Abstract
RNA localization is emerging as a general principle of sub-cellular protein localization and cellular organization. However, the sequence and structural requirements in many RNA localization elements remain poorly understood. Whereas transcription factor-binding sites in DNA can be recognized as short degenerate motifs, and consensus binding sites readily inferred, protein-binding sites in RNA often contain structural features, and can be difficult to infer. We previously showed that zebrafish squint/nodal-related 1 (sqt/ndr1) RNA localizes to the future dorsal side of the embryo. Interestingly, mammalian nodal RNA can also localize to dorsal when injected into zebrafish embryos, suggesting that the sequence motif(s) may be conserved, even though the fish and mammal UTRs cannot be aligned. To define potential sequence and structural features, we obtained ndr1 3′-UTR sequences from approximately 50 fishes that are closely, or distantly, related to zebrafish, for high-resolution phylogenetic footprinting. We identify conserved sequence and structural motifs within the zebrafish/carp family and catfish. We find that two novel motifs, a single-stranded AGCAC motif and a small stem-loop, are required for efficient sqt RNA localization. These findings show that comparative sequencing in the zebrafish/carp family is an efficient approach for identifying weak consensus binding sites for RNA regulatory proteins.
INTRODUCTION
Sub-cellular RNA localization restricts protein localization within cells, contributing to the formation of sub-cellular domains, and cellular asymmetry. It is a remarkably common phenomenon, with ∼70% of Drosophila RNAs found to be localized in a large number of different patterns during embryogenesis (1). Significantly, many RNAs localize to the same place in the cell as the protein they encode. RNA can be localized by various mechanisms, including active transport, which typically involves large RNP molecules containing RNA-binding proteins, adaptor molecules and motor proteins, often along the actin or microtubule cytoskeleton (2). Although some of the trans-acting factors and cis-elements have been identified, understanding of the molecular mechanisms remains fragmentary. In particular, RNA cis-elements and the cognate RNA-binding proteins are largely unknown.
A number of cis-acting elements have been identified in localized RNAs, and these elements can be composed of sequence, or structure, or both. For instance, the vegetally localized Xenopus RNAs encoding VegT and Vg1 contain multiple copies of two localization motifs, the YYCAC-containing E2 motif that is bound by VgIRBP (homologous to ZBP1), and the YYUCU motif (VM1) bound by VgRBP60/hnRNP I [reviewed in (3)]. In contrast, the bicoid RNA localization element is a large complex structure, and its cognate-binding proteins (such as Staufen) are thought to recognize structure, rather than sequence (4). The Drosophila fs(1)K10 (5) and orb (6,7) Transport/Localization Signals (TLS), and the wg Localization Element 3 [WLE3, (6)] are short stem-loops, in which the structure, and a part of the sequence, is required for localization.
Previous studies have tried to extract sequence/structure consensuses, either by mutagenesis, or by comparing groups of elements that can mediate the same localization pattern, with the assumption that the elements are bound by the same protein(s). For instance, the E2 and VM1 motifs in VegT and Vg1 RNA are short, repeated sequence motifs, reminiscent of clusters of transcription factor-binding sites in cis-regulatory modules (7). In cases where the signal includes structure, it is more difficult to identify a consensus. A number of transcripts are localized apically in fly embryos, including the pair rule transcripts, ftz and hairy (8) and wg (9,10). Additionally, bicoid (11), fs(1)K10 (12) and orb transcripts (13) are localized anteriorly in oocytes, but can also be localized apically when injected into embryos (14). While the orb and K10 TLS signals are similar to each other (5), there is no obvious similarity between these and the complex bicoid localization element, or the wingless localization elements, WLE1 and WLE2, or the ftz and hairy localization elements. Recently, a third element, WLE3, has been described as having putative similarities to the orb and K10 TLSs (6). From these three stem-loops, the authors inferred a weak consensus. Similarly weak putative sequence/structure consensuses have been extracted computationally from RNAs localized to the yeast bud tip (15), yeast mitochondria (16) and colocalized during Drosophila development (17). These consensus structures might be weak because the signals are inherently weak. For instance, the K10 TLS confers only a modest bias in the direction of RNA transport (14), and is recognised with low specificity by Egalitarian (the cognate RNA-binding protein) in in vitro binding studies (18). Identifying and confirming such weak consensus motifs remains a challenge.
The zebrafish embryo is an excellent system for testing the function of regulatory elements such as these, since DNA and RNA injections are facile, and embryos are transparent, allowing simple live imaging of fluorescent molecules. We have shown that in zebrafish, RNA encoding Squint/Nodal-related 1 (Sqt/Ndr1), an activin/TGFβ-like morphogen, is localized to the future embryonic dorsal in a microtubule-dependent manner (19). A localization element was mapped to the first 50 nt of the 3′-UTR. Remarkably, the orthologous mammalian NODAL 3′-UTR can also localize in zebrafish embryos, indicating that the fish machinery may recognize conserved localization signals in NODAL RNA (19). Furthermore, the NODAL 3′-UTR does not align to the zebrafish sqt 3′-UTR, suggesting that the element(s), if conserved, may be quite degenerate.
In order to define sequence and structure motifs in the sqt/ndr1 Dorsal Localization Element (DLE), we sequenced the ndr1 3′-UTR from a large number of species related to zebrafish. We reasoned that using a large number of closely related species would allow us to identify even weakly conserved elements. We identified three motifs in the sqt DLE; two apparently single-stranded motifs, AAACCCNRAA and AGCAC, and a short predicted stem-loop. By injecting various sqt RNA deletion mutants we show that both the AGCAC motif and predicted stem-loop are required for efficient localization to future dorsal in zebrafish embryos. These findings suggest that one or both of these motifs may correspond to the functional DLE.
MATERIALS AND METHODS
Amplification of ndr1 3′-UTR regions
Fish were collected from a local fish farm, together with Linnean names, and DNA was extracted according to standard protocols. In order to amplify the ndr1 3′-UTR from the various species, we designed non-degenerate primers to sequences in the coding regions of ndr1, and the downstream gene, eif4ebp1 that are highly conserved between zebrafish and Fugu (UCSC browser, http://genome.ucsc.edu/) (Figure 1B). We placed one primer in the adjacent gene to avoid amplification of the paralogs cyclops and southpaw, since neither gene is flanked by an eif4ebp paralog. The primers amplified the ndr1 3′-UTR from the majority of the ∼80 otophysan species we obtained (Figure 1A). We used the Primer3 program (20) (http://frodo.wi.mit.edu/) to design non-degenerate primers (sqtCF1 5′ AGAAGGTGGATATGTGGGTGGACT 3′, sqtCF2 5′ TCAGATTGGTTGGAGCGACTGGAT 3′, and eifCR1 5′ ATGCCCAGTTTGAAATGGACATC 3′) to these blocks. We performed touchdown PCR (94°C 2 min, then 12 cycles of 94°C, 30 s; A(–0.5°C/cycle)°C, 30 s; 68°C 1 min; 72°C 2 min (where the initial annealing temperature, A, was varied between 50 and 60°C), then 23 cycles of 94°C 30 s; 58°C 30 s; 72°C 2 min) with ∼10–100 ng of genomic DNA, 200 mM dNTPs, 1 µM primers, 75 mM Tris–HCl (pH 8.8 at 25°C), 20 mM (NH4)2SO4, 1 mg/ml BSA and 0.1% (v/v) Tween 20 with 1U Taq polymerase in 20 µl. The PCR product was gel purified and sequenced with the same primers. Sequences were aligned with CLUSTALX (21), and the alignments manually edited to remove spurious gaps. Species names and Genbank Accession Numbers are: Hampala macrolepidota GU390605, Puntius titteya GU390606, Puntius ticto GU390607, Puntius denisonii GU390609, Probarbus jullieni GU390610, Puntius narayani GU390611, Puntius conchonius GU390612, Puntius tetrazona GU390614, Puntius rhomboocellatus GU390615, Balantiocheilos melanopterus GU390616, Crossocheilus siamensis GU390617, Epalzeorhynchos sp. GU390618, Cyprinus carpio GU390620, Barbonymus schwanenfeldii GU390621, Cyprinella lutrensis GU390622, Leptobarbus hoevenii GU390623, Danio nigrofasciatus GU390624, Danio dangila GU390625, Puntius sachsii GU390626, Botia almorhae GU390627, Botia kubotai GU390628, Botia striata GU390629, Botia dario GU390630, Botia lohachata GU390631, Botia dario GU390632, Gyrinocheilus aymonieri GU390633, Pseudoplatystoma fasciatum GU390601, Pangasianodon hypophthalmus GU390602, Synodontis euptera GU390603, Kryptopterus bicirrhis GU390604, Cichlasoma salvini GU390634, Heros severus GU390635, Archocentrus sajica GU390636, Aequidens rivulatus GU390637, Melanochromis auratus GU390638, Pseudotropheus elongatus GU390639, Cyrtocara moorii GU390640, Sciaenochromis ahli GU390642, Labeotropheus fuelleborni GU390644, Copadichromis borleyi GU390645, Dimidiochromis compressiceps GU390646, Hemichromis bimaculatus GU390647, Paratilapia polleni GU390648, Thorichthys ellioti GU390649, Hyphessobrycon megalopterus GU390650, Paracheirodon innesi GU390651, Hyphessobrycon bentosi GU390653, Nematobrycon laioreti GU390654.
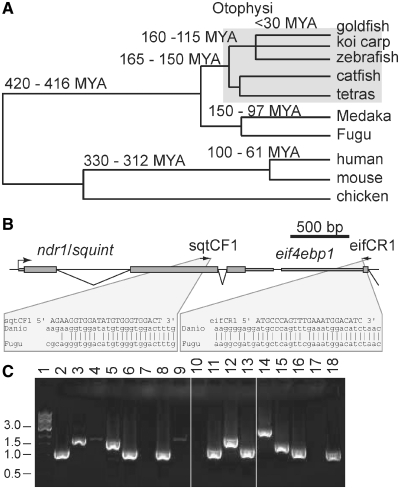
Figure 1.
Amplification of ndr1/squint (sqt) 3′-UTRs from various fish. (A) Phylogenetic tree, showing relationships between Otophysan fish and other model organisms. Divergence times are from (47), except for the cypriniforme-catfish, characin divergence, which is from (48), and the cyprinid last common ancestor, from (27,28). (B). Schematic representation of the sqt locus showing the positions of PCR primers (sqtCF1 and eifCR1, arrows) in sqt and the flanking gene, eif4ebp1. (C) Representative agarose gel, showing amplified fragments from a number of cypriniformes, catfish and tetras. Lane 1, 1 kb DNA ladder; 2, Crossocheilos siamensis; 3, Hyphessobrycon bentosi; 4, Synodontis eupterus; 5, Megalamphodus sweglesi; 6, Inlecyprius auropurpurous; 7, Nematobrycon laioreti; 8, Chela dadyburjori; 9, Astyanax spp.; 10, Nematobrycon palmeri; 11, Puntian narayani; 12, Popondichthys furcata; 13, Puntius fasciatus; 14, Pelteobagrus ornatus; 15, Kryptopterus bicirrhis; 16, Puntius denisoni; 17, Hemmigrammus bleheri; 18, Cyprinella lutrensis. 2, 6, 8, 11, 13, 16 and 18 are cypriniforms; 4, 14 and 15 are catfish; and 3, 5, 7, 9, 10 and 17 are characiforms (which includes tetras).
Structure predictions
Structures were predicted with the programmes Mfold (22), UTRScan (23); (http://www.ba.itb.cnr.it/BIG/UTRScan/) and RNAalifold (24,25).
Fluorescent mRNA injections
Primer sequences for sqt mutant constructs are available upon request. For transcription, constructs were linearized with Not I. Fluorescein or Alexa 488 labelled capped RNA was transcribed with SP6 RNA polymerase (Promega) in a reaction containing 0.5 mM rGTP, rATP and rCTP, 0.375 mM unlabelled rUTP (Roche) and 0.125 mM Chromatide Alexa 488 rUTP (Molecular Probes, Invitrogen), following the manufacturers instructions. Purified mRNA was injected into one-cell embryos, and live embryos were imaged at the four-cell stage using a Zeiss Axioplan2 upright microscope and CoolSNAP Photometrics camera (Roper Scientific). Breeding pairs of fish were pre-screened for high fertilization rates. Asymmetric localization was scored visually, and independently, by two individuals for each construct. Injected embryos that were not fertilized or did not cleave normally were discarded. Only embryos that had discrete puncta in one or two cells on one side of the embryo were considered to have asymmetric localization. For antisense morpholino oligo injections, fluorescent RNA was co-injected with 4 ng of the appropriate morpholino. The sqt TP miR430 and control morpholinos are as described (26). The sequence of the DLE morpholino is 5′ aaggagcatatccaaagtgc 3′.
RESULTS
Evolutionary conservation in zebrafish sqt DLE identifies discrete conserved blocks
In order to identify putative conserved elements at high resolution in zebrafish sqt DLE, we sequenced this region from a collection of closely related fish. Zebrafish belong to the order Cypriniformes (Figure 1A), along with goldfish, carp, minnows and barbs. This is a species-rich clade, containing ∼2600 species (out of a total of ∼20 000 teleosts); similarly, its sister orders, Siluriformes (catfish) and Characiformes (characins, tetras), are also speciose with ∼2300 and 1300 species, respectively. Cypriniformes belong to Otophysi, which is rapidly evolving, and contains a large number of families, genera and species [∼95% of all freshwater fish, and ∼25% of all teleosts (27)]. Since many of these fish can be obtained through the aquarium trade, we sequenced and aligned the ndr1 3′-UTR from a large number of more and less closely related species.
In order to amplify the ndr1 3′-UTR, we designed primers to highly conserved sequences in the coding regions of ndr1, and the downstream gene, eif4ebp1 (Figure 1B and C). We obtained ndr1 sequences from ∼30 cyprinids, six tetras and five catfish (See accession number section). Upon manual inspection of the alignments, we noted that ndr1 sequences aligned well within the orders, but tetra and catfish ndr1 sequences aligned poorly with cyprinid sequences. We also sequenced ndr1 from a number of other fish, including ∼15 cichlids, but these were too divergent to align to the cyprinid sequences, and too similar to each other to be informative, since most of the cichlids come from a single lake, Lake Malawi.
Alignments between species within one genus [e.g. Botia (a genus of loaches) and Danio], were too close, and did not provide any information. Sequences from different orders aligned only very weakly, and were also not informative. The most informative alignment is within the family cyprininae (last common ancestor ∼30 MYA) (27,28). In this alignment, the nucleotide similarity is high, and much of the divergence is due to insertion/deletion variants. Thus, the most striking feature of the alignment is that it breaks up into well-conserved blocks, separated by multiple, independent insertion/deletion variants or ‘indels’ (arrows in Figure 2A). The discrete blocks are consistent with functional elements separated by ‘linker’ sequences, which may allow small variations in spacing. The conserved blocks might correspond to protein-binding sites, or conserved stems in stem-loop structures, or both. Interestingly, almost the whole UTR consists of conserved blocks, including a highly conserved long block (∼100 nt) in the 3′ region of the UTR (Supplementary Figure S1). In injection assays, the full-length sqt RNA construct is more tightly localized than the minimal construct containing only the first 50 nt of the 3′-UTR. This suggests that the downstream conserved elements shown in the alignment may function cooperatively to confer precise localization. These additional elements are also interesting in view of the exquisite dose-sensitivity of the embryo to sqt RNA. The factors and mechanisms that regulate translation of sqt mRNA are not known.
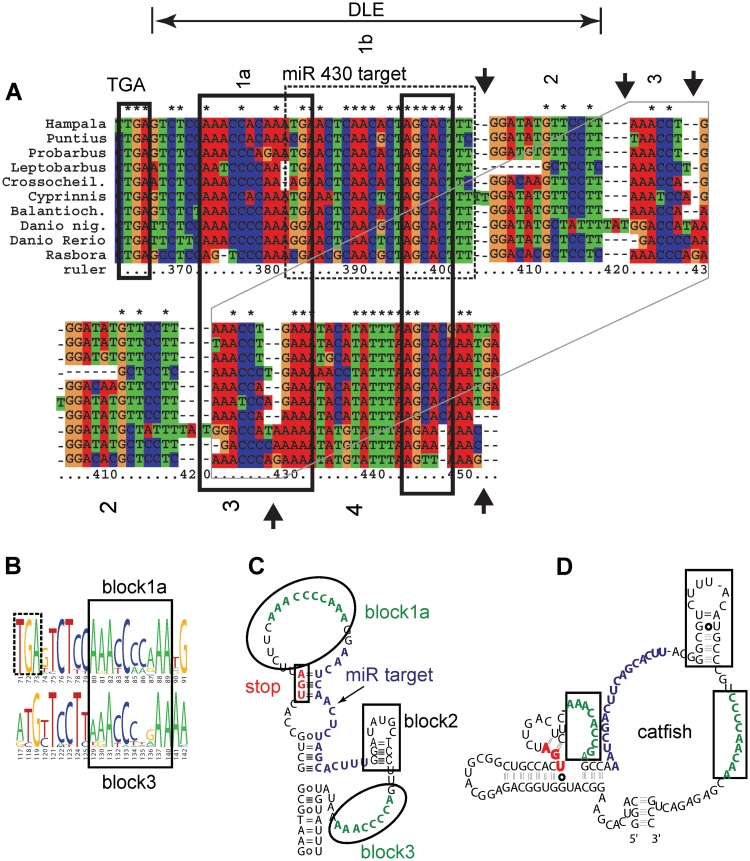
Figure 2.
Conservation in ndr1 3′-UTRs defines short conserved blocks and a predicted conserved secondary structure. (A) The DLE region is indicated above the alignment, and the stop codon is indicated (TGA). Strikingly, short, well conserved blocks of sequence (boxed, blocks 1–4) are separated by multiple independent insertion/deletion variants (‘indels’, arrows; gaps are indicated by dashes). Part of the alignment, blocks 2–4, is duplicated below the DLE region, to show the repeat of the putative AAACCCNRAA(N∼15)AGCAC motif (black boxes). Block 1b (dashed box) corresponds to a miR430 target (29). (B) A repeated sequence motif. The alignment was submitted to Weblogos (31,32) (http://weblogo.berkeley.edu/logo.cgi) to produce sequence logos, and inspected for repeated sequence motifs. Blocks 1a and 3 correspond to the consensus, AAACCCN(G/A)AA. (C and D) Conserved structures predicted from alignments by RNAalifold. The more divergent cyprininae sequences were used for the input alignment (Hampala macrolepidota GU390605, Puntius tetrazona GU390614, Probarbus jullieni GU390610, Leptobarbus hoevenii GU390623, Crossocheilus siamensis GU390617, Cyprinus carpio GU390620, Balantiocheilos melanopterus GU390616, Danio nigrofasciatus GU390624, D. rerio, Rasbora heteromorpha DQ080243.1) (C) Conserved secondary structure from Cyprinid alignment, overlapping the DLE, redrawn with the corresponding zebrafish sequence. Blocks 1a, 2 and 3 from the alignment are indicated on the structure. (D) Similar structure and sequence elements are present in the corresponding region of catfish ndr1, although catfish ndr1 3′-UTR sequences are somewhat divergent from zebrafish/cyprinids.
In the first 50 nt of the sqt 3′-UTR, corresponding to the minimal DLE sequences (19), two conserved blocks are evident (Figure 2A). When more divergent sequences are added to the alignment, block 1 resolves into two blocks, 1a and 1b (Figure 2A). Block 1b corresponds to a target site for miR430, which, in zebrafish, is responsible for degrading maternally deposited mRNAs at the mid-blastula transition (29), and similar to Xenopus (30). The spacing between blocks 1a and 1b is changed by only 1 nt by the observed indels. This spacing may be conserved possibly because block 1a overlaps with the miRNA target site. These conserved blocks are candidate components of the DLE.
Identification of putative protein-binding sequence motifs
We next asked whether these blocks contain any repeated sequence motifs. Cis-regulatory elements in DNA are typically composed of clusters of repeated binding sites for one or more transcription factors (7). Transcription factor-binding sites are typically degenerate, and are often represented by consensus sequences, position weight matrices, or graphically as sequence logos. It is possible that protein-binding sites in RNA have similar properties. In order to identify potential repeated motifs, we submitted the alignment to the web-based application, WEBLOGO [http://weblogo.berkeley.edu/logo.cgi (31)] which represents motifs as ‘sequence logos’ (32), in which the height of the each letter reflects the frequency with which the corresponding base occurs at that position. We observed a repeated degenerate motif, AAACCCNRAA (Figure 2B), which corresponds to blocks 1a and 3 (Figure 2A). It is present in, and just adjacent to, the DLE, and is a candidate component of the DLE. We did not observe an obvious reverse complement of the AAACCCNRAA motif, which suggests that ‘block 1’ may correspond to a single-stranded RNA sequence motif. It is possible that the element corresponding to block 1a overlaps with the miRNA target site. Interestingly, there is a conserved sequence, AGCAC, present in an apparently conserved spacing, downstream of the two AAACCCNRAA motifs, i.e. in both the miRNA target (specifically, in the seed sequence), and in block 4. The element might, therefore, correspond to the consensus AAACCCNRAA(N∼15)AGCAC. Such a motif might be bound by a protein with independent RNA-binding domains, or by a protein complex. Since the second AGCAC is missing in some species (zebrafish, for instance), this arrangement cannot be strictly required, and may perhaps simply increase binding affinity to some protein or complex.
Prediction of conserved DLE structure
RNA secondary structures are important in recognition of many RNA elements by proteins. We used a number of programs to visualize putative conserved secondary structures. One such program, RNAalifold (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi), uses alignments to predict conserved secondary structures (25). We submitted various alignments containing closely or distantly related sequences, and with varying lengths of flanking sequence. As expected, the more divergent the sequences were, the more they constrained structure prediction. RNAalifold stably predicts a structure that overlaps the DLE (Figure 2C), which contains two single-stranded loops, each of which corresponds to an instance of the AAACCCNRAA motif. The miR430/AGCAC motif has no clear secondary structure, being predicted as single stranded in one instance, and partly single stranded in the other. There is a predicted short internal stem-loop between the two loops, which corresponds to ‘block 2’ from the alignment in Figure 2A. A similar arrangement is predicted in catfish ndr1, overlapping the first 50 nt of the 3′-UTR, containing conserved single-stranded miR430/AGCAC sequence, and single-stranded AC rich motifs flanking a small stem-loop with a GC-rich stem (Figure 2D). Thus, DLE conservation in cyprinids and catfish suggests three candidate DLE elements: a single-stranded motif, AAACCCNRAA; the single-stranded AGCAC motif; and a short stem-loop.
Functional analysis of identified motifs
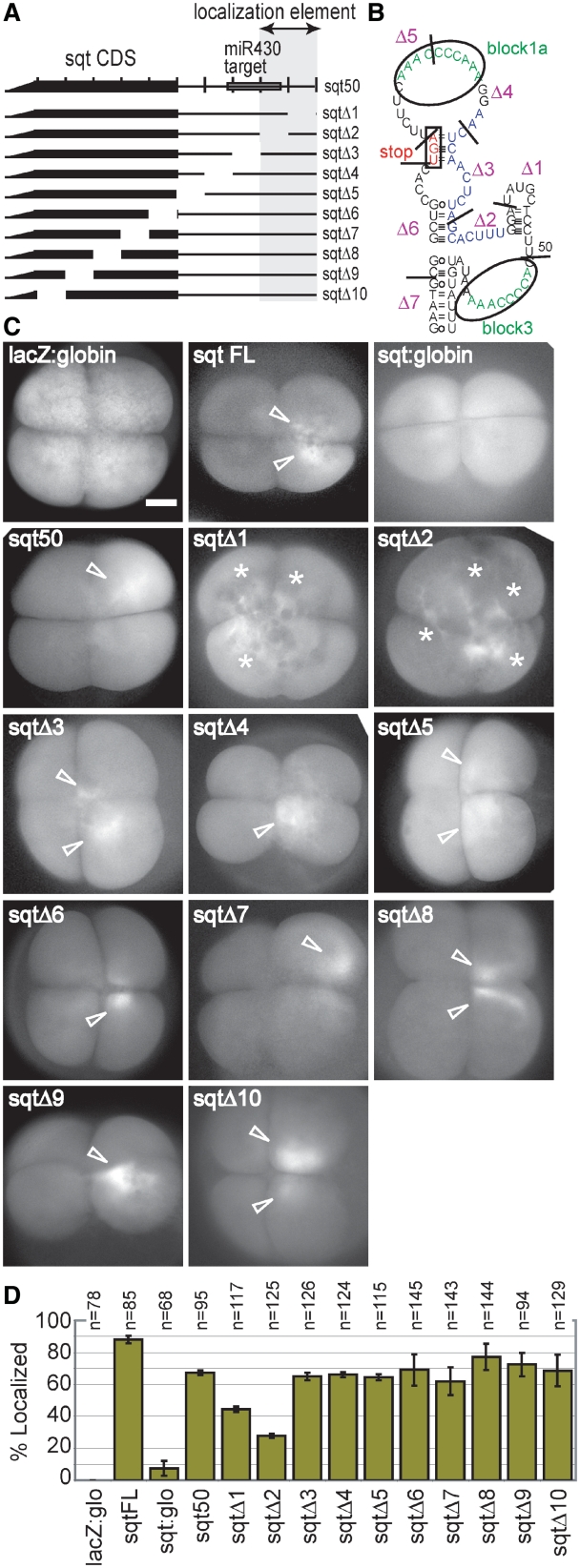
To investigate the role of the conserved DLE elements in localization, we injected fluorescently labelled RNAs, including a series of 10-nt deletions in the DLE region (Figure 3A and B, and Supplementary Table S1), into one-cell zebrafish embryos and monitored localization. The negative control, lacZ:globin RNA, is uniformly distributed in the cytoplasm of all four cells at the four-cell stage (Figure 3C and D). Similarly, in embryos injected with sqt:globin, in which the non-localizing globin 3′-UTR is substituted for the sqt 3′-UTR, the RNA is usually distributed uniformly (67%, n = 68), sometimes un-localized in a stringy pattern (25%, n = 68), and is only rarely localized (7%, n = 68, Figure 3C and D). In contrast, sqt FL, which contains the full-length 3′-UTR, is clearly asymmetric, being restricted to only one or two cells (88%, n = 85 Figure 3C and D). RNA synthesized from the minimal localizing construct, sqt50, which contains only the first 50 nt of the 3′-UTR (19), is also localized, albeit at a lower frequency (67%, n = 95; Figure 3C and D). Deletions Δ3 to Δ10 have little or no effect on localization (sqtΔ3–sqtΔ10, Figure 3C and D). Since sqtΔ3–7 together lack most of the main stem, and sqt50, which localizes, lacks the distal part of the stem, this predicted stem is dispensable for localization. Deletion 4 (sqtΔ4, Figure 3B–D), which removes most of the block 1a AAACCCNRAA motif, has no discernable effect on localization, possibly because it functions in some other process than localization, or perhaps because it is redundant with the stem-loop and AGCAC sequence motifs.
Figure 3.
Deletion analysis identifies localization sequences. (A) Schematic diagram of the minimal localizing construct, sqt50, and deletion mutants. (B) A predicted structure of the DLE region, showing the position of the deletions. (C). Fluorescently labelled sqt RNAs and deletion mutants sqtΔ1–sqtΔ10 shown in (A), were injected at the one-cell stage, and imaged from the animal pole at the four-cell stage. (D) Graph showing frequency of localization of the constructs. Note that the negative control RNA, lacZ:globin, is uniformly distributed in the cytoplasm, and that the minimal localizing construct sqt50 is asymmetrically localized (arrowheads). In contrast, sqtΔ2 and sqtΔ1 are frequently ectopically localized to ‘stringy’ structures in the cytoplasm (asterisks). Scale bar represents 100 µm.
In contrast, deletions 1 and 2 (sqtΔ1 and sqtΔ2, Figure 3C and D) markedly reduce the frequency of correct localization (44%, n = 117, P < 0.01; and 28%, n = 125, P < 0.01, respectively). Remarkably, these two RNAs have a new patchy distribution pattern, and are frequently restricted to stringy patches in the cytoplasm (asterisks in Figure 3C), but fail to localize asymmetrically. This may be because these RNAs do not have a high enough affinity for the localization machinery, or may have failed to bind some component, and consequently either cannot be properly localized, or fail to complete some stage in localization. It is also notable that neither deletion completely abolishes localization of the RNA. sqtΔ2 lacks the AGCAC motif, which is in the seed sequence of the miR430 target, together with the first 3 nt of the conserved stem-loop in block 2 (Figure 3B). sqtΔ1 lacks all but 3 nt of block 2. Neither deletion completely abolishes localization, suggesting that these two elements function cooperatively.
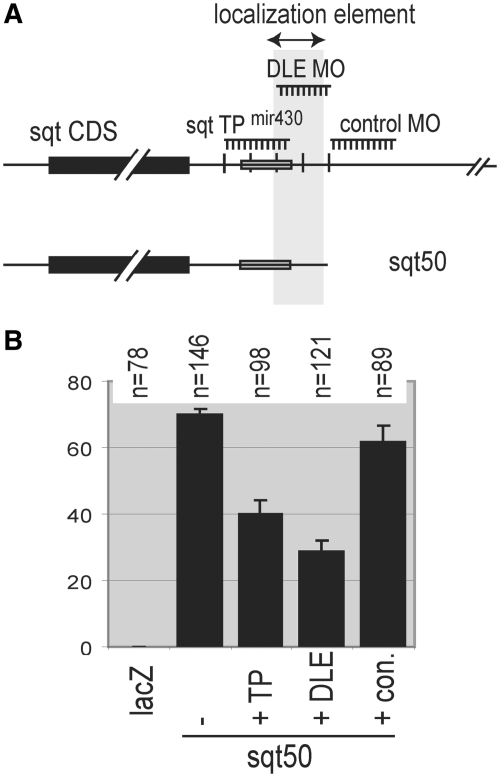
Intriguingly, the localization element appears to overlap with the miR430 target site. It has been shown that an antisense morpholino oligo complementary to the target site can block miR430 regulation of sqt RNA (26). This morpholino also has the potential to occlude the localization element, so we wondered whether it would affect localization. To test this possibility, we injected the minimal sqt50 RNA with the Target site Protector (sqt TP miR430) or control morpholino, and a morpholino, DLE MO, designed to block the region corresponding to the sqtΔ1 and sqtΔ2 deletions (Figure 4, Supplementary Table S2). The control morpholino, which is not complementary to sqt50 RNA, has little or no effect on localization (62%, n = 89). In contrast, the DLE morpholino (28%, n = 71, P < 0.01), and to a lesser extent, the miR430 target site protector (40%, n = 98, P < 0.01), both reduce localization (Figure 4B, Supplementary Table S2). This effect is likely due to blocking of DLE-binding factors, rather than interference with miR430 binding, because the DLE blocking morpholino has a stronger effect than the miR430 target protector, and because miR430 is not present or active until the mid blastula transition [(29,33), about 3h after the four-cell stage].
Figure 4.
Antisense morpholinos to the Dorsal Localization Element block localization. (A) Schematic diagram, showing the sqt RNA, the minimal sqt50 construct and position of morpholinos. The miR430 target site protector (sqt TP miR430), and control are as described (26). (B) Morpholinos that bind to the DLE reduce localization, in contrast to the control morpholino. The DLE morpholino fully covers the DLE, and significantly reduces localization more strongly than the miR430 target site protector (P < 0.01).
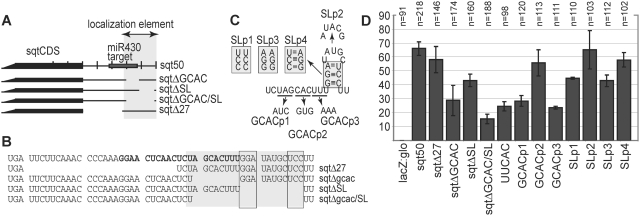
To more precisely test the elements, we made deletions of the AGCAC and stem-loop separately, and together (Figure 5 and Supplementary Table S3). Deletion of the AGCAC sequence (sqtΔGCAC, Figure 5A, B and D) strongly reduces localization (29%, n = 174, P < 0.01), and deletion of the predicted stemloop (sqtΔSL, Figure 5A, B and D) mildly reduces localization (43%, n = 160, P < 0.01). Deletion of both together (sqtΔGCAC/SL, Figure 5A, B and D) further reduces localization (15%, n = 188; significantly different from sqtΔGCAC, P < 0.05), but does not abolish it, similar to deletion of the entire 3′-UTR from sqt (sqt:glo, 7% localized, n = 68, Figure 3C and D). In contrast, deletion of all the 3′-UTR except for the AGCAC motif and predicted stem-loop (sqtΔ27, Figure 5A, B and D), has little effect on localization (58%, n = 146), indicating that the AGCAC and stem-loop motifs are necessary and sufficient for efficient localization of the sqt coding sequence.
Figure 5.
Mutagenesis defines localization motifs. (A) Schematic diagram of the deletion mutants. The localization element is indicated by grey shading. (B) Sequence of deletions, with the miR430 target highlighted in bold, the localization element in grey shading, and the base-paired stem residues indicated by shaded boxes in (B) and (C). (C) Schematic diagram of the predicted single-stranded motif and stem-loop, showing the mutations. Mutated residues correspond to the grey shading in (A) and (B). (D) Graph showing frequency of localization of deletion and point mutant RNAs. Deletion of the first 27 nt of the 3′-UTR has little or no effect on localization. Deletion of the AGCAC motif strongly reduces localization. Deletion of the predicted stem-loop reduces localization to a lesser extent. Simultaneous deletion of the AGCAC motif and the predicted stem-loop reduces localization further, but does not abolish it, indicating that the AAACCCAAA motif, or elements in the coding sequence, makes a slight contribution. Point mutations in the AGCAC motif, including changing the motif to the match the ventral localization motif, ‘UUCAC’, strongly reduce localization. Point mutations provide support for the predicted stem-loop, since mutations to either strand of the predicted stem-loop reduce localization, whereas compensatory mutations that restore the stem-loop restore localization to near wild-type levels.
To test the predicted stem-loop motif, we mutated the predicted loop, and made disruptive and compensatory mutations to the predicted stem. Mutations in the predicted loop (SLp2, Figure 5A, C and D) have little or no effect on localization (65%, n = 103). In contrast, mutation of either strand of the stem (SLp1 and SLp3, Figure 5A, C and D) reduces localization (45%, n = 110, P < 0.01 and 41%, n = 68, P < 0.01, respectively). The compensatory mutation that restores base pairing (SLp4, Figure 5A, C and D) rescues localization to near wild-type levels (58%, n = 102; significantly different from SLp1, P < 0.01, and SLp3 P < 0.01), indicating that the structure, and possibly not the sequence, of the stem-loop contributes to localization.
Previously, CAC-containing motifs were identified as being over-represented in localized chordate RNAs (34). More specifically, a GCAC or GCACUU motif is over-represented in maternal RNAs localized to the germ plasm, and a UUCAC motif is over-represented in maternal RNAs localized to the vegetal pole of the oocyte. The wild-type sequence of the sqt motif is UAGCACUUU. Mutation of the central CAC to UAGgtgUUU (GCACp2, Figure 5A, C and D) reduces localization weakly (55%, n = 113, p < 0.05). In contrast, mutation of the flanking sequence to aucCACUUU (GCACp1, Figure 5 A, C and D) or UAGCACaaa (GCACp3, Figure 5A, C and D) strongly reduces localization (28%, n = 120, P < 0.01 and 23%, n = 82, P < 0.01, respectively). Mutation of the motif to UuuCACUUU (UUCAC, Figure 5A, C and D), to match the vegetal localization motif, also strongly reduces localization (24%, n = 98, P < 0.01). Taken together, these results show that the consensus of the sqt motif is approximately AGCACUUU, similar to the germ plasm localization motif, and distinct from the UUCAC vegetal localization motif.
DISCUSSION
We have shown that an AGCACUUU motif is prominently involved in localization of the sqt RNA. CAC-containing motifs have previously been shown to be over-represented in localized chordate RNAs (34). A UUCAC motif is overrepresented in RNAs localized to the vegetal pole of Xenopus oocytes (34), and is required for localization [reviewed in (35)]. The UUCAC motif is localized via binding to Vg1RBP (36,37). A GCAC motif is required for localization of germ plasm RNAs (38,39). The sqt AGCACUU DLE motif is intriguingly similar to the germ cell localization motif GCAC, raising the question of whether the same protein binds both motifs. It will be interesting to see whether these two localization pathways share machinery, including RNA-binding proteins.
If the dorsal localization motif and germ plasm motif are indeed the same, in the sense of being recognized by the same protein, the question of specificity arises. Localization of sqt RNA has not been detected in primordial germ cells [(19,40,41), Gilligan,P.C. et al., unpublished data]. Furthermore, as shown in Figure 3, we did not detect any fluorescent sqt signal in the furrows in four-cell stage embryos. In contrast, zebrafish vasa RNA is localized to the germ plasm at the cleavage furrows in four-cell stage embryos (42). Other germ plasm RNAs, such as nanos, are distributed throughout the blastoderm, and selectively protected from degradation in the germ plasm (42). It is not possible to see the injected fluorescent sqt RNA past the 16-cell stage, but injection of GFP:sqt3′-UTR RNA shows bright fluorescence at later stages in anterior tissues, but not in germ cells [Gilligan,P.C. et al., unpublished data; see also Figure 3b in (29)]. Therefore, neither endogenous sqt nor ectopically injected sqt RNA localizes to the germ plasm.
If the dorsal and germ plasm elements are the same, one possibility is that specificity is achieved by a combinatorial code. It is conceivable that the AGCAC motif plus the conserved stem-loop drives localization to future dorsal, whereas the AGCAC motif plus some undetermined motif, might drive localization to germ plasm. The possibility of an overlapping localization code is intriguing. In flies, germ cell RNAs are initially localized to the posterior pole, then to primordial germ cells, which arise at the posterior pole (35,43). Similarly, in frogs and fish, germ cell RNAs are initially localized (via the METRO structure in frogs) to the vegetal pole which will become the posterior end of the embryo, and then after fertilization, these RNAs are re-localized to prospective primordial germ cells (35,42). In both fish and frogs, there are maternal dorsal determinants [wnt11 RNA and associated proteins in frogs (40,41), and unknown determinants in Fundulus (44) and zebrafish (19)] that are initially localized to the vegetal pole, and later to future dorsal. Thus, there are RNAs that ultimately localize to the vegetal pole, germ cells and future dorsal, that transit via the vegetal pole of the oocyte, possibly consistent with overlapping machinery.
In a striking molecular parallel, in Drosophila, the posteriorly localized oskar RNA contains clusters of UUUAY motifs (45), similar to the UUCAC motifs in vegetally localized Xenopus RNAs. The oskar UUUAY motifs are required for translation and posterior anchoring of the oskar RNA, and the Drosophila Vg1RBP homolog binds these repeats in vivo (45).
Since the AGCAC motif overlaps the miR430 target site, it is formally possible that miR430 is involved in sqt RNA localization. For instance, maternal dazl, tudor and nanos RNAs are translationally repressed and degraded in the somatic blastoderm via a miR430 target site (which contains the GCACUUU motif), but are specifically protected from degradation in the germ plasm (33,46). In contrast, sqt is actively transported, rather than selectively protected from degradation, as we were previously unable to detect any significant degradation during localization (19). Furthermore, although the miR430 target site protector morpholino (which overlaps the localization element) reduces localization, the DLE morpholino reduces localization more effectively (Figure 4B). In addition, our mutant AGgtgUU, has the weakest effect on localization, and is very similar to a mutation, AGgtCUU, reported to abolish binding of miR430, resulting in increased GFP expression from a GFP:sqt3′-UTR chimeric RNA (29). Finally, sqt localization takes place by the four-cell stage, when miR430 does not appear to be present or active (29). Taken together, it seems unlikely that miR430 is directly involved in sqt RNA localization.
Another possibility is that the DLE-binding protein and the miR430 compete for the site. In this scenario, the protein that binds to the DLE motif in the miR430 seed sequence, might also bind to the miR430 target site in the germ plasm RNAs, and may contribute to their protection from degradation, or to their localization within the primordial germ cells, to the nuage. However, this is speculative, and the proteins Deadend (Dnd1) and DAZL have already been shown to be required for protection of these RNAs (33,46).
Post-transcriptional regulation of RNA, such as sub-cellular localization, is in general poorly understood, and it is becoming increasingly clear that RNA localization is a pervasive phenomenon. In particular, protein-binding sites in RNA are poorly understood. In order to gain insight into RNA recognition by proteins, it is highly desirable to understand what features in RNA-binding sites are functionally constrained. To do this, we performed high resolution ‘phylogentic footprinting’ of the sqt 3′-UTR from a number of species closely related to zebrafish. This turns out to be a straightforward and efficient strategy since the relevant species of fish are readily available in aquarium shops, and all the PCR products in this study were directly sequenced after gel purification. Interestingly, there are many conserved elements in the sqt 3′-UTR, which supports the idea that many 3′-UTRs are likely to function as ‘cis-regulatory modules’ of RNA, analogous to promoters and enhancers in DNA. It would be interesting to test these elements for localization or translation regulation activity.
In addition to the DLE, there are a number of other conserved elements downstream in the sqt 3′-UTR. These may be involved in localization and/or translation regulation. Consistent with this idea, full-length sqt RNA localizes more efficiently than sqt RNA containing only the minimal DLE. This suggests that there are additional localization elements in the UTR. With regard to translational regulation, it appears that many or most localized RNAs are translationally repressed, at least until they are localized. Also, since the zebrafish embryo is exquisitely sensitive to the dose of Sqt, it is possible that there are additionally, unknown mechanisms to regulate Sqt activity via translational regulation. It will be important in future to map these also.
The short predicted stem-loop also appears to be distinct from known RNA elements. Taken together, this suggests that the trans-factor(s) that recognize the sqt DLE may be distinct from known RNA-binding factors. This, together with the demonstration in flies that a majority of transcripts have some restricted intracellular distribution, further implies that there still remain additional RNA localization machineries to be discovered. Identification and refinement of more elements such as these should eventually allow reliable prediction of localization elements, and localized RNAs.
ACCESSION NUMBERS
GU390601–GU390607, GU390609–GU390612, GU390614–GU390618, GU390620–GU390640, GU390642, GU390644–GU390651, GU390653, GU390654.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Temasek Life Sciences Laboratory, Singapore (to K.S.); Singapore Millennium Foundation (to P.C.G.); Temasek Life Sciences Laboratory (to P.C.G.). Funding for open access charge: Temasek Life Sciences Laboratory.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Sampath laboratory for suggestions and discussions. The authors thank Byrappa Venkatesh for critical reading of the manuscript.
REFERENCES
- 1.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chabanon H, Mickleburgh I, Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief. Funct. Genomic. Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- 4.Macdonald PM, Kerr K. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol. Cell Biol. 1998;18:3788–3795. doi: 10.1128/mcb.18.7.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serano TL, Cohen RS. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- 6.dos Santos G, Simmonds AJ, Krause HM. A stem-loop structure in the wingless transcript defines a consensus motif for apical RNA transport. Development. 2008;135:133–143. doi: 10.1242/dev.014068. [DOI] [PubMed] [Google Scholar]
- 7.Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- 8.Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- 9.Simmonds AJ, dosSantos G, Livne-Bar I, Krause HM. Apical localization of wingless transcripts is required for wingless signaling. Cell. 2001;105:197–207. doi: 10.1016/s0092-8674(01)00311-7. [DOI] [PubMed] [Google Scholar]
- 10.Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 11.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7:1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung HK, Serano TL, Cohen RS. Evidence for a highly selective RNA transport system and its role in establishing the dorsoventral axis of the Drosophila egg. Development. 1992;114:653–661. doi: 10.1242/dev.114.3.653. [DOI] [PubMed] [Google Scholar]
- 13.Lantz V, Ambrosio L, Schedl P. The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding proteins and has localized transcripts in ovaries and early embryos. Development. 1992;115:75–88. doi: 10.1242/dev.115.1.75. [DOI] [PubMed] [Google Scholar]
- 14.Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- 15.Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu JM, Liu DR. Discovery of a mRNA mitochondrial localization element in Saccharomyces cerevisiae by nonhomologous random recombination and in vivo selection. Nucleic Acids Res. 2007;35:6750–6761. doi: 10.1093/nar/gkm777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rabani M, Kertesz M, Segal E. Computational prediction of RNA structural motifs involved in posttranscriptional regulatory processes. Proc. Natl Acad. Sci. USA. 2008;105:14885–14890. doi: 10.1073/pnas.0803169105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23:1546–1558. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gore AV, Maegawa S, Cheong A, Gilligan PC, Weinberg ES, Sampath K. The zebrafish dorsal axis is apparent at the four-cell stage. Nature. 2005;438:1030–1035. doi: 10.1038/nature04184. [DOI] [PubMed] [Google Scholar]
- 20.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pesole G, Liuni S. Internet resources for the functional analysis of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Trends Genet. 1999;15:378. doi: 10.1016/s0168-9525(99)01795-3. [DOI] [PubMed] [Google Scholar]
- 24.Fekete M, Hofacker IL, Stadler PF. Prediction of RNA base pairing probabilities on massively parallel computers. J. Comput. Biol. 2000;7:171–182. doi: 10.1089/10665270050081441. [DOI] [PubMed] [Google Scholar]
- 25.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 2002;319:1059–1066. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
- 26.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 27.Nelson J. Fishes of the World. New York: John Wiley and Sons; 1994. [Google Scholar]
- 28.Zardoya R, Doadrio I. Molecular evidence on the evolutionary and biogeographical patterns of European cyprinids. J. Mol. Evol. 1999;49:227–237. doi: 10.1007/pl00006545. [DOI] [PubMed] [Google Scholar]
- 29.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 30.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 31.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 35.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol. Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 36.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12:1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 1998;8:489–496. doi: 10.1016/s0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 38.Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol. Biol. Cell. 2004;15:4669–4681. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choo S, Heinrich B, Betley JN, Chen Z, Deshler JO. Evidence for common machinery utilized by the early and late RNA localization pathways in Xenopus oocytes. Dev. Biol. 2005;278:103–117. doi: 10.1016/j.ydbio.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 40.Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev. Biol. 1998;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- 41.Erter CE, Solnica-Krezel L, Wright CV. Zebrafish nodal-related 2 encodes an early mesendodermal inducer signaling from the extraembryonic yolk syncytial layer. Dev. Biol. 1998;204:361–372. doi: 10.1006/dbio.1998.9097. [DOI] [PubMed] [Google Scholar]
- 42.Raz E. Primordial germ-cell development: the zebrafish perspective. Nat. Rev. Genet. 2003;4:690–700. doi: 10.1038/nrg1154. [DOI] [PubMed] [Google Scholar]
- 43.Wilson JE, Macdonald PM. Formation of germ cells in Drosophila. Curr. Opin. Genet. Dev. 1993;3:562–565. doi: 10.1016/0959-437x(93)90091-3. [DOI] [PubMed] [Google Scholar]
- 44.Oppenheimer JM. The development of isolated blastoderms of Fundulus heteroclitus. J. Exp. Zool. 1936;72:247–269. [Google Scholar]
- 45.Munro TP, Kwon S, Schnapp BJ, St Johnston D. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J. Cell Biol. 2006;172:577–588. doi: 10.1083/jcb.200510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS ONE. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benton MJ, Donoghue PC. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 48.Briggs JC. The biogeography of otophysan fishes (Ostariophysi: Otophysi): a new appraisal. J. Biogeogr. 2005;32:287. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.