Abstract
Retroviruses require both spliced and unspliced RNAs for replication. Accumulation of Rous Sarcoma virus (RSV) unspliced RNA depends upon the negative regulator of splicing (NRS). Its 5′-part is considered as an ESE binding SR proteins. Its 3′-part contains a decoy 5′-splice site (ss), which inhibits splicing at the bona fide 5′-ss. Only the 3D structure of a small NRS fragment had been experimentally studied. Here, by chemical and enzymatic probing, we determine the 2D structure of the entire RSV NRS. Structural analysis of other avian NRSs and comparison with all sequenced avian NRSs is in favour of a phylogenetic conservation of the NRS 2D structure. By combination of approaches: (i) in vitro and in cellulo splicing assays, (ii) footprinting assays and (iii) purification and analysis of reconstituted RNP complex, we define a small NRS element retaining splicing inhibitory property. We also demonstrate the capability of the SR protein 9G8 to increase NRS activity in vitro and in cellulo. Altogether these data bring new insights on how NRS fine tune splicing activity.
INTRODUCTION
For all retroviruses, alternative splicing plays a key role in the modulation of gene expression. After integration within the host genome, the proviral genome is transcribed as a primary transcript by RNA polymerase II. A large number of the transcribed molecules remain unspliced. They are needed to serve as genomes for new virions and as messenger RNAs for translation of the Gag–Pol protein precursor. The remaining part of the primary transcript undergoes alternative splicing in the nucleus to generate mRNAs for production of other retroviral proteins. Rous Sarcoma virus (RSV), which was the first discovered replication competent oncogenic retrovirus, exemplified this situation (1). This simple avian retrovirus belongs to the avian sarcoma/leucosis virus (ASLV) family. It contains the gag, pol and env genes common to all retroviruses and codes the oncogenic protein v-Src. ASLV viruses deserve interest because of the damage they generate in poultry breeding. In addition, RSV is an interesting, already well-studied model, to investigate mechanisms involved in the regulation of retroviral alternative splicing (2). Its RNA contains a single 5′-splice site (ss) (position 398 as referred to the transcription start site) and two 3′-ss in competition one with the other, the env and src 3′-ss (positions 5078 and 7054, respectively). Coupling of the env 3′ or src 3′-ss with the 5′-ss leads to the production of the env and src mRNAs, respectively.
The low level of RSV RNA splicing (20–30%) is due to the presence of suboptimal splicing sites and to the presence of strong splicing inhibitory elements. The branch point sequence of the env 3′-ss is not canonical and the polypyrimidine tract of src 3′-ss is suboptimal (3–6). Splicing at the src 3′-ss is specifically down regulated by an upstream sequence named SSS for suppressor of src splicing (SSS) (7). SSS seems to bind an unknown factor specific to chicken cells and its mechanism of action is not elucidated. A dr1 direct repeat flanks the src gene. It was first identified as a constitutive transport element (CTE) promoting the accumulation of unspliced viral RNA into the cytoplasm (8,9). This element may in addition contain a second silencing element acting specifically on the src 3′-ss (10,11). A third negative cis-splicing regulatory sequence, called the negative regulator of splicing (NRS), prevents splicing at both 3′-ss (12–15). NRS (positions 703–930) is located in the gag gene ∼300 nt downstream from the 5′-ss.
NRS was found to inhibit splicing of a heterologous myc intron as well in vitro as in cellulo. Deletion of an internal 76-nt long segment (nucleotides 799–874) (NRS Δ76) had a limited effect on NRS activity (13). Thus, NRS Δ76 was defined as the minimal sequence required for splicing inhibition activity. Two segments of the NRS were described as having an important role for splicing inhibition: the NRS 5′-part (positions 700–800, NRS-5′) was found to have splicing enhancer activity in an heterologous context, the NRS 3′-terminal part (positions 801–930, NRS-3′) contains two overlapping 5′-ss sequences. One is capable of binding U1 snRNA (major site), the other one binds the U11 snRNP (minor site) (16–18). U1 snRNP binding does not induce splicing and is required for the inhibition activity, suggesting that the degenerated major 5′-ss (five out of eight matches fit to the 5′-ss consensus) is a decoy 5′-ss (17). Unlike U1 snRNP, U11 snRNP binding to the minor 5′-ss is dispensable for splicing inhibition and is just expected to have a moderate negative effect on U1 snRNP binding to the major site (17,18). NRS-5′-had been shown to bind SR proteins and hnRNP H (19–21). Binding of SR proteins to the NRS-5′ segment was proposed to enhance U1, but not U11 snRNP recruitment (18). The NRS appears to function as a non-productive decoy 5′-ss, which competes with the authentic viral 5′-ss, and forms abortive spliceosomal complexes when coupled to one of the two 3′-ss. In vitro analysis demonstrated that human U5-220kD protein (hPrp8) does not cross link to the NRS pseudo 5′-ss, highlighting the fact that the abortive complex cannot switch into a catalytic complex (22).
A computer assisted 2D structure model of the NRS element was proposed long time ago (13). In this model, the NRS-5′ and NRS-3′ regions are paired together. Our earlier chemical and enzymatic probing of part of the NRS structure in collaboration with the Beemon group showed that the NRS-3′ region, encompassing nucleotides from positions G910 to U926, forms an individual stem–loop structure and this was the basis for an NMR-3D structure analysis of this NRS fragment (23). Formation of the identified stem–loop structure is incompatible with the proposed 2D structure of entire NRS. An experimental analysis of the 2D structure of the entire NRS element was therefore needed to get a clear answer on how NRS folds. As determination of RNA 3D structure by NMR can only be performed on short RNAs, only the RNA probing method based on utilization of chemical and enzymatic probes was adapted to this purpose. It should be pointed out that RNA 2D structure was already shown to play an important role in pre-mRNA splicing regulation (24–26). Therefore, determination of the NRS structure was expected to bring some insight on how NRS inhibits splicing.
In this article, we performed an experimental analysis of the entire RSV and ALV LR9 NRSs (27,28) 2D structures. The data provide evidence for a cruciform structure different from the previously proposed computer-based 2D structure. In the new cruciform structure, NRS-3′ forms an individual stem–loop structure, which is in accordance with previous NMR data (23). The NRS-5′ segment that binds ASF/SF2 also forms a stem–loop structure. Based on these data and in order to test for phylogenetic conservation of the NRS 2D structure, we undertook a comparative analysis of the possible 2D structures of avian retrovirus NRSs using available avian retrovirus nucleotide sequences. In parallel, we tested the NRS activity of truncated forms of the RSV NRS by in vitro splicing assays using a hybrid construct derived from HIV-1. By this approach, we identified a small discontinuous 114-nt element as the minimum element required for splicing inhibition. Then, we used a new affinity chromatography procedure (29) to define which of the produced truncated RSV NRSs can recruit the SR proteins ASF/SF2, SC35 and 9G8. Our finding of a strong binding of the SR protein 9G8 on WT and some of the truncated NRSs stimulated us to test the activation role of this SR protein on RSV NRS activity. By in vitro and in cellulo assays, we demonstrated a strong stimulatory effect of protein 9G8 on RSV NRS activity and we identified its binding site on RSV NRS by footprinting assays.
MATERIALS AND METHODS
DNA constructs
The 2D structure of NRS WT, NRS Δ76, LR9 and ΔLR9 RNAs was analysed on transcripts produced from PCR products. PCR products were amplified from the earlier described pRSVNeo-Int, pΔ76Neo-Int, pLR9 and pΔLR9 plasmids (13,28). Plasmids pRSVNeo-Int, pΔ76Neo-Int contain DNA sequences of the WT and Δ76 RSV NRSs from strain Prague C (PrC) (13), respectively. The sense primer (O-2954, Supplementary Table S1) contained a T7 RNA polymerase promoter and the antisense primer (O-2955, Supplementary Table S1) contained the unrelated AAATT sequence at its 5′-end (Figure 1A).
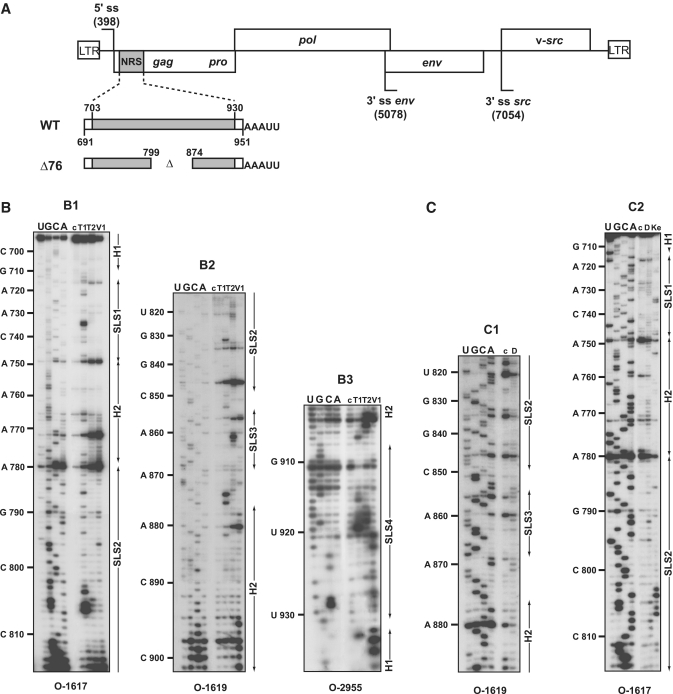
Figure 1.
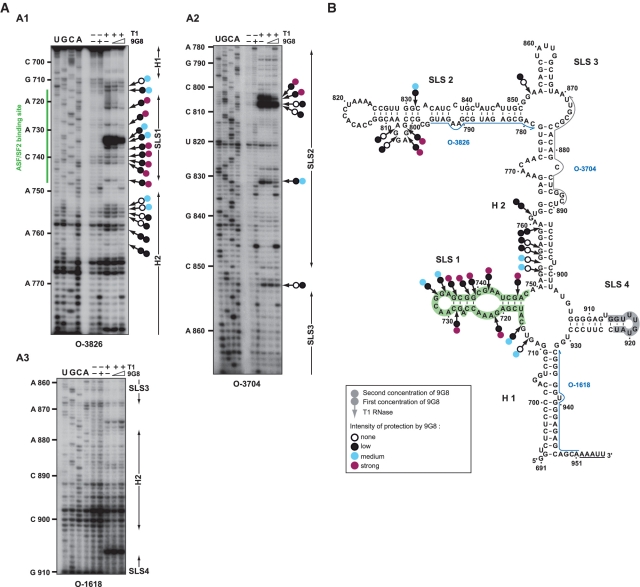
(A) Schematic representation of the RSV Prague C (PrC) genome and of the NRS RNAs used for secondary structure analyses. The architecture of the RSV PrC genome is shown on top of the Figure. The 5′-ss and 3′-ss are indicated and boxes represent the open reading frames. The grey box depicts the NRS element (nucleotides 703–930) located in the gag gene used for secondary structure experiments. Viral sequences belonging to the RSV segments located upstream and downstream from NRSs are in white. The non-viral AAAUU sequence at the 3′-end of the transcripts is shown. Delimitations of NRS are indicated according to NRS nucleotide numbering (48). (B and C) Primer extension analyses of enzymatic cleavages and chemical modifications of NRS WT. (B) Primer extension analyses of enzymatic cleavages in NRS WT (position 691– 951) (B1–B3), (C) primer extension analyses of chemical modifications in NRS WT (position 710 to 890) (C1 and C2). T1, T2, V1, D and Ke above the lanes indicate T1, T2, V1 RNase digestions and DMS or Kethoxal modifications, respectively. Conditions for digestions, modifications and reverse transcription are given in ‘Materials and Methods’ section. Lanes c correspond to control experiments with untreated RNA; lanes U, G, C and A correspond to the sequencing ladder. Primers used for reverse transcription are indicated below each autoradiogram (see Supplementary Table S1 for their nucleotide sequences). Nucleotide numbering according to (48) is indicated on the left side of the autoradiograms. Positions of the stem–loop structures (SLS1, SLS2, SLS3 and SLS4) and the helices (H1 and H2) are shown on the right side of the autoradiograms.
To get constructs containing WT and Δ76 NRS coding sequences under the control of a T7 RNA polymerase promoter, which are fused to a DNA sequence coding 3′-binding sites for the coat protein of phage MS2, the MS2 DNA sequences were excised from plasmid pADML-M3 (30) by BamHI–HindIII digestions and were subcloned into plasmid pUC18 (pMS2 plasmid). To obtain the control pT7MS2 plasmid, a T7 promoter was generated by annealing primers O-4672 and O-4673 and the resulting DNA was inserted between the BamHI and SacI sites located upstream of the MS2 DNA in plasmid pMS2. To get pNRS-MS2 and pNRS Δ76-MS2 plasmids, the NRS WT and NRS Δ76 DNA fragments were PCR amplified with O-4666 (containing a T7 promoter sequence) and O-4667 and inserted into a BamHI/SacI digested pMS2 plasmid. Other NRS variant DNAs, NRS 22, NRS 24, NRS 26 and NRS 28 were obtained by PCR mutagenesis using the primers pairs O-5422/O-5423, O-5424/O-5425, O-5426/O-5427 and O-5428/O-5429, respectively (Supplementary Table S1).
For in vitro splicing assays, plasmid pC2-NRS and its derivatives were produced by insertion of the NRS DNA sequences in the sense (pC2-NRS and derivatives) or antisense orientations (pC2-NRS Inv) into the intron of the earlier described C2 HIV construct built with HIV-1 BRU cDNA (31). These constructions were made in two steps using PCR primers described in Supplementary Table S1. First, the HIV sequence from position 4871 to 5053 was PCR amplified and cloned between the PstI and EcoRI sites of plasmid pUC18. Second, a DNA fragment containing a T7 RNA polymerase promoter followed by the HIV sequence including exon 1 (nucleotides 1–289) and the intron from position 290 to 340 and a second fragment corresponding to RSV NRS WT were PCR amplified, ligated together using BglII sites introduced at their extremities and cloned into the HindIII and PstI sites of plasmid pC2 to obtain pC2-NRS. The same strategy was used to insert the NRS sequence in the reverse orientation (NRS Inv) and the NRS Δ76 sequence. Other NRS variant DNAs, NRS 22, NRS 24, NRS 26 and NRS 28 were obtained by PCR mutagenesis as described above.
To produce plasmids used for transfection assays, fragments from pC2, pC2-NRS WT and pC2-NRS Inv constructs were amplified using O-5824 and O-5825 and cloned between the NcoI and XbaI sites of plasmid pGL3c (Promega), so that, C2-NRS DNA sequences replaced the luciferase gene and were under the control of the SV40 promoter (pGL3-C2, pGL3-C2 NRS WT and pGL3-C2 NRS Inv, respectively). The SV40 poly(A) signal sequence was located at the other extremity. Plasmid pXJ42-9G8 (32) was used for overexpression of the 9G8 SR protein. The empty pXJ42 plasmid was used in control experiments. Plasmid phRGB (Promega) was used to verify the efficiency of transfection during overexpression experiments. The pBluescript SK-II (Stratagene) was used in order to reach a final amount of 2 and 4 µg of plasmid during the overexpression and siRNA experiments, respectively.
RNA transcription
For RNA secondary structure analysis and RNP purification using MS2-RNAs, unlabelled RNAs were transcribed using 1.7–3 pmols of PCR products and T7 RNA polymerase in conditions described earlier (33). Transcripts were purified by electrophoresis on 5% polyacrylamide gel as described earlier (33). For in vitro splicing assays, 5 µg of plasmid were linearized by EcoRI and used as the template for the synthesis of unlabelled capped RNAs as described earlier, except that [α-32P] UTP was omitted (31). After digestion of the DNA matrix with 5 µl of TURBO DNase (Ambion), RNAs were purified using the GenElute kit (Sigma-Aldrich) according to manufacturer’s recomendations.
RNA secondary structure analysis
RNA secondary structure analyses were performed on 100 ng of RNA transcript, in the presence of 3.125 mM MgCl2 in buffer D (73 mM KCl, 0.15 mM EDTA, 0.35 mM DTT, 0.18 mM PMSF, 15% glycerol, 15 mM HEPES–KOH pH 7.9) and in the presence of 5 µg of yeast tRNA mixture. RNA was pre-incubated for 5 min at 65°C in 15 µl of buffer D and after slow cooling to 30°C, 0.5 U of RNase T1 (Roche), 0.25 U of RNase T2 (Invitrogen) or 5 × 10−5 U of RNase V1 (Kemotex) were added and incubation was continued for 10 min at 30°C. RNase V1 hydrolyses were stopped by addition of 1 µl of 100 mM EDTA and 20 µg of yeast tRNA. RNase T1 and T2 hydrolyses were stopped by addition of 20 µg of yeast tRNA, followed by phenol extraction and ethanol precipitation. Kethoxal and DMS modifications were performed as described earlier (34). Positions of enzymatic and chemical cleavages were identified by primer extension analyses using Avian Myeloblastosis virus reverse transcriptase (Q Biogene) and the 5′-end labelled primers described in Supplementary Table S1. Primer 5′-end-labelling, annealing and extensions were performed as described earlier (34). The sequencing ladder was obtained as usual, using the RNA sequencing method (34) with the same primer as for identification of cleavage and modification positions.
RNA secondary structure prediction and phylogenetic analysis
Predictions of secondary structure were made with the Mfold software version 3.2 (available at http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) (35,36). The Mfold calculated thermodynamic values correspond to RNA in 1 M NaCl at 37°C. Results of experimental analyses were introduced in the computer search, in order to define the most stable secondary structure which had the best fit with the experimental data.
The GenBank accession number of RSV and related ASLV sequences used in the comparative secondary structure analysis are: L29198 (Schmidt Ruppin A), AF052428 (Schmidt Ruppin B), DQ365814 [ALV MQNCSU (A)], AY027920 (ALV ADOL-7501), AB303223 (ALV Stain Tym_S90), AF033810 (Fujinami Sarcoma Virus), V01170 (Y53 Sarcoma Virus).
Footprinting assays on complexes formed with recombinant 9G8 protein
To form 9G8-NRS RNP complexes, 100 ng of cold NRS WT RNA transcript (1.17 pmol) and 5 µg of yeast tRNA were mixed in 15 µl of buffer D containing 3.125 mM MgCl2. NRS WT RNA transcript was denatured for 5 min at 65°C and after slow cooling at 30°C, recombinant protein was added. Then, 0.7 or 1.5 µg (28.5 and 57 pmol, respectively) of purified recombinant 9G8 protein expressed in baculovirus and purified as described earlier (37) were added. Complexes were formed by incubation for 10 min at 30°C and then, 0.75 U of T1 RNase was added for 10 min at 30°C. The reaction was stopped by addition of 20 µg of yeast tRNA mixture and by rapid cooling to 0°C, immediately followed by phenol extraction. As a control, enzymatic digestion was performed with the same amount of NRS WT RNA transcript and yeast tRNA incubated in the same conditions except that 9G8 protein was missing. Positions of enzymatic cleavages were identified by extension of 5′-end-labelled primers (Supplementary Table S1), in conditions described earlier (34). The sequencing ladder was obtained as usual, using the RNA sequencing method (34) with the same primer as for identification of cleavage and modification positions. Experiments were all repeated several times using different batches of RNA transcripts.
MS2 affinity chromatography
The procedure used was an adaptation of the one described earlier (29). MS2-tagged purified WT or variant NRS RNAs (250 pmol) were denatured, renatured and incubated with a 5-fold molar excess of purified MS2-MBP fusion protein at 4°C for 30 min. This protein was prepared as described earlier (38). The RNA–MS2–MBP complexes formed were incubated for 30 min at 4°C, with 4.3 mg of HeLa nuclear extract in a 1.1 ml of buffer D containing 60 µg of yeast tRNA. The HeLa nuclear extract was purchased from CILBiotech. Then, 100 µl of amylose beads (GE Healthcare) equilibrated in buffer D were incubated with the complexes for 1 h at 4°C under constant agitation. Three successive washes with buffer D were performed, and RNP complexes were eluted by incubation with 100 µl of buffer D containing 15 mM maltose (30 min at 4°C). About 10% of the proteins contained in the eluted RNP complexes were fractionated by 10% SDS–PAGE for western blot analysis. Another 10% of the eluted material was phenol extracted and RNAs were fractionated on a 10% denaturing polyacrylamide gel for northern blot analysis. Finally, for WT NRS-MS2 and control MS2–RNP complexes, the remaining part of the eluate was fractionated by SDS–PAGE and the gel was systematically sliced. Then, proteins extracted from the bands were analysed by nano-liquid chromatography–mass spectrometry experiments, using a CapLC capillary liquid chromatography system (Waters, Milford, MA, USA) coupled to a hybrid quadrupole time-of-flight mass spectrometer (Q-TOF II, Waters) according to standard protocols (39).
In vitro splicing assays
Twenty fmol of purified pre-mRNA that contained the WT NRS sequence, its reverse sequence or variant NRS sequences inserted in the C2 HIV-1 pre-mRNA were incubated for 2 h at 30°C in a 22 µl standard reaction containing 8 µl of HeLa cell nuclear extract (31). To study the influence of protein 9G8 on NRS activity, splicing reactions were performed with a 1:1 mixture of HeLa cell cytoplasmic extract and HeLa cell nuclear extract and 180 or 270 ng of purified recombinant human 9G8 protein (produced in baculovirus) or 500 ng of commercial purified BSA used as a control. Splicing reactions were stopped by addition of 20 µg of Proteinase K, followed by phenol/chloroform extraction. RNAs were ethanol precipitated and dissolved in 20 µl of bi-distilled H2O. Reverse transcriptions were performed by addition of 2 µl of this solution in 20 µl of 1 × MMLV buffer (Promega) containing 5 pmol of O-5825 primer, 1 mM of each dNTP, 1 µl of RNAguard (GE Healthcare) and 200 U of MMLV reverse transcriptase (Promega). Incubation was for 1 h at 37°C. In order to detect spliced RNAs, PCR amplifications were performed with the O-6246/O-6519 primer pair. To detect both spliced and unspliced RNAs, the PCR amplification was performed with the O-6519/O-6187 primer pair. PCR reactions were performed in semi-quantitative conditions using 50 µl of 1 × GoTaq buffer containing 1 µl of reverse transcriptase products, 200 µM of each dNTP, 1 µM of each primers and 1.25 U of GoTaq polymerase (Promega). The following protocol was used: initial denaturation for 5 min at 94°C, followed by 30 cycles of amplification for analysis of spliced plus unspliced RNAs (Sp+UnSp) or 35 cycles of amplification for analysis of spliced RNAs (Sp). Each cycle included incubations for 30 s at 94°C, 45 s at 46°C and 45 s at 72°C. After agarose gel electrophoresis, ethidium bromide fluorescence of RT–PCR products was quantified using a digital ccd camera (GeneGenius, SynGene) and the SynGene GeneTools version 3.08 software. Each in vitro splicing experiment was repeated three times using different batches of RNAs and for each assay, RT–PCR amplifications were performed in triplicate. The mean values of the three RT–PCR experiments and their standard deviations were established. This allowed us to estimate the mean values of splicing efficiencies and their standard deviations. Student’s t-paired test was used for statistical analysis.
In cellulo splicing assays
HeLa cells were grown in Dulbecco modified Eagle's medium (DMEM Sigma-Aldrich) supplemented with 10% of fetal calf serum and 2 mM glutamine. About 24 h before transfection, cells were seeded in six well plates at a density of 200 000 cells per well. To test for the effect of 9G8 expression, cells were cotransfected with 0.5 µg of plasmid pGL3-C2, pGL3-C2 NRS WT or pGL3-C2 NRS Inv expressing the hybrid pre-mRNAs, 0.5 µg of pXJ42-9G8 expressing 9G8 and 0.9 µg of empty pXJ42 plasmid, together with 0.1 µg of phRGB plasmid encoding the renilla luciferase, this plasmid was used to verify transfection efficiency. In control experiments the pXJ42-9G8 plasmid was omitted and 1.4 µg of empty pXJ42 plasmid was used in order to maintain the same total amount of plasmid. Transfections were performed with the JetPEI (Polyplus-transfection kit) in a DNA/JetPEI ratio of 1:2, according to manufacturer’s recommendations.
For siRNA experiments, HeLa cells were grown in OptiMEM medium (Invitrogen), supplemented with 5% fetal calf serum, in the absence of antibiotics. As above, 24 h before transfection, cells were seeded in six well plates at a density of 200 000 cells per well. Cells were transfected using lipofectamine 2000 (Invitrogen) according to the manufacturer’s recommendations. Two successive transfections of 100 nM of siRNA directed either against 9G8 mRNA [si9G8, (40)] or against the Renilla luciferase transcript of plasmid phRGB [siLuc, (41)] were carried out at 24 h interval. Cells were transfected 24 h after the second siRNA treatment with the following plasmid mixture: 0.5 µg of either pGL3-C2, pGL3-NRS WT or pGL3-C2 NRS Inv and 3.5 µg of pBSK plasmid per well.
To estimate the in cellulo splicing efficiencies, total cellular RNA was extracted with the GenElute kit (Sigma-Aldrich), 48 h after the unique transfection step for overexpression experiments and 24 h after the third transfection step for siRNA experiments. Then, 4 µg of total cellular RNA were treated with 4 U of TURBO DNase (Ambion) in a 20 µl of TURBO DNase buffer 1 × for 30 min at 37°C. The reaction was stopped by adding 20 µl of 30 mM EDTA and heating at 75°C for 15 min. Finally, 1 µg of DNase treated RNA was subjected to reverse transcription, using 0.5 pmol of oligo(dT)18 as primer and PCR amplification and quantification were performed as described above. Two independent experiments were performed for the 9G8 overexpression and siRNA experiments, respectively. In each experiment, total cellular proteins were extracted and analysed by SDS–PAGE followed by western blot analysis, using antibodies specific for 9G8 and β-tubulin (Sigma-Aldrich) as described earlier (41).
RESULTS
Experimental analysis of the RSV NRS 2D structure
To analyse experimentally the 2D structure of the entire RSV NRS in solution, we used RNases T1 and T2 in conditions such that they only cleaved single-stranded segments and V1 RNase which cleaves specifically double-stranded and stacked RNA regions. Inspection of the NRS bordering sequences in ASLV retroviruses revealed the conservation of a C-rich segment 5′ to NRS and of a G-rich segment 3′ to NRS. These two segments showed conserved complementarity in the ASLV family. We hypothesized that they may form a helix closing the NRS. Therefore, our structural analyses were performed on an in vitro transcript produced by T7 RNA polymerase that encompassed the RSV RNA segment from position 691 to 951 (NRS encompasses nucleotides 703–930) (Figure 1A). The cleaved NRS residues were identified by primer extension analyses. Efficient binding of RT primers requires their base pairing with single-stranded residues. Hence, an AAAUU sequence, which was not expected to interfere with the helix predicted to be formed by segments 691–700 and 940–947, was added at the 3′-end of the transcript (Figure 1A). The RT primer (O-2955) complementary to this additional sequence and segment 939–951 (Supplementary Table S1) was efficient to identify cleavages located between positions 901 and 936. We defined two other primers (O-1619 and O-1617), which allowed us to identify RNase cleavage positions in the entire NRS region (Supplementary Table S1). The 2D structure probing experiments were performed after RNA refolding in conditions described in ‘Materials and Methods’ section. Experiments were repeated several times and reproducible results were obtained. Examples of primer extension analyses are shown in Figure 1B (B1–B3). Enzymatic probing experiments were completed by chemical probing experiments. We used DMS, which methylates single-stranded adenosines at position N-1 and single-stranded cytosines at position N-3, and kethoxal that modifies unpaired guanine residues at position N-3. Location of modifications was also achieved by primer extension analyses, representative examples are shown in Figure 1C (C1 and C2).
Computer assisted determination of the RSV NRS structure
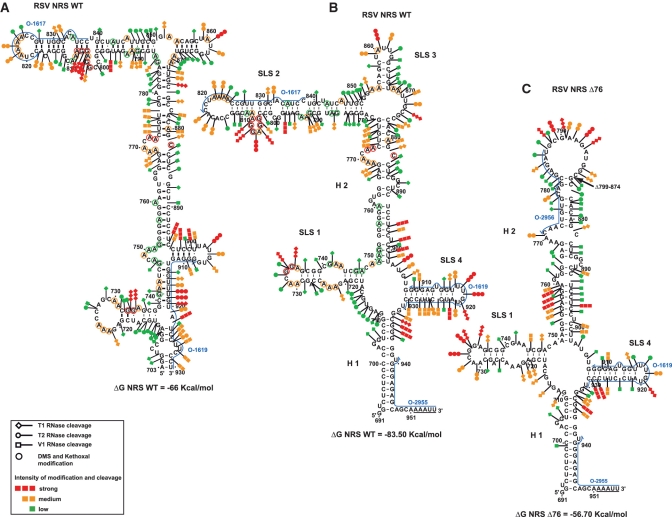
The previously proposed computer predicted structure for RSV NRS WT (13) could not explain the results of our chemical and enzymatic probing experiments (Figure 2A). To search for the most stable 2D structure in agreement with our experimental data, we used the Mfold software (35,36) and introduced our data as constraints. The structure identified (Figure 2B) has a very low free energy (−83.5 kcal/mol). In agreement with our previous probing data, which has served as the basis for NMR analysis of the NRS-3′ structure (23), segment 907–929 containing both the U1 and U11 binding sites, forms an individual stem–loop structure (SLS4). In the established structure, segment 717–747, which was previously found to bind the ASF/SF2 SR protein (19), also folds into an individual stem–loop structure (SLS1). As expected, the 3′- and 5′-termini of the NRS form an irregular helix (H1) that is extended by the complementary sequences bordering the NRS. The segment from position 750 to 778, which was previously proposed to bind several SR proteins and hnRNP H, forms a very long irregular helix (H2) by interaction with segment 877–902. The sequence from position 781 to 850, which is largely deleted in the functional NRS Δ76 truncated element, is involved in formation of a long irregular SLS2 stem–loop structure, and fragment 854 to 868 which is also deleted in NRS Δ76, forms a small stem–loop structure (SLS3).
Figure 2.
Schematic representation of the experimental data on the previously proposed structure for RSV NRS WT (13) in (A), and on the secondary structure models that we established for RSV WT and RSV Δ76 NRSs (B and C, respectively). Cleavages by enzymes and modifications by chemical reagents are depicted as shown in the inset (modified residues are circled, phosphodiester bonds cleavages by T1, T2 or V1 RNases are indicated by arrows surmounted by lozenges, circles or squares, respectively). The intensities of cleavages and modifications are indicated by colors (red, orange and green for strong, medium and low, respectively). Positions of nucleotides are according to (48). Stem–loop structures (SLS) and helices (H) are numbered. The free energies of the proposed structures as calculated by the Mfold software (37°C, in 1 M NaCl) (35,36) are given below each structure. The hybridisation sites used for primer extensions are indicated by blue arrows. The underlined sequences at the 3′-end of the RNAs correspond to the added unrelated nucleotides.
The main structural features of NRS are conserved in the NRS Δ76 variant
As the variant NRS Δ76 is functional, we hypothesized that its 2D structure should not be drastically modified. To verify this hypothesis, we probed its structure experimentally. The NRS Δ76 transcript contained the same additional sequences at the 5′- and 3′-extremities as the full length NRS transcript used above, and was analysed as the full length NRS. The data obtained are in perfect agreement with formation of helices H1 and H2 and stem–loops SLS1 and SLS4 in NRS Δ76 (Figure 2C and Supplementary Figure S1). The parts of the SLS2 and SLS3 stem–loops, which are remaining in NRS Δ76, form two base pairs extending helix H2 and a large terminal loop. Therefore, the 2D structure elements formed by sequences which are important for binding of U1 snRNA, U11 snRNA (SLS4), SR and hnRNP proteins (SLS1, H1 and H2, respectively) are conserved in the NRS Δ76.
NRSs from viruses of the ASLV family can fold according to the structure established for RSV NRS
LR9 is an RSV-related recombinant ALV virus which contains the gag, pol and env genes derived from the UR2-associated virus (UR2AV) and the LTR of ring-necked pheasant virus (RPV) (27). Its NRS element is highly similar to RSV NRS (10 base substitutions) and shows similar efficiency in an heterologous context (27). To test whether the presence of these 10 base substitutions are compatible with the 2D structure established for the RSV NRS, we produced an LR9 NRS transcript homologous to that used for RSV NRS and probed its structure with T1, T2 and V1 RNases. As shown in Supplementary Figure S2, base substitutions are either located in single strands, or reinforce the stability of helix H2. Therefore, the possibility to form the structure is preserved. Note that when repeating the same experiment on the defective LR9 NRS deletion mutant (ΔLR9), which is missing the segment 735 to 776 (27), we observed strong 2D structure alterations (Supplementary Figure S3): the SLS1 structure disappeared and, helix H2 and SLS2 were remodelled. Therefore, both the sequence and 2D structure of this defective NRS are profoundly altered.
Then, for a larger investigation on the possible phylogenetic conservation of the NRS 2D structure, we took advantage of the available sequences for several ASLVs genomic RNAs (42). As shown in the Supplementary Figure S4, A–G, the nucleotide sequences involved in formation of helix H1, SLS1 and SLS4 stems are highly conserved. Only one half of helix 2 has a conserved sequence. The various base substitutions, insertions or deletions in the viruses that we compared have no expected detrimental effect on the RNA 2D structure, which is in favour of a phylogenetic conservation of the NRS 2D structure.
A minimal NRS element including helix H1 and stem–loops SLS1 and SLS4 is active in vitro
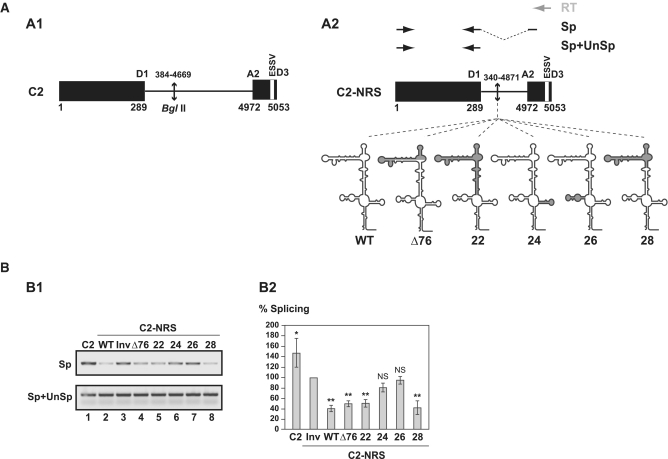
Then, based on the established structure, we generated internal truncation in the NRS in order to define the minimal NRS sequence required for NRS activity. To this end, we used a heterologous construct derived from the HIV-1 C2 construct (Figure 3A1) (31) which produces a pre-mRNA containing a short intron bordered by the HIV-1 strong D1 5′-ss and the A2 3′-ss. Site A2 has a limited efficiency due to the presence of the ESSV splicing silencer that binds hnRNP A1 (43), and up to now, only an activation by the SR protein ASF/SF2 was detected at this site (44). Therefore, we hypothesized, that the C2 construct might be a favourable model to compare activities of WT and variant NRSs. The NRS WT coding sequence was inserted in the middle of the C2 intron (Figure 3A2). In these conditions, the NRS pseudo 5′-ss was located 278 nt downstream from the HIV-1 D1 5′-ss. This distance was selected as it was previously shown to be optimal for NRS activity (13). As in vitro splicing efficiency is dependent on the length of the intron to be spliced, we compared the splicing efficiency of C2-NRS WT RNA to a control C2 RNA containing the NRS sequence in the reverse orientation (C2-NRS Inv RNA). Transcripts were spliced in a Hela cell nuclear extract, as described in ‘Materials and Methods’ section. The ratio of spliced (Sp) versus unspliced plus spliced (Sp+UnSp) products was quantified by RT–PCR as described in ‘Materials and Methods’ section (Figure 3A2, and Supplementary Table S1). For each transcript, splicing assays were performed in triplicate and highly reproducible results were obtained as evidenced by data in the Supplementary Figure S5A. As expected, insertion of a non-related 266-nt long sequence in C2 RNA decreased the in vitro splicing efficiency by ∼40% (C2-NRS Inv RNA compared with C2 RNA). However, the decrease observed for the C2-NRS WT RNA was reproducibly more drastic (80%), demonstrating NRS activity in the C2 context. We thus considered that this model is valid and could be used to test the effects of deletions in NRS on splicing efficiency.
Figure 3.
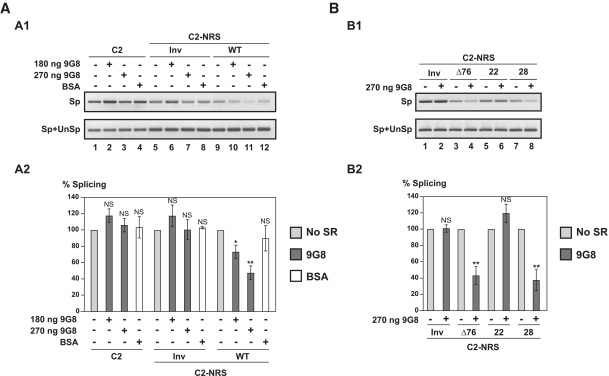
Comparison of splicing inhibition properties of NRS WT and Δ76, 22, 24, 26 and 28 NRS variants in the C2 pre-mRNA context. (A) Schematic representation of the HIV C2 pre-mRNA (A1) (31) and C2-NRS heterologous pre-mRNAs (A2). Black boxes and thin lines represent exons and intron, respectively. Numbering of C2 RNA is according to (49). End-joining of the two HIV-1 sequences in the C2 intron is depicted by an arrow. The D1 5′-ss and A2 HIV-1 3′-ss as well as the ESSV regulatory element are shown (43). Heterologous C2-NRS constructs, created by insertion of the WT and variant Δ76, 22, 24, 26 and 28 NRSs in the intron, are schematically represented below. Segments deleted in the variant NRS elements are in grey. The RT (in grey), and PCR (in black) primers used for evaluation of the yields of C2 and C2-NRS pre-mRNAs splicing efficiencies are represented by arrows above the C2-NRS construct. Yields of spliced RNAs (Sp) and of spliced plus unspliced products (Sp+UnSp) were analysed by PCR amplifications using the pairs of primers marked by Sp and Sp+UnSp, respectively. (B) RT–PCR analysis of in vitro splicing products of the C2 and heterologous C2-NRS pre-mRNAs. In vitro splicing assays were performed as described in ‘Materials and Methods’ section. Experiments were repeated three times using different batches of RNA (Supplementary Figure S5A). The relative splicing efficiencies of the various constructs were estimated by semi-quantitative RT-PCR performed in triplicate (Supplementary Figure S5A). A representative example of cDNA fractionation obtained for each of the tested pre-mRNAs is shown (B1). Identities of RT–PCR products spliced (Sp) or spliced plus unspliced (Sp+UnSp) products are indicated. The identity of the pre-mRNA used in the assay is given above the lane. The Sp/Sp+UnSp pre-mRNA ratio obtained for the C2-NRS Inv construct was taken as the reference (100%). The Sp/Sp+UnSp ratios established for the other constructs are given as a percentage of this reference. The mean values of the Sp/Sp+UnSp percentages determined for three independent splicing reaction and their standard deviations are given (B2). *P < 0.05, **P < 0.01, NS: not significant.
As a positive control, we used the functional NRS Δ76 mutant and we produced four other variants (Figure 3A2). In variant 28 both stem–loops SLS2 and SLS3, as well as a very small portion of helix H2, were deleted. In variant 22, helix H2 and stem–loops SLS2 and SLS3 were deleted. In this variant, only helix H1 and the stem–loops SLS1 and SLS4 are present and they are joined by a 7-nt long single-stranded segment. Stem–loop SLS4 was deleted in variant 24 and SLS1 was absent in variant 26. As expected NRS Δ76 was as efficient in splicing inhibition as the NRS WT (Figure 3B1 lane 4 and B2). Interestingly, NRS 28, which is smaller than NRS Δ76, had a splicing inhibitory property superior to that of NRS Δ76 (Figure 3B1 lane 8 and B2). No splicing inhibition was detected for NRS 24, which confirmed the requirement of the U1 snRNA binding site for NRS activity (Figure 3B1 lane 6 and B2). The same absence of inhibition was observed for NRS 26, showing that deletion of stem–loop SLS1 (position 717–747) has the same negative effect on NRS activity as deletion of stem–loop SLS4 (Figure 3B1 lane 7 and B2). Accordingly, previous data (20) showed a strong loss of NRS activity with a deletion which included part of the SLS1 sequence (deletion from position 720 to 744). Importantly, NRS 22, which only contained stem–loops SLS1 and SLS4 and helix H1, had a negative effect on splicing similar to that of NRS Δ76. Therefore, our data reveal that a very small truncated NRS variant containing only stem–loop structures SLS1 and SLS4 and helices H1 and H2 is sufficient to inhibit splicing in vitro.
Sequences corresponding to SLS1 and SLS4 are required for recruitment of SR proteins
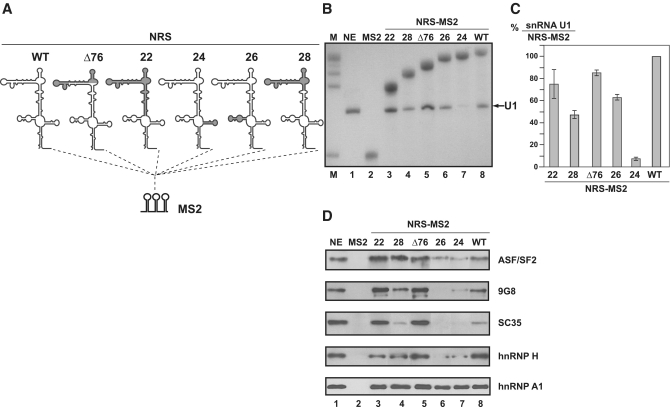
As a complement to the in vitro splicing inhibition study of truncated NRSs described above, we attempted to identify the nuclear proteins recruited by these NRSs mutant. To this end, we used an RNP purification approach that is based on the fusion of MS2 sequences to the studied RNA (29,30). The WT, Δ76, 22, 24, 26 and 28 NRS variant RNAs, described above, were transcribed with three binding sites for the MS2 coat protein fused at their 3′-ends (Figure 4A). Each of these RNAs was bound to a recombinant fusion protein containing the MS2 coat protein RNA binding domain and the E. coli maltose binding protein (MBP). The RNA–protein complexes obtained were then incubated with a Hela cell nuclear extract competent for in vitro splicing. The WT and variant NRS RNPs formed were bound to amylose beads, washed and eluted by addition of maltose, as described in ‘Materials and Methods’ section.
Figure 4.
Only functional NRS variants bind SR proteins. (A) Schematic representation of NRS-MS2 and NRS-MS2 derived RNAs used in affinity selection experiments. The three binding sites for the MS2 coat protein (MS2 RNA) were fused to the 3′-end of the NRS RNAs. Formation of RNP complexes and their purification is described in ‘Materials and Methods’ section. (B and C) Northern blot analysis of RNA from purified RNP complexes, using 5′-end-labelled U1 snRNA (O-6524) and MS2 RNA (O-6534) specific probes (Supplementary Table S1). The RNAs used to form the RNP complexes are indicated above the lanes. M, is a mixture of MS2 and all NRS-MS2 RNAs used as size markers. NE corresponds to RNAs extracted from cell nuclear extract without addition of exogeneous RNA. Intensities of the bands were quantified and the ratios of U1 snRNA versus NRS-MS2 RNA are given in C. The U1 snRNA/NRS-MS2 RNA ratio obtained for the WT NRS RNA is taken as the 100% reference and ratios obtained for the other NRS-MS2 RNA are given as a percentage of this reference. The mean values and standard deviations of two experiments are shown. (D) Western blot analyses of SR and hnRNP proteins contained in the purified RNP complexes formed with the various NRS-MS2 RNAs. Proteins extracted from equal amounts of purified complexes were analysed by western blot using antibodies specific for the SR and hnRNP proteins indicated on the right side of the blot. The identity of the NRS-MS2 RNA used in the assay is given above the gel. Two µg of total proteins of HeLa nuclear extract (NE) were loaded as a reference.
Both the RNA and the protein composition of the purified complexes were analysed. The presence of U1 snRNA and MS2 tagged RNAs in the purified complexes was evidenced by northern blot analysis using specific oligonucleotides (Supplementary Table S1), and quantified as described in Figure 4. As expected, only the deletion of the SLS4 sequence had a marked negative effect on U1 snRNA association (Figure 4B and C). Proteins present in the RNP complexes formed with the WT NRS were subjected to SDS–PAGE, the bands were cut and analysed by mass spectrometry. We detected proteins already found to bind the entire NRS element (ASF/SF2, hnRNP H) (19,21,45) among a large number of other proteins. For a comparative analysis of SR and hnRNP H and A1 proteins in RNP complexes formed with the WT and variant NRSs, we turned to the western blot approach using specific antibodies (Figure 4D). As expected the SR proteins ASF/SF2, 9G8 and SC35 and the hnRNP H proteins were detected in complexes formed with NRS WT. They were also detected with NRSs Δ76, 22 and 28. In contrast, weak amounts of ASF/SF2 and only traces of 9G8 and SC35 were detected in complexes formed with NRSs 24 and 26, suggesting that both the SLS1 and SLS4 sequences are required for efficient binding of SR proteins. The strong binding of hnRNP A1 to each of the truncated forms of NRSs (Figure 4D) suggested a possible involvement of this protein in regulation of NRS activity.
The SR protein 9G8 has a stimulatory effect on NRS activity
Our observation of a strong binding of the SR protein 9G8 on NRS together with previous description of its association with NRS (21) encouraged us to test for a possible effect of this protein on NRS activity. First, we used the in vitro splicing assay based on C2 derived contructs. The C2-NRS WT RNA was spliced in HeLa cell nuclear extract in the absence or the presence of increasing amounts of recombinant 9G8 protein, which had been produced in baculovirus. As illustrated in Figure 5A and Supplementary Figure S5B, the NRS inhibitory property was significantly and reproducibly increased by addition of the recombinant 9G8 protein, whereas addition of BSA had no significant effect. For the C2-NRS WT RNA only 50% of the splicing activity was detected upon addition of 270 ng of 9G8.
Figure 5.
Protein 9G8 enhances NRS splicing inhibition properties in vitro. (A) WT NRS activity is strongly enhanced by 9G8 protein in vitro. C2 (depicted in Figure 3A1), C2-NRS WT and C2-NRS Inv pre-mRNAs (depicted in Figure 3A2) were incubated for 2 h in a 1:1 mixture of HeLa cell cytoplasmic and nuclear extracts in the absence or the presence of purified recombinant human 9G8 protein (180 and 270 ng) or BSA (500 ng). RNAs were subjected to RT–PCR analysis (in triplicate) as in Figure 3. One representative example of cDNA fractionation is shown (A1). Splicing experiments were repeated three times using different batches of RNA. For each tested pre-mRNA, the Sp/Sp+UnSp ratio obtained in the absence of 9G8 or BSA addition was taken as the 100% reference and the Sp/Sp+UnSp ratios obtained in the presence of 180 or 270 ng of 9G8 or 500 ng of BSA were expressed as a percentage of this reference. The data for the three splicing experiments are shown in Supplementary Figure S5B. The mean values and the standard deviations of these experiments are given in A2. *P < 0.05, **P < 0.01, NS: not significant. (B) 9G8 increases the in vitro activity of the Δ76 and 28 variant NRSs but not of the 22 variant NRS. The C2-NRS Inv, C2-NRS Δ76, C2-NRS 22 and C2-NRS 28 pre-mRNAs were incubated in a 1:1 mixture of HeLa cell cytoplasmic and nuclear extract without (9G8−) or with (9G8+) 270 ng of 9G8 protein and were subjected to RT–PCR analysis as described above. Three independent experiments were performed and a representative example of fractionation of the RT–PCR products is shown in B1. The Sp/Sp+UnSp ratios were quantified as in Figure 3 and the Sp/Sp+UnSp ratio obtained in the absence of 9G8 over-expression were taken as the 100% reference. The Sp/Sp+UnSp ratios obtained in the presence of 270 ng of 9G8 were expressed as a percentage of this reference. The percentages established for the three independent experiments are shown in Supplementary Figure S5, D. Their mean values and standard deviations are given in B2. *P < 0.05, **P < 0.01, NS: not significant.
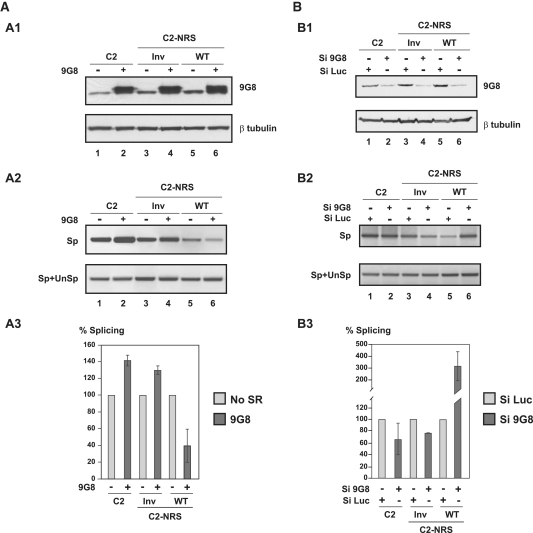
To verify that 9G8 also stimulates the NRS inhibitory property in cellulo, the C2, C2-NRS WT and C2-NRS Inv DNA sequences from the in vitro constructs were inserted within plasmid pGL3c, in such a way that they replaced the luciferase gene. The resulting plasmids, pGL3-C2, pGL3-C2 NRS WT or pGL3-C2 NRS Inv were co-transfected in HeLa cells with either the empty plasmid pXJ42 or plasmid pXJ42-9G8 expressing 9G8 protein and 48 h after transfection, the amounts of spliced (Sp) and spliced plus unspliced (Sp+UnSp) transcripts were estimated by RT–PCR as described in Figure 6. The over-expression of protein 9G8 by a factor of about three reproducibly decreased splicing of the C2-NRS WT RNA, but not of splicing of C2 and C2-NRS Inv transcripts (Figure 6A and Supplementary Figure S5C). Therefore, the stimulatory effect of 9G8 on the NRS activity that we discovered by in vitro assays also exists in cellulo.
Figure 6.
NRS activity in cellulo depends on the level of protein 9G8 expression. (A) Protein 9G8 also increases NRS activity in cellulo. DNAs of the C2, C2-NRS WT and C2-NRS Inv constructs described in Figure 3A2 were cloned into plasmid pGL3c. HeLa cells were co-transfected with these constructs and a vector expressing the 9G8 SR protein (9G8+) or an empty vector as the control (9G8−); 48 h after transfection, total cellular proteins and RNAs were extracted as described in ‘Materials and Methods’ section and analysed by western blot after SDS–PAGE analysis (A1) and by RT–PCR (A2 and A3), respectively. The presence of equal amounts of extracted proteins in each assay was controlled by western blots using β-tubulin antibody and the level of 9G8 overexpression was evaluated by western blot using a specific 9G8 antibody. For RT–PCR analysis of the extracted RNAs, the same primers as in Figure 3A2 were used, except that the RT primer was an oligo(dT) (Supplementary Table S1). Two independent experiments were performed and a representative example of cDNA fractionation is shown in A2. For each of the tested constructs, the Sp/Sp+UnSp ratios obtained in the absence of 9G8 overexpression was used as the 100% reference and the Sp/Sp+UnSp ratios obtained in the presence of 9G8 overexpression are expressed as a percentage of this reference. The percentages established for the two independent experiments are shown in Supplementary Figure S5C. Their mean values and standard deviations are shown in A3. (B) NRS activity is strongly reduced upon decreasing the cellular 9G8 protein concentration. HeLa cells were treated with siRNA targeting the 9G8 mRNA (si9G8) or with control siRNA (targeting luciferase mRNA, siLuc) in conditions described in ‘Materials and Methods’ section. They were then transfected with plasmids pGL3-C2, pGL3-C2 NRS Inv or pGL3-C2 NRS WT. Total proteins and RNAs were extracted 24 h after transfection and were analysed by western blot (B1) and RT–PCR (B2 and B3), respectively, as described for the (A). Two independent experiments were performed and a representative exemple of cDNA fractionation is shown in B2. RT–PCR were performed in triplicate and quantified, as described above. For each construct, the Sp/Sp+UnSp ratios obtained for the control siLuc siRNA was taken as the 100% reference and the Sp/Sp+UnSp ratios obtained for the si9G8 siRNA are expressed as a percentage of this reference. The mean values of the two independent experiments and their standard deviations are shown in B3.
To confirm the existence of this stimulatory effect in cellulo, we then diminished the level of 9G8 in HeLa cells by using a si9G8 directed against the endogeneous 9G8 mRNA (si9G8) (40). As a control we used a siRNA directed against the Renilla luciferase (siLuc) (41). Cells were transfected twice with siRNAs and 48 h later were transfected by the C2, C2-NRS Inv or C2-NRS WT constructs for 24 h. Two independent experiments were performed and in our transfection conditions, the 9G8 concentration was decreased to a moderate extent (factor of about 2) (Figure 6B and B1). In these conditions, C2 RNA splicing was slightly decreased and we reproducibly detected an increased splicing of C2-NRS WT RNA (factor of about 2.5) while no increase was detected in the control experiment performed with the siLuc siRNA (Figure 6B, B2 and B3). Therefore, we concluded that the strength of splicing regulatory activity of RSV NRS depends upon the yield of expression of protein 9G8.
Helix H2 is required for stimulation of the NRS splicing inhibition by 9G8
Then, we used again in vitro splicing assays in order to define which NRS regions are important to get the 9G8 stimulatory effect. Tests were performed three times on each of the NRS Δ76, 22 and 28 variants (Figure 5B and Supplementary Figure S5D). Strong increases of the Δ76 and 28 NRS inhibitory properties were reproducibly detected in the presence of recombinant 9G8. In contrast, addition of 9G8 protein did not stimulate the splicing inhibitory activity of the NRS 22 variant, showing that in spite of the strong association of 9G8 with SLS1, the helix H2 sequence which is missing in NRS 22 is required for stimulation of the RSV NRS activity by 9G8. This suggested a direct binding of protein 9G8 on sequences in helix H2.
Therefore, for a better delineation of the 9G8 binding sites on RSV NRS, we then performed footprinting assays on RNP complexes formed by incubation of RSV WT NRS transcripts with the recombinant 9G8 protein at two distinct [9G8]/[RNA] ratios 25 and 50, respectively, as described in ‘Materials and Methods’ section. Formation of RNP complexes was verified by EMSA experiments (data not shown). The patterns of T1 RNase digestion of free and complexed RNA were compared by primer extension analysis followed by gel electrophoresis. Examples of cDNA fractionation are shown in Figure 7A, A1–A3. Neither SLS3 nor SLS4 are protected. At the lowest [9G8]/[RNA] ratio, protections are mainly detected in SLS1 and its bordering sequences and in the purine rich large bulged loop in SLS2 (Figure 7B). Upon increasing 9G8 concentration, protections in these regions were reinforced and new protections are detected in helix H2 (Figure 7B). Interestingly, these data are in agreement with our finding of a requirement of the SLS1 sequence for efficient association of 9G8 to RSV NRS. They are also in agreement with previous data suggesting that there are 9G8 protein binding sites in the RSV NRS segment extending from positions 740 to 798 (21). However, our data reveals that 9G8 binding sites in RSV NRS are not restricted to this segment.
Figure 7.
Enzymatic footprinting of 9G8 binding sites on RSV NRS RNA. (A1–A3) Primer extension analyses of RSV NRS cleaved by T1 RNase in the absence or the presence of protein 9G8. RNA-protein complexes were formed at [9G8]/[RNA] molar ratios of 25 and 50, respectively. The absence of protein addition is indicated by (−). Enzymatic reactions were performed as described in ‘Materials and Methods’ section. Lanes U, G, C and A correspond to the sequencing ladder and primers used for reverse transcription are indicated below each autoradiogram. Nucleotide numbering in the RSV NRS RNA and positions of the ASF/SF2 (19) binding site are indicated on the left side of the autoradiogram. Positions of helices (H) and stem–loop structures (SLS) are shown on the right side of the autoradiograms. Cleavages by T1 RNase and protection generated by protein 9G8 are represented by arrows with two circles. Colours of the first and second circles represent the level of protection obtained at the 25 and 50 [protein]/[RNA] molar ratios, respectively (white, black, blue and purple for none, low, medium and strong protection, respectively). (B) Schematic representation of the protections generated by protein 9G8 is depicted on the RSV NRS RNA 2D structure using the same rules as in A. Primers are indicated by blue arrows. The ASF/SF2 and snRNA U1 binding sites according to references (17,19) are highlighted in green and grey, respectively.
DISCUSSION
Here, we describe the first experimental analysis of the 2D structure of the entire RSV NRS and based on a proteomic analysis, followed by in vitro splicing assays, we show that the SR protein 9G8 stimulates the RSV NRS splicing inhibition property.
Each of the NRS functional sequences belongs to a defined 2D structure motif and the possibility to form these motifs is conserved in ASLVs
Retroviruses generally have highly structured genomic RNAs in order to resist to digestion by RNases of the host cells. The RSV NRS element does not escape to this rule. It folds into a tight 2D structure. Other ASLV NRSs can adopt this structure due to compensatory and semi-compensatory base substitutions in the stems and the location of several point mutations and insertion of nucleotide stretches in single-stranded segments.
The structural motif established by NMR for a small RSV NRS segment containing the pseudo 5′-ss (23) is included in the established structure. This motif corresponds to the apical part of the SLS4 stem–loop structure. Importantly, formation of this apical part of SLS4 was previously shown to be needed for efficient binding of U1 snRNP to the pseudo 5′-ss (23).
Our data change the conception of NRS architecture which was based on a previous computer prediction (13). We found that the ASF/SF2 binding region, which is also shown here to bind protein 9G8, folds into a stem–loop structure facing the stem–loop structure containing the U1 and U11 binding sites. In the previous computer model, these two sequences were in part base paired together (13). Absence of protection by protein 9G8 in the SLS4 stem–loop, while sequences in SLS1 are protected, is in agreement with the formation of two distinct structural elements by these two NRS sequences. However, as the sequences bordering the SLS1 and SLS4 stem–loops form a large internal loop, one can imagine that the flexibility of this internal loop facilitates the establishment of interactions between partners of SLS1 and SLS4: protein ASF/SF2 and/or 9G8 bound to SLS1 and the U1 snRNP bound to SLS4. The nucleotide sequences of these two stem–loop structures are highly conserved. The possibility to form helix H1 was not taken into consideration in the previous model. Based on the present data, one of its functions may be to stabilize the NRS structure. In addition, helix H1, whose primary sequence is highly conserved, contains one of the G tracts (nucleotides 935–942) which are needed to bind hnRNP H and this binding is important for U11 snRNP association (46). Conservation of this G tract in ASLVs is also in accordance with the presence of similar G tracts downstream from several of the minor 5′-ss that bind U11 snRNP (46). Sequences involved in helix H2 formation were previously proposed to bind SR proteins (47). Here, we show that this helix is required for activation of NRS by protein 9G8 and delineated the H2 segment protected by this SR protein.
Noticeably, the strong sequence conservations of SLS1, SLS4, helix H1 and the bottom part of helix H2 can not be simply explained by specific needs for coding capacity, since all the NRS sequences code for a single polypeptide (the Gag protein). Consequently, the strong conservations of some parts of the NRS sequence and of its overall 2D structure altogether likely reflect specific needs for NRS activity.
SLS1, SLS2 linked together by helix H1 are sufficient for splicing inhibition
The HIV-1 derived construct that we designed for in vitro and in cellulo assay of the NRS activity turned to be efficient. It shows that utilization of the pair of HIV-1 D1 and A2 sites can be efficiently down-regulated by the heterologous NRS sequence. Accordingly, like the RSV env and src 3′-ss, the HIV-1 site A2 is poorly efficient due to the presence of: (i) suboptimal polypyrimidine tract and branch site sequences and (ii) an ESSV inhibitory element (31,43). By using the HIV-1-NRS heterologous construct, we confirmed the requirement of sequences contained in SLS1 for NRS activity and showed that these sequences are as much important for NRS activity as sequences in SLS4. This strongly suggest that the previously identified ASF/SF2 binding sites in SLS1 (19,20) and/or the presently identified 9G8 binding sites in SLS1 is/are needed to get NRS activity. These data are consistent with the absence of activity of the NRS ΔLR9 which is missing the sequence from position 735 to 776 (27).
The NRS Δ76 variant was previously defined as the minimal functional NRS element (13). Interestingly, although shorter in length than NRS Δ76, variant NRS 28, designed on the basis of the RNA 2D structure, inhibited in vitro splicing as efficiently as the NRS Δ76. Most importantly, we could design a very short NRS variant (NRS 22), which is still able to inhibit splicing in vitro. This is the first observation of the activity of a very small NRS variant, which only contains SLS1 linked to SLS4 by helix H1.
Increased protein 9G8 concentration reinforces the NRS inhibitory property
Whereas the SR protein 9G8 was previously shown to bind NRS, its activity on this element had never been tested directly. Here, by in vitro and in cellulo splicing assays, we show the dependence of the NRS activity on the 9G8 protein concentration. The effect on NRS activity observed upon addition of 9G8 protein in nuclear extract was almost opposite to that observed upon decreasing the 9G8 concentration by siRNAs in cellulo. Importantly also, addition of extra 9G8 protein in nuclear extract increased the NRS WT, Δ76 and 28 activity, but had no effect on the NRS 22 inhibition property, in spite of the presence of 9G8 binding sites in SLS1. We propose the following interpretation of our splicing data. First, by binding to SLS1, protein 9G8 may have in combination with ASF/SF2 a positive basic effect on NRS activity. Secondly, at higher concentration, by binding to sequences in helix H2 which are present in NRS Δ76 and 28 but are missing in NRS 22, protein 9G8 may have a supplementary stimulatory effect on NRS activity.
Altogether, our data bring strong support to the implication of protein 9G8 in modulation of NRS activity. Obviously, these data were obtained by using human nuclear extracts and recombinant human SR proteins, which are more easily available than splicing competent avian nuclear extracts. However, based on the strong sequence conservation of SR proteins and components of the splicing machinery in mammalian and avian organisms, we propose that activation of NRS by the 9G8 avian counterpart protein also takes place in avian cells.
Deletions of SLS1 or SLS4 both have a negative effect on SR binding to NRS in nuclear extract
Our adaptation of the MS2 based RNP purification method to the purification of RNP complexes formed by WT or variant NRSs upon incubation in nuclear extract led to an unexpected result: binding of SR proteins to NRS is found to depend on both SLS1 and SLS4, while binding of U1 snRNP is strictly dependent on the presence of SLS4. The dependence of the binding of the SR proteins ASF/SF2 on the presence of SLS1 is in agreement with previous data showing its direct binding to a segment overlapping SLS1 (19,20). Finding of a requirement of SLS1 for recruitment of proteins 9G8 is in accord with our 9G8 footprinting assays (Figure 7A and B).
Our finding of a strong negative effect of SLS4 deletion on SR protein binding is unexpected. Firstly, because we found no significant protection of SLS4 in RNP complexes formed with protein 9G8 (Figure 7A and B), secondly because the NRS segment extending from position 715 to 748 binds alone the recombinant ASF/SF2 protein (19). One possible explanation is that, in nuclear extracts containing a high level of protein hnRNP A1, binding of the U1 snRNP or other splicing factors on SLS4 may help SR protein to compete with hnRNP A1 and to bind to SLS1. Accordingly, we detected a strong binding of hnRNP A1 on all the truncated NRSs that were tested, which was confirmed by footprinting assays (data not shown).
One model to explain the overall data presented in this paper is that the role of SR proteins in NRS activity is probably not to reinforce U1 snRNP binding on NRS but to couple the NRS pseudo 5′-ss to the RSV 3′-ss, which is needed for splicing inhibition. SR proteins and hnRNP A1 likely compete for binding to NRS and in future studies the possible role of hnRNP A1 will have to be investigated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique (CNRS); the French Ministry de l’Enseignement Supérieur et de la Recherche; French National Agency for Research (ANR) (ANR-05-BLAN-0261-03); European Alternative Splicing Network of excellence (LSHGCT 2005 518238); Région Lorraine (Bioingénierie project); French Ministère de la Recherche et de l’Enseignement (to A.B., V.M. and G.K.). Funding for open access charge: European Alternative Splicing Network of excellence (LSHGCT 2005 518238).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
K. Beemon (John Hopkins, Baltimore) is strongly thanked for her generous gift of plasmids pRSVNeo-Int, pΔ76Neo-Int, pLR9 and pΔLR9 and helpful discussions. R. Lührmann is acknowledged for providing plasmid pADML-M3 containing the three MS2 coat protein binding sites and plasmid pMAL containing the MS2-MBP coding sequence. R. Lührmann and K. Hartmuth. (Max Planck Institute, Göettingen) are strongly thanked for providing MS2 RNP chromatography affinity protocol and technical formation in their lab. L. Fouillen, S. Sanglier and A. Van Dorsselaer. (Laboratoire de Spectrométrie de Masse Bio-Organique, Strasbourg) are thanked for mass spectrometry analyses. L. Ayadi and F. Oillo-Blanloeil are thanked for their participation to footprinting assays. J. Ugolini is thanked for her excellent technical assistance. CILBiotech society (Mons, Belgium) is thanked for providing nuclear and cytoplasmic extracts from HeLa cells.
REFERENCES
- 1.Coffin JM, Hughes SH, Varmus HE. Retroviruses. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory; 1997. [PubMed] [Google Scholar]
- 2.McNally MT. RNA processing control in avian retroviruses. Front Biosci. 2008;13:3869–3883. doi: 10.2741/2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz RA, Skalka AM. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol. Cell. Biol. 1990;10:696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu XD, Katz RA, Skalka AM, Maniatis T. The role of branchpoint and 3′-exon sequences in the control of balanced splicing of avian retrovirus RNA. Genes Dev. 1991;5:211–220. doi: 10.1101/gad.5.2.211. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Stoltzfus CM. A suboptimal src 3′ splice site is necessary for efficient replication of Rous sarcoma virus. Virology. 1995;206:1099–1107. doi: 10.1006/viro.1995.1033. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Simpson SB, Stoltzfus CM. Selection and characterization of replication-competent revertants of a Rous sarcoma virus src gene oversplicing mutant. J. Virol. 1996;70:3636–3644. doi: 10.1128/jvi.70.6.3636-3644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amendt BA, Simpson SB, Stoltzfus CM. Inhibition of RNA splicing at the Rous sarcoma virus src 3′ splice site is mediated by an interaction between a negative cis element and a chicken embryo fibroblast nuclear factor. J. Virol. 1995;69:5068–5076. doi: 10.1128/jvi.69.8.5068-5076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogert RA, Lee LH, Beemon KL. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J. Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simpson SB, Guo W, Winistorfer SC, Craven RC, Stoltzfus CM. The upstream, direct repeat sequence of Prague A Rous sarcoma virus is deficient in mediating efficient Gag assembly and particle release. Virology. 1998;247:86–96. doi: 10.1006/viro.1998.9233. [DOI] [PubMed] [Google Scholar]
- 10.Simpson SB, Zhang L, Craven RC, Stoltzfus CM. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J. Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo W, Winistorfer SC, Stoltzfus CM. Selective inhibition of splicing at the avian sarcoma virus src 3′ splice site by direct-repeat posttranscriptional cis elements. J. Virol. 2000;74:8513–8523. doi: 10.1128/jvi.74.18.8513-8523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arrigo S, Beemon K. Regulation of Rous sarcoma virus RNA splicing and stability. Mol. Cell. Biol. 1988;8:4858–4867. doi: 10.1128/mcb.8.11.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNally MT, Gontarek RR, Beemon K. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology. 1991;185:99–108. doi: 10.1016/0042-6822(91)90758-4. [DOI] [PubMed] [Google Scholar]
- 14.McNally MT, Beemon K. Intronic sequences and 3′ splice sites control Rous sarcoma virus RNA splicing. J. Virol. 1992;66:6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Sullivan CT, Polony TS, Paca RE, Beemon KL. Rous sarcoma virus negative regulator of splicing selectively suppresses SRC mRNA splicing and promotes polyadenylation. Virology. 2002;302:405–412. doi: 10.1006/viro.2002.1616. [DOI] [PubMed] [Google Scholar]
- 16.Gontarek RR, McNally MT, Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 17.Hibbert CS, Gontarek RR, Beemon KL. The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA. 1999;5:333–343. doi: 10.1017/s1355838299981347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNally LM, McNally MT. U1 small nuclear ribonucleoprotein and splicing inhibition by the rous sarcoma virus negative regulator of splicing element. J. Virol. 1999;73:2385–2393. doi: 10.1128/jvi.73.3.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNally LM, McNally MT. SR protein splicing factors interact with the Rous sarcoma virus negative regulator of splicing element. J. Virol. 1996;70:1163–1172. doi: 10.1128/jvi.70.2.1163-1172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNally LM, McNally MT. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol. Cell. Biol. 1998;18:3103–3111. doi: 10.1128/mcb.18.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fogel BL, McNally MT. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 2000;275:32371–32378. doi: 10.1074/jbc.M005000200. [DOI] [PubMed] [Google Scholar]
- 22.Giles KE, Beemon KL. Retroviral splicing suppressor sequesters a 3′ splice site in a 50S aberrant splicing complex. Mol. Cell. Biol. 2005;25:4397–4405. doi: 10.1128/MCB.25.11.4397-4405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabello-Villegas J, Giles KE, Soto AM, Yu P, Mougin A, Beemon KL, Wang YX. Solution structure of the pseudo-5' splice site of a retroviral splicing suppressor. RNA. 2004;10:1388–1398. doi: 10.1261/rna.7020804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varani L, Hasegawa M, Spillantini MG, Smith MJ, Murrell JR, Ghetti B, Klug A, Goedert M, Varani G. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl Acad. Sci. USA. 1999;96:8229–8234. doi: 10.1073/pnas.96.14.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donahue CP, Muratore C, Wu JY, Kosik KS, Wolfe MS. Stabilization of the tau exon 10 stem loop alters pre-mRNA splicing. J. Biol. Chem. 2006;281:23302–23306. doi: 10.1074/jbc.C600143200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Z, Cote J, Kwon JM, Goate AM, Wu JY. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 2000;20:4036–4048. doi: 10.1128/mcb.20.11.4036-4048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith MR, Smith RE, Dunkel I, Hou V, Beemon KL, Hayward WS. Genetic determinant of rapid-onset B-cell lymphoma by avian leukosis virus. J. Virol. 1997;71:6534–6540. doi: 10.1128/jvi.71.9.6534-6540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polony TS, Bowers SJ, Neiman PE, Beemon KL. Silent point mutation in an avian retrovirus RNA processing element promotes c-myb-associated short-latency lymphomas. J. Virol. 2003;77:9378–9387. doi: 10.1128/JVI.77.17.9378-9387.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol. 2010;8:e1000276. doi: 10.1371/journal.pbio.1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damier L, Domenjoud L, Branlant C. The D1-A2 and D2-A2 pairs of splice sites from human immunodeficiency virus type 1 are highly efficient in vitro, in spite of an unusual branch site. Biochem. Biophys. Res. Commun. 1997;237:182–187. doi: 10.1006/bbrc.1997.7091. [DOI] [PubMed] [Google Scholar]
- 32.Bourgeois CF, Popielarz M, Hildwein G, Stevenin J. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol. Cell. Biol. 1999;19:7347–7356. doi: 10.1128/mcb.19.11.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mougin A, Gregoire A, Banroques J, Segault V, Fournier R, Brule F, Chevrier-Miller M, Branlant C. Secondary structure of the yeast Saccharomyces cerevisiae pre-U3A snoRNA and its implication for splicing efficiency. RNA. 1996;2:1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 34.Jacquenet S, Ropers D, Bilodeau PS, Damier L, Mougin A, Stoltzfus CM, Branlant C. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 2001;29:464–478. doi: 10.1093/nar/29.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 37.Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jurica MS, Moore MJ. Capturing splicing complexes to study structure and mechanism. Methods. 2002;28:336–345. doi: 10.1016/s1046-2023(02)00240-2. [DOI] [PubMed] [Google Scholar]
- 39.Miguet L, Pacaud K, Felden C, Hugel B, Martinez MC, Freyssinet JM, Herbrecht R, Potier N, van Dorsselaer A, Mauvieux L. Proteomic analysis of malignant lymphocyte membrane microparticles using double ionization coverage optimization. Proteomics. 2006;6:153–171. doi: 10.1002/pmic.200500133. [DOI] [PubMed] [Google Scholar]
- 40.Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 41.Dreumont N, Hardy S, Behm-Ansmant I, Kister L, Branlant C, Stevenin J, Bourgeois CF. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2009;38:1353–1366. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bilodeau PS, Domsic JK, Mayeda A, Krainer AR, Stoltzfus CM. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 2001;75:8487–8497. doi: 10.1128/JVI.75.18.8487-8497.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ropers D, Ayadi L, Gattoni R, Jacquenet S, Damier L, Branlant C, Stevenin J. Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J. Biol. Chem. 2004;279:29963–29973. doi: 10.1074/jbc.M404452200. [DOI] [PubMed] [Google Scholar]
- 45.Cook CR, McNally MT. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 46.McNally LM, Yee L, McNally MT. Heterogeneous nuclear ribonucleoprotein H is required for optimal U11 small nuclear ribonucleoprotein binding to a retroviral RNA-processing control element: implications for U12-dependent RNA splicing. J. Biol. Chem. 2006;281:2478–2488. doi: 10.1074/jbc.M511215200. [DOI] [PubMed] [Google Scholar]
- 47.Fogel BL, McNally LM, McNally MT. Efficient polyadenylation of Rous sarcoma virus RNA requires the negative regulator of splicing element. Nucleic Acids Res. 2002;30:810–817. doi: 10.1093/nar/30.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz DE, Tizard R, Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983;32:853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- 49.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich B, Josephs SF, Doran ER, Rafalski JA, Whitehorn EA, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.