinet F. Implication of Ref-1 in the repression of renin gene transcription by intracellular calcium. J. Hypertens. 2003;21:327–335. doi: 10.1097/00004872-200302000-00024. [DOI] [PubMed] [Google Scholar]
- 53.Ziel KA, Campbell CC, Wilson GL, Gillespie MN. Ref-1/Ape is critical for formation of the hypoxia-inducible transcriptional complex on the hypoxic response element of the rat pulmonary artery endothelial cell VEGF gene. FASEB J. 2004;18:986–988. doi: 10.1096/fj.03-1160fje. [DOI] [PubMed] [Google Scholar]
- 54.Fantini D, Vascotto C, Deganuto M, Bivi N, Gustincich S, Marcon G, Quadrifoglio F, Damante G, Bhakat KK, Mitra S, et al. APE1/Ref-1 regulates PTEN expression mediated by Egr-1. Free Radic. Res. 2008;42:20–29. doi: 10.1080/10715760701765616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pines A, Bivi N, Romanello M, Damante G, Kelley MR, Adamson ED, D'Andrea P, Quadrifoglio F, Moro L, Tell G. Cross-regulation between Egr-1 and APE/Ref-1 during early response to oxidative stress in the human osteoblastic HOBIT cell line: evidence for an autoregulatory loop. Free Radic. Res. 2005;39:269–281. doi: 10.1080/10715760400028423. [DOI] [PubMed] [Google Scholar]