Abstract
Protocadherins are a group of transmembrane proteins with homophilic binding activity, members of the cadherin superfamily. Apart from their role in adhesion, the cellular functions of protocadherins are essentially unknown. Protocadherin (PCDH)12 was previously identified in invasive trophoblasts and endothelial and mesangial cells in the mouse. Invalidation studies revealed that the protein was required for optimal placental development. In this article, we show that its human homolog is abundantly expressed in various trophoblast subtypes of the human placenta and at lower levels in endothelial cells. We demonstrate that PCDH12 is shed at high rates in vitro. The shedding mechanism depends on ADAM10 and results in reduced cellular adhesion in a cell migration assay. PCDH12 is subsequently cleaved by the γ-secretase complex, and its cytoplasmic domain is rapidly degraded by the proteasome. PCDH12 shedding is regulated by interlinked intracellular pathways, including those involving protein kinase C, PI3K, and cAMP, that either increase or inhibit cleavage. In endothelial cells, VEGF, prostaglandin E2, or histamine regulates PCDH12 shedding. The extracellular domain of PCDH12 was also detected in human serum and urine, thus providing evidence of PCDH12 shedding in vivo. Importantly, we observed an increase in circulating PCDH12 in pregnant women who later developed a pre-eclampsia, a frequent pregnancy syndrome and a major cause of maternal and fetal morbidity and mortality. In conclusion, we speculate that, like in mice, PCDH12 may play an important role in human placental development and that proteolytic cleavage in response to external factors, such as cytokines and pathological settings, regulates its activity.
Keywords: ADAM ADAMTS, Cell-Cell Interaction, Endothelium, Metalloprotease, Signal Transduction, Placenta
Introduction
The cadherin superfamily is composed of membrane proteins with Ca2+-dependent cell adhesion properties and homologous sequence repeats located in their extracellular domains (1–3). These repeats are responsible for the homophilic adhesive behavior of members of the cadherin family. Four groups of cadherins have been identified so far: the “classical” type I and type II cadherins; the desmosomal cadherins; and the most recently discovered protocadherin family. Protocadherins were initially discovered by PCR cloning using cadherin homology sequences (4). With genome sequencing, more than 70 different protocadherin genes have been identified altogether. This makes protocadherins the largest subgroup within the cadherin superfamily (5). Most protocadherin genes are clustered in three loci: protocadherin (Pcdh)3 α, β, and γ. Clustered protocadherins are mainly expressed in the brain, whereas unclustered protocadherins display original tissue distributions.
The functions of classical cadherins in cell-cell adhesion, their links to the cytoskeleton, and their participation in intracellular signaling have all been extensively studied (1). In contrast, the function(s) of protocadherins remain elusive. Although all of the protocadherins studied so far were found to display homophilic adhesion properties, their cytoplasmic domains do not interact with the cytoskeleton or do so very weakly (2, 3). What is known of their functions derives mainly from in vivo studies. Protocadherins have been shown to be involved in cellular migration and cell sorting (6–10), establishment of neuronal circuits and synapses (11), cell survival (12, 13), and inhibition of cell growth (14, 15). Mutations of some protocadherin genes have been linked to human diseases. This is the case for PCDH15, mutations of which cause Usher syndrome type 1F (16, 17), PCDH19, for which mutations were directly linked to epilepsy and cognitive impairment (18), and PCDH10, whose deletion was linked to some forms of autism (19).
However, the cellular and molecular functions of protocadherins are essentially unknown and are suspected to be highly variable between protocadherins. In this paper, we focus on PCDH12 (also called VE-cadherin 2). This protein was originally described in mouse endothelioma cell lines where it concentrates at cell-cell junctions (20). Its extracellular domain contains five cadherin domain repeats that promote homophilic adhesion. Its cytoplasmic domain is not homologous to classical cadherins or other protocadherin family members and does not interact with the cytoskeleton. Unlike classical cadherins, Pcdh12 ectopic expression was unable to inhibit cell migration from a confluent monolayer of transfected cells (20). In the mouse, Pcdh12 is highly expressed in several cell types: (i) in a specific type of trophoblast called the “glycogen cells” that emerge in the placenta and invade the maternal decidua; (ii) in the mesangial cells of kidney glomeruli; and (iii) in angiogenic endothelial cells, whereas its expression is reduced in resting endothelium (21, 22). Pcdh12-deficient mice were alive and fertile but showed alterations in placental development that resulted in embryonic growth retardation (23). In the absence of Pcdh12, placentas were smaller and showed limited angiogenesis and mis-segregation of the labyrinthine and intermediate layers, likely attributable to cell migration defects.
In the present study, we characterized human PCDH12. We observed its expression in several tissues and organs, including villous and extravillous trophoblasts. In transfected CHO cells, we observed a high degradation rate caused by a cascade of proteolytic events. This proteolysis was initiated by the cleavage of PCDH12 by ADAM10 (a disintegrin and metalloprotease 10) metalloprotease, resulting in shedding of its extracellular domain. Shedding was followed by secondary cleavage by the γ-secretase complex and the subsequent proteasomal degradation of its cytoplasmic domain. We were able to show that PCDH12 shedding in endothelial cells was regulated by VEGF, histamine, and prostaglandin E2 (PGE2) probably through PKC and cAMP pathways. PCDH12 degradation has functional implications because it modulates the cell migration rate in a monolayer wound assay. In addition, we provide evidence that these phenomena are also in play in vivo, as evidenced by the detection of circulating PCDH12 in both serum and urine. We found that seric levels of PCDH12 were up-regulated in pre-eclampsia (PE), which is a leading cause of maternal and fetal mortality and morbidity that complicates 2–8% of pregnancies (24, 25). Altogether, our results indicate that PCDH12 cell surface expression is highly dependent on protease-dependent shedding, a process regulated by cytokines and probably modified in pathological pregnancies.
EXPERIMENTAL PROCEDURES
Reagents
Phorbol 12-myristate 13-acetate (PMA), the γ-secretase inhibitor L685,458, 1,2-dipalmitoylphosphatidylinositol 3,4,5-trisphosphate, LY294002, genistein, forskolin (Fsk), PGE2, and histamine were from Sigma-Aldrich. VEGF was from Peprotech (Rocky Hill, NJ). The metalloprotease inhibitor GM6001 was from BioMol (Villeurbanne, France). The ADAM inhibitors GI254023X and GW280264X were generous gifts from Prof. Andreas Ludwig (Aachen, Germany).
Cell Culture and Treatments
CHO cells were grown in α-minimum essential medium (Invitrogen) with 10% FCS (Lonza, Levallois-Perret, France). HEK-293 were grown in Dulbecco's modified Eagle's medium with 5% FCS. HUVECs were purchased from Lonza, grown in complete EBM-2 medium (Lonza), and used at passages 3–5. All of the pharmacological molecules were solubilized in Me2SO, which was added at similar concentrations in control experiments. Molecules were applied on confluent cells in serum-free medium.
Plasmid Construction and Cell Transfection
The human PCDH12 cDNA, obtained from ImaGenes (Berlin, Germany), was cut by SpeI and NheI and inserted into pCDNA3.1 Hygro (Invitrogen) linearized by XbaI. The resulting construct was transfected into CHO cells. Stable transfectants were selected with 200 μg/ml hygromycin, and individual clones were picked.
Antibodies
For production of anti-PCDH12 EC antibody, the entire extracellular domain of human PCDH12 (amino acids 1–718) was produced in 293-EBNA cells according to described procedures (26) and injected into rabbits. Briefly, PCDH12 cDNA was amplified by PCR, which allowed the addition of a sequence coding for six C-terminal histidine residues and cloned into the episomal PCEP4 expression vector. After transfection, the cells were selected with hygromycin, and conditioned serum-free medium was collected for PCDH12 EC purification using DEAE ion exchange followed by nickel affinity chromatography. After immunization, PCDH12 antibodies were purified from antisera using antigen affinity chromatography. For production of PCDH12 Cter antibody, a peptide (KSRGSSSSSRCL), corresponding to PCDH12 C terminus sequence, was coupled to ovalbumin and injected into rabbits. PCDH12 Cter antibodies were purified using peptide affinity chromatography. β-Catenin antibodies were from Transduction Laboratories. N-cadherin antibodies for immunoprecipitation (clone 32) and for Western blot (3B9) were from BD Biosciences (Pont de Claix, France) and from Zymed Laboratories Inc. (San Francisco, CA), respectively. Tubulin antibodies were from Sigma-Aldrich. ADAM10 antibodies were from Merck-Calbiochem (Nottingham, UK). C3 polyclonal antibodies were a generous gift from MB Villiers.
Immunoprecipitation and Western Blot
Cell or tissue extracts were obtained using a lysis buffer containing 0.5% Nonidet P-40. Protein concentration was measured using the MicroBCA kit from Thermo Scientific (Rockford, IL). Cell supernatants were collected after culture of confluent cells in serum-free medium. HUVEC supernatants were concentrated on Amicon Ultra 4 (Millipore, Billerica, MA). Immunoprecipitation experiments were performed using standard procedures. Briefly, proteins were extracted from placenta (1 mg) or CHO-PCDH12 (300 μg) with 1% Nonidet P-40 and 0.25% sodium deoxycholate. The extracts were incubated with 4 μg of PCDH12 Cter or N-cadherin antibodies, and the complex was coupled to protein G-Sepharose beads (Sigma-Aldrich). The proteins were electrophoresed in polyacrylamide gels and transferred onto Immobilon-P membranes (Millipore, Bedford, MA). Dot blots were performed with a PR600 from Hoefer Scientific Instruments (San Francisco, CA). The membranes were incubated in presence of primary antibodies and horseradish peroxidase-coupled secondary antibodies (Jackson Immunoresearch Laboratories, Suffolk, UK). Antibody interactions were revealed using the luminol substrate ECL-Plus (PerkinElmer Life Sciences). Signals were revealed by autoradiography, films were scanned, and signal intensity was measured using Bio-Profil software from Vilber Lourmat (Marne-la-Vallée, France). Alternatively, signals were captured with a Fusion-FX7 camera from Vilber-Lourmat.
Immunofluorescence
12-week-old placentas from nonpathological abortions were obtained from Grenoble University Hospital after full consent of the patients and approval of the university hospital ethics committee. The placentas were embedded in OCT compound and snap frozen. 10-μm cryosections were prepared using a cryomicrotome (Leica Microsystems, Wetzlar, Germany). The cells were grown on coverslips. Tissue sections or cells were fixed with formaldehyde, permeabilized with Triton X-100 0.5%, saturated with 2% bovine serum albumin, and incubated with primary antibodies and Cy3- or alexa 488-conjugated secondary antibodies from Jackson Immunoresearch Laboratories and Molecular Probes (Cergy Pontoise, France), respectively. The samples were further incubated for 3 min in Hoechst 33258 (Sigma) for nuclear staining. Fluorescent slides were mounted in Fluorsave reagent (Calbiochem). Immunofluorescence labeling was observed with an Axioplan 2 Zeiss microscope or a confocal microscope (Leica Microsystems TCS SP2).
siRNA
For RNA interference of ADAM10 expression, the following siRNAs from Dharmacon were used: 5′-GAAGGAAGCUUUAGUCAUG-3′, 5′-CCAAAGUCUCUCAUAUUA-3′, 5′-GCAGAGAGAUACAUUAAAG-3′, and 5′-GAAUUGCCCUGAUCAUGUU-3′ as an equimolar mixture of all. For negative control, an siRNA was used that does not target any known mammalian gene (5′-UAGCGACUAAACACAUCAA-3′). CHO-PCDH12 were transfected with 1.6 μg of siRNA using nucleofection (Amaxa Biosystems) according to the manufacturer's instructions. The cells and conditioned media were harvested at 48 h post-transfection.
Subcellular Fractionation
Lipid raft isolation was performed using established procedures. Briefly, CHO-PCDH12 cells were lysed in 50 mm Hepes, pH 7.4, 150 mm NaCl, 5 mm EGTA, 1% Triton X-100. After a brief centrifugation, the lysates were adjusted to 40% sucrose, placed at the bottom of a 5–30% sucrose gradient, and centrifuged for 17 h at 40,000 rpm. Twelve 1-ml fractions were collected from the top and analyzed by Western blotting with anti-PCDH12 Cter and anti-flotillin antibodies.
Cell Migration Assay
Assay was performed as described previously (27). Confluent cells were scratched, and either Me2SO or GM6001 was added to complete medium. Phase contrast images were obtained under a Cell-R inverted microscope (Olympus, Hamburg, Germany) with controlled temperature and CO2, and using 10× objective. Measurement of cell invasion in the wounded area was performed with ImageJ software. Statistic differences were established through Student's t test.
Human Sera and Urine
Human sera of healthy donors were obtained from the Grenoble blood center (Etablissement Français du Sang). Sera were depleted in albumin using the Vivapure anti-human serum albumin, affinity resin from Sartorius (Aubagne, France). Urine from a healthy donor was concentrated 40 times, using Amicon ultrafiltration devices; retentate was subjected to two cycles of dilution with water and reconcentration to remove salts. Sera from pregnant women were collected at Grenoble University Hospital and kept frozen until use. The study protocol was approved by the university hospital ethics committee, and written informed consent was obtained from each participant. Serum samples were obtained once the legal conservatory period of the sanitary bank of maternal serum markers was ended (see Table 1 for clinical records). PE was defined as systolic pressure of ≥140 mm Hg and/or diastolic blood pressure of 90 mm Hg, occurring beyond the 20th week of amenorrhea associated with a proteinuria (>0.3 g/24 h) also arising beyond the 20th week of amenorrhea.
TABLE 1.
Clinical characteristics of normal and pathological pregnancies
| Maternal characteristics | Normal gestations (n = 27) | Pre-eclamptic pregnancies (n = 13)a |
|---|---|---|
| Maternal age (years) | 29.9 (19–41) | 30.9 (23–44) |
| Gestation age at sampling (weeks+days) | 15+5 (12+5- to 18+1) | 15+5 (14+2 to 18+3) |
| Systolic blood pressure (mm Hg) | 110 (90–130) | 157 (145–170) |
| Diastolic blood pressure (mm Hg) | 80 (50–90) | 97 (90–110) |
| Proteinuria (g/24 h) | 0 | 1.52 (0.42–3.97) |
a See “Experimental Procedures” for a description of criteria for PE symptoms.
RESULTS
Human PCDH12 Characterization
Human PCDH12 was characterized in various tissues at the mRNA (supplemental Fig. S1), as well as the protein levels (supplemental Fig. S2), using antibodies directed either against its extracellular domain “PCDH12 EC” or against the 12-amino acid C-terminal peptide “PCDH12 Cter.” Both antibodies revealed a single band at 145 kDa in PCDH12-transfected cells; no reactivity was observed in control cells, as expected (supplemental Fig. S2, A and B).
PCDH12 was detectable in HUVECs (endothelial cells) by both Western blot and RT-PCR (supplemental Fig. S2, C and D) but not by immunofluorescence (not shown). These results indicate that PCDH12 is only weakly expressed by this cell type. PCDH12 expression was not detectable by RT-PCR or Western blotting in other cell types of epithelial origin, including MCF7, HeLa, and Caco2 cells (supplemental Fig. S2D and data not shown).
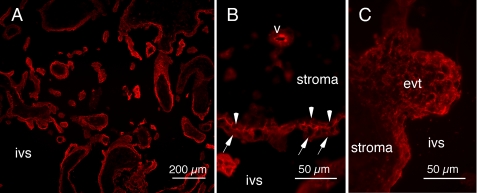
PCDH12 was also detected in human term placenta (supplemental Fig. S2E). Cryosections of 12-week human placenta were strongly immunostained, in particular at the borders of syncytiotrophoblast and cytotrophoblast cells, which make up the outer layer of placental microvilli (Fig. 1, A and B). A more diffuse staining pattern was detected in extravillous trophoblasts, forming cell columns at the end of placental villi (Fig. 1, A and C). Some rare intravillous vessels were also labeled (Fig. 1B).
FIGURE 1.
PCDH12 localization in human placenta. Human 12-week placenta was cryosectioned and immunolabeled with anti-PCDH12 Cter antibody and observed at low magnification (A) or high magnification (B, C). Syncytiotrophoblasts (arrows) and villous cytotrophoblasts (arrowheads) exhibited strong labeling at the cell membrane. Extravillous cytotrophoblasts (evt) showed rather diffuse staining, whereas intravillous vessels (v) harbored only occasional staining. Controls with secondary antibody alone yielded no signals (data not shown). ivs, intervillous space.
As opposed to the mouse, human PCDH12 could not be detected by immunofluorescence in either glomeruli or endothelium of the kidney (not shown). Altogether, PCDH12 appeared to be weakly expressed by endothelial cells and strongly by syncytiotrophoblasts and cytotrophoblasts.
In PCDH12-transfected CHO cells (CHO-PCDH12), PCDH12 was located at cell-cell junctions, in membrane ruffles, or in intercellular filopodia (supplemental Fig. S3A). This is consistent with the homophilic binding activity of cadherin superfamily members.
Evidence of PCDH12 Cleavage and the Mechanism Involved
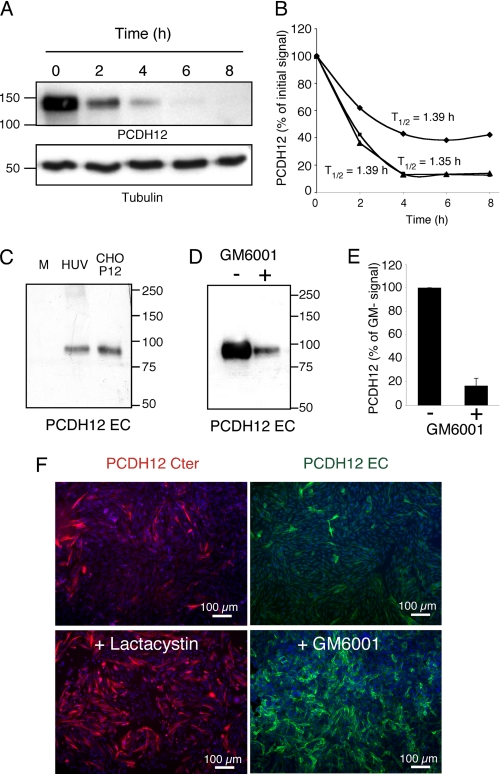
PCDH12 stability was investigated by incubating CHO-PCDH12 with the translation blocking drug cycloheximide. Our data revealed a very short half-life in the range of 1.35–1.39 h (Fig. 2, A and B), thereby suggesting a very high degradation rate. Because PCDH12 instability at the cell membrane may be related to external proteolytic cleavage, we analyzed the release of its extracellular domain into cell supernatants. Fig. 2C shows that PCDH12 extracellular domain was indeed detected in CHO-PCDH12-conditioned medium. A similar fragment was also observed in HUVEC supernatants but only after concentration, indicating that it is present at very low levels (Fig. 2C). The apparent molar mass of the protein (90 kDa) revealed by PCDH12 EC antibody indicates that the protein is probably cleaved close to the membrane. Release of PCDH12 extracellular domain into the cell culture medium was significantly inhibited by the metalloprotease inhibitor GM6001 (Fig. 2, D and E). In addition, incubation of CHO-PCDH12 with either GM6001 or the proteasome inhibitor lactacystin resulted in a striking enhancement of PCDH12 immune labeling in these cells (Fig. 2F). Thus, these data suggest that PCDH12 is cleaved by metalloproteases and that the proteasome plays a role in its degradation. The γ-secretase complex may also play a role in PCDH12 degradation. It is known to cleave target proteins within their transmembrane domain, liberating the cytoplasmic domain into the cytosol, where it is subsequently degraded by the proteasome (28).
FIGURE 2.
Instability of PCDH12 protein at the cell surface. CHO-PCDH12 were incubated in presence of cycloheximide (100 μg/ml). A, cells were harvested at indicated times, and protein extracts were analyzed by Western blotting using PCDH12 and tubulin antibodies. B, intensities of PCDH12 signals from three independent experiments were plotted, and protein half-lives (T½) were deduced. C, cell conditioned media of confluent CHO-PCDH12 (CHO-P12; 40 μl) or HUVECs (HUV; 40 μl after 40× concentration) were harvested after 24 h of incubation and analyzed by Western blot with anti-PCDH12 EC antibody. Unconditioned medium (M) was run as control. D, cell conditioned media of CHO-PCDH12, cultured for 24 h in presence (+) or absence (−) of 10 μg/ml of metalloprotease inhibitor GM6001, were analyzed as in C but with a longer exposure of autoradiographic film. E, quantitative analysis of three experiments performed as in D. Histogram shows the means ± S.D.; p < 0.001. F, images show PCDH12 immunofluorescence of CHO-PCDH12 cultured in regular medium or treated with either lactacystin 5 μm or GM6001 10 μg/ml, as indicated.
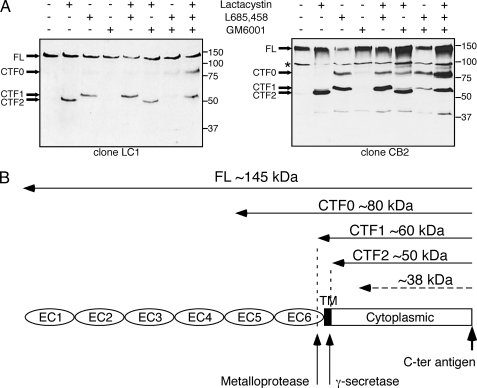
To determine the precise molecular mechanisms of PCDH12 cleavage, the cells were incubated with inhibitors of metalloproteases (GM6001), of γ-secretase (L685,458), and of the proteasome (lactacystin), either separately or in combination (Fig. 3A). Two clones of CHO-PCDH12 with different PCDH12 expression levels were used. Protein extracts were analyzed by Western blotting with PCDH12 Cter antibody to detect C-terminal fragments (CTFs). Lactacystin blocked the degradation of a 50-kDa fragment (CTF2), which probably corresponds to the entire PCDH12 cytoplasmic tail. When γ-secretase was inhibited, two larger fragments were detected at 60 (CTF1) and 80 (CTF0) kDa. The electrophoretic pattern for these fragments was identical with or without lactacystin treatment. It is therefore likely that these fragments escaped degradation by the proteasome because they were still embedded in the membrane. Thus, γ-secretase converts the 80-kDa (CTF0) and 60-kDa (CTF1) fragments to a 50-kDa (CTF2) fragment that can then be rapidly degraded in the proteasome. Inhibition of metalloproteases alone did not modify the electrophoretic profile of C terminus-containing fragments compared with untreated cells. However, when γ-secretase and metalloprotease inhibitors were added together, CTF0 intensities remained similar, but CTF1 levels were significantly reduced compared with when cells were treated by the γ-secretase inhibitor alone. Thus, metalloprotease cleavage mainly yields CTF1, whereas CTF0 is due to a different cleavage mechanism; nevertheless, both fragments are substrates for γ-secretase. Results on PCDH12 cleavage are summarized in Fig. 3B.
FIGURE 3.
Sequential processing of PCDH12 protein. Two different clones of CHO-PCDH12, called LC1 and CB2, were treated for 16 h with 5 μm lactacystin, 5 μm L685,458, and 10 μg/ml GM6001, either alone or in combination, as indicated. A, protein extracts were analyzed by Western blotting using anti-PCDH12 Cter antibody to visualize the full-length (FL) protein and the CTF, called CTF0, 1, and 2, that accumulate after inhibition of proteases. The fragment at 100 kDa (*), not regulated by pharmacological inhibitors, is an artifact of secondary antibody. The low abundance fragment at 38 kDa may represent a minor proteolytic event. B, scheme of PCDH12 proteolytic cleavage summarizing the above data.
ADAM10 Cleaves the Extracellular Domain of PCDH12
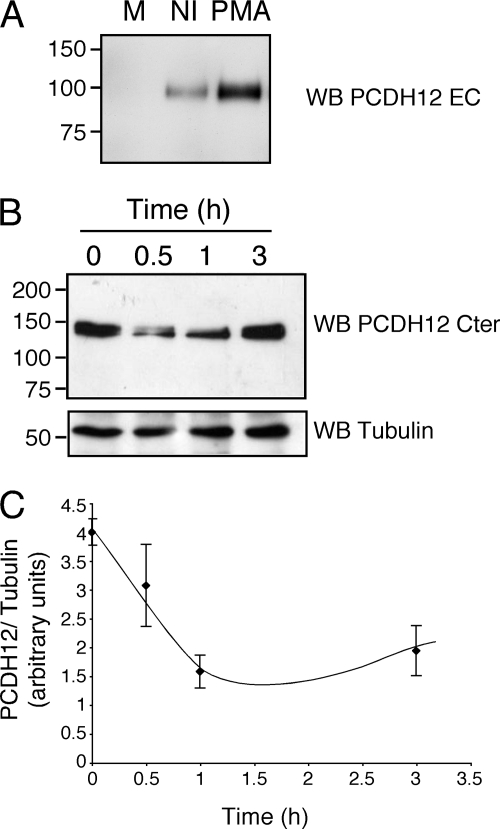
Activation of PKC by phorbol esters has been shown to promote shedding of several transmembrane proteins. This shedding seems to be a characteristic of metalloproteases from the ADAM family (29, 30). PMA treatment enhanced PCDH12 release into the cell supernatant (Fig. 4A). Furthermore, PMA rapidly and transiently reduced the cellular PCDH12 concentration in CHO-PCDH12 (Fig. 4, B and C). Thus, activation of PKC increases PCDH12 proteolytic cleavage.
FIGURE 4.
Induction of PCDH12 cleavage by phorbol esters. A, cell supernatant of CHO-PCDH12 was harvested after 24 h of treatment with PMA (0.1 μg/ml) and analyzed by Western blotting (WB, PCDH12 EC) together with that of noninduced cells (NI) and unconditioned medium (M). B, PCDH12 content in cell extracts of CHO-PCDH12 treated with PMA and harvested at indicated time. The cell extracts were analyzed by Western blotting using PCDH12 Cter and tubulin antibodies. C, three independent experiments were performed as in B. The signals were quantified and plotted as PCDH12: tubulin signal ratio versus time. The data represent the means ± S.D.
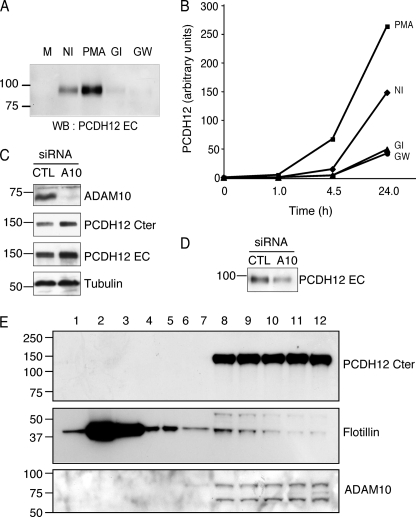
Among metalloproteases, ADAM10 and ADAM17 are the principal sheddases (29, 30). To test whether these proteases might be responsible for PCDH12 shedding, we used two selective pharmacological inhibitors: GW280264X, which has been shown to block both ADAM10 and ADAM17; and GI254023X, which specifically inhibits ADAM10 (31). Both inhibitors dramatically inhibited PCDH12 release into the cell supernatant (Fig. 5, A and B), which suggests that PCDH12 metalloprotease cleavage is mainly mediated by ADAM10.
FIGURE 5.
Reduction of PCDH12 shedding after ADAM10 inhibition. A and B, CHO-PCDH12 were treated with either Me2SO (NI), PMA 0.1 μg/ml, GI254923 5 μm (GI), or GW280264 5 μm (GW). Aliquots of cell supernatant were removed at different time points. A, analysis of cell supernatants (20 μl) at 24 h post-induction by Western blotting (WB) with anti-PCDH12 EC. M, unconditioned cell medium. B, dot blot analysis of cell supernatants at different time points, incubated with same antibody. The optical densities were measured on low exposure films. The data are representative of three independent experiments. C, CHO-PCDH12 cells were transfected either with siRNAs against ADAM10 (A10) or control (CTL) siRNAs and grown without PMA. The cell extracts were analyzed by Western blot using antibodies against ADAM10 to prove siRNA efficiency, against PCDH12 Cter and EC to examine PCDH12 stability and against tubulin as loading control. D, effect of ADAM10 siRNAs in PCDH12 release in cell supernatants. E, subcellular fractionation of CHO-PCDH12 using sucrose density gradient. Fractions 1–12 (top to bottom) were analyzed by Western blot with PCDH12 Cter, flotillin, and ADAM10 antibodies. Flotillin, a lipid raft protein marker, is mainly located in fractions 2 and 3, whereas PCDH12 is concentrated in the nonlipid raft fractions (fractions 8–12), together with ADAM10.
To further demonstrate the participation of ADAM10 in PCDH12 cleavage, we transfected cells with siRNAs to knock down ADAM10 expression. PCDH12 stability in cell extracts and its release into cell supernatants were then analyzed. ADAM10 siRNAs dramatically decreased ADAM10 expression levels compared with control siRNAs (Fig. 5C). This correlated with an increase in PCDH12 levels in cell extracts (Fig. 5C) and a decrease of PCDH12 EC in cell supernatants (Fig. 5D). Therefore, ADAM10 knockdown strikingly improves PCDH12 stability.
ADAM17 is sequestered in lipid rafts, and its substrates seem to be restricted to proteins present in these microdomains. In contrast, the majority of ADAM10 is excluded from lipid rafts (32). We therefore examined the membrane localization of PCDH12 by subcellular fractionation using sucrose density gradients. Although flotillin, a lipid raft component, was mainly located in fractions 2 and 3, PCDH12 and ADAM10 were exclusively located in fractions 8–12, containing the nonlipid raft proteins (Fig. 5E). Likewise, PCDH12 was found in the same membrane compartment as ADAM10, but not in the same as ADAM17. Taken together, these data indicate that PCDH12 is cleaved by ADAM10.
Signaling Molecules Regulate PCDH12 Cleavage
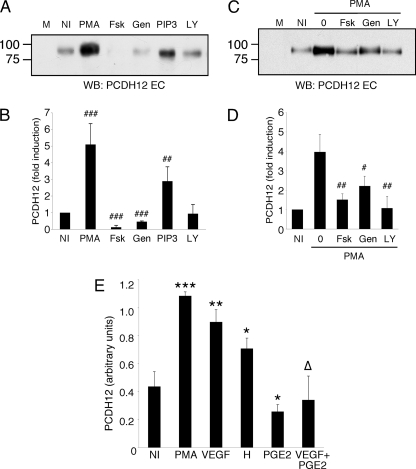
We next wondered whether PCDH12 processing was regulated in response to the activation of various signal transduction pathways. To study this, diverse pharmacological activators or inhibitors of general signaling pathways were used (Fig. 6, A and B). Both Fsk, an activator of adenylate cyclase that raises intracellular levels of cAMP, and the general tyrosine-kinase inhibitor genistein inhibited PCDH12 shedding. In contrast, 1,2-dipalmitoylphosphatidylinositol 3,4,5-trisphosphate, which activates the PI3K-Akt pathway, resulted in increased PCDH12 release, although release was lower than that observed with PMA. Conversely, inhibition of PI3K with LY294002 did not modify PCDH12 release compared with mock conditions. Altogether, these results indicate that PKC and PI3K pathways enhance PCDH12 shedding, whereas cAMP, possibly through activation of protein kinase A, decreases basal cleavage. Furthermore, protein-tyrosine kinases may be involved in basal PCDH12 cleavage.
FIGURE 6.
PCDH12 shedding is regulated by various transduction pathways. A and B, CHO-PCDH12 were treated with Me2SO (NI), 0.1 μg/ml PMA, 10 μm Fsk, 50 μm genistein (Gen), 10 μm 1,2-dipalmitoylphosphatidylinositol 3,4,5-trisphosphate (PIP3), or 10 μm LY294002 (LY). The cell supernatants were harvested at 24 h and analyzed by Western blot, as illustrated in A. M stands for unconditioned cell medium. B, supernatants of three independent experiments were also analyzed by dot blot, and the signal intensities were measured. The data represent the means ± S.D. of signals normalized with signals of controls (NI). Statistical analysis compares controls versus treated cell signals. C and D, CHO-PCDH12 were treated with Me2SO (NI) or PMA alone (0) or PMA together with either forskolin, genistein, or LY294002. Cell supernatant were harvested and analyzed as above. Statistical analysis compares double treatments to PMA alone. ###, p < 0.001; ##, p < 0.01; #, p < 0.02. E, HUVECs were incubated with Me2SO (NI), 0.1 μg/ml PMA, 200 ng/ml VEGF, 1 mm histamine (H), 100 nm PGE2, or VEGF and PGE2. The supernatants were harvested at 24 h, and 100 μl were used for dot blot analysis. The data are the means ± S.D. of triplicates and are representative of three independent experiments. Significance with NI: ***, p < 0.001; **, p < 0.01; *, p < 0.05. Significance with VEGF alone: Δ, p < 0.01.
To examine possible interconnections between signaling pathways leading to PCDH12 cleavage, the cells were treated with PMA together with Fsk, genistein, or LY294002. All three compounds inhibited PMA-activated PCDH12 shedding, returning it to levels close to those found in untreated conditions (Fig. 6, C and D). Altogether, our data show that signaling pathways may have opposing and interdependent effects on PCDH12 shedding.
We then investigated whether natural agonists of endothelial cells, known to activate PKC or to induce cAMP elevation, could modulate PCDH12 shedding. HUVECs were challenged either with VEGF that activates PKC, with PGE2 that stimulates cAMP synthesis, or with histamine, which activates both pathways through H1 and H2 receptors, respectively (33, 34). Both VEGF and histamine significantly increased PCDH12 shedding, and PGE2 diminished basal PCDH12 shedding (Fig. 6E). Interestingly, the VEGF-enhanced release of PCDH12 was abolished by simultaneous addition of PGE2 (Fig. 6E), thus indicating that this latter molecule has a high impact on PCDH12 cleavage. In conclusion, our data strongly suggest that PCDH12 cleavage and activity are regulated by cytokines produced in pathophysiological settings.
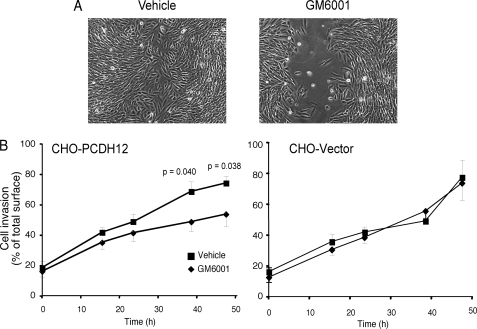
Preventing PCDH12 Cleavage Delays Cell Migration
Because of their role in cell-cell adhesion, cadherins are known to delay cell migration and wound closure of a scratched cell monolayer. However, no significant difference in a wound assay was observed between PCDH12- or mock-transfected CHO cells (Fig. 7). In contrast, a significant decrease in cell migration was measured when CHO-PCDH12 was incubated in the presence of a metalloprotease inhibitor (Fig. 7). This effect was not seen when the inhibitor was applied to mock-transfected cells (Fig. 7). We conclude that PCDH12 stabilization at the cell membrane increased cell-cell adhesion, which slows cell migration into the wounded area.
FIGURE 7.
Effect of metalloprotease inhibitor GM6001 on CHO-PCDH12 migration. CHO-PCDH12 and CHO empty vector confluent monolayers were scraped with a sterile cell scraper to create scratch wounds. After injury, the cells were washed and incubated with culture medium in presence of either GM6001 or vehicle alone. The cells were observed with a videomicroscope, and the images were analyzed with ImageJ software to measure cell invasion in the wounded area. A shows CHO-PCDH12 at 48 h after the wound and the addition of Me2SO or GM6001. B shows graphs representing kinetics of wound closure in each condition (n = 9 for each condition). The error bars represent S.E.
PCDH12 Extracellular Domain Is Detectable in Human Serum and Urine
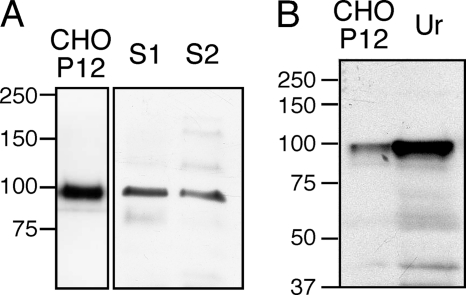
Because our in vitro data showed that PCDH12 was cleaved at high rates, we wondered whether the PCDH12 extracellular domain could be detected in vivo in biological fluids. Human serum from two healthy donors was albumin-depleted before Western blot analysis with anti-PCDH12 EC antibody. A signal at 90 kDa, co-migrating with PCDH12 extracellular domain from CHO-PCDH12 supernatant, could be detected in both sera (Fig. 8A). Interestingly, the PeptideAtlas database indicates that a specific tryptic peptide of PCDH12, located within its extracellular domain (34YQVSEEVPSGTVIGK48), was previously identified by proteomic analysis of two different human plasma samples. These data demonstrate the existence of PCDH12 in human plasma and serum.
FIGURE 8.
Presence of soluble PCDH12 in human serum and urine. A, sera from two individuals (S1 and S2) were depleted in albumin, and 50 μg of protein were electrophoresed and immunoblotted with anti-PCDH12 EC. CHO-PCDH12 supernatant (CHO P12) was loaded as control. B, urine (Ur) was concentrated, and 90 μg of proteins were loaded on SDS-PAGE together with CHO-PCDH12 cell supernatant.
Similarly, PCDH12 extracellular domain is detectable by Western blot in human urine, as illustrated in Fig. 8B. This was confirmed by MS/MS analysis performed on total human urine proteome, in which seven specific peptides of PCDH12 were identified (supplemental Fig. S4). In conclusion, PCDH12 is cleaved in vivo, and its extracellular domain is detectable by two complementary approaches in both serum and urine.
PCDH12 Seric Content of Pregnant Women Is Increased in Pathological Gestations
We next examined whether circulating levels of PCDH12 were altered in pregnant women exhibiting pre-eclampsia (PE), a frequent placental disorder, compared with seric levels of women with nonpathological gestations. PE is currently diagnosed by proteinuria and hypertension and is caused by abnormal placentation, trophoblast invasion deficiency, and subsequent maternal response to placenta-derived circulating stress factors (25). Histological modifications of the placenta, as well as variations of maternal seric markers, have been described in the first trimester of gestation, before the clinical symptoms (35, 36).
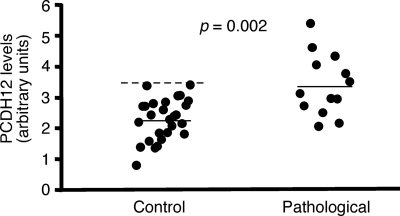
In our study, maternal sera from first trimester gestations were collected at Grenoble University Hospital, before women were diagnosed for PE, and stored. Sera from women who later developed a PE were retrieved, together with sera from nonpathological gestations (n = 27 for nonpathological gestations and n = 13 for gestations with PE; see Table 1 for the clinical characteristics of the pregnant women). The sera were analyzed for their PCDH12 content by Western blotting, as illustrated in supplemental Fig. S5. The results are summarized in Fig. 9 and show a significant increase in mean PCDH12 levels, as well as a shift in the range, when sera were obtained from women who later developed a PE. Thus, the steady state level of circulating PCDH12 is modified in women with PE, which may be related to increased cleavage efficiency, as discussed below.
FIGURE 9.
Comparison in PCDH12 content of maternal sera in normal and pathological gestations. Sera from pregnant women in their first trimester of pregnancy were collected (see Table 1 for gestation characteristics) and analyzed by Western blot for their PCDH12 content (see supplemental Fig. S4 for a representative example of serum analysis). PCDH12 signals were quantified and plotted. The solid lines represent the means, and the dashed line is the 2-fold S.D. value for nonpathological gestations.
DISCUSSION
Human PCDH12 expression is similar to that of mouse Pcdh12: weak in endothelial cells and high in specialized trophoblast cells, i.e. glycogen cells in the mouse and syncytiotrophoblasts as well as villous and extravillous cytotrophoblasts in humans. Because of the extent of anatomical differences between human and mouse placenta, the localization of PCDH12 in human placenta was not self-evident. The high level of expression in this organ, as shown by immunohistology data, is in favor of the protein playing an important role in human placenta, as is the case in the mouse (23).
A striking characteristic of PCDH12 is the high rate at which it is degraded. In this article, we show that PCDH12 is sequentially cleaved by ADAM10 and γ-secretase. Added to this shedding of the extracellular domain, the cytoplasmic domain of PCDH12 is also rapidly degraded by the proteasome.
Sheddase activity is regulated in response to a wide range of transduction pathways and thus participates in numerous physiological processes, such as the regulation of epithelial receptor pathways, Notch signaling, or amyloid precursor protein processing (29, 30). Here, we show that constitutive PCDH12 shedding is enhanced by activation of PKC and PI3K pathways, whereas it is reduced by raising intracellular cAMP or inhibiting tyrosine kinases. PMA-induced shedding could also be inhibited by elevated cAMP and repression of tyrosine phosphorylation, as well as by PI3K inhibition, suggesting that PCDH12 cleavage is regulated by multiple interconnected pathways. Furthermore, endothelial cells regulate PCDH12 shedding in response to cytokines known to induce the cAMP or the PKC pathways. Endothelial cells mainly express PGE2 receptors EP2 and EP4, two G protein-coupled receptors that both stimulate adenylate cyclase via stimulatory subunit Gs, whereas VEGF activates the phospholipase C-PKC pathway through VEGFR2 (33, 34). As expected, VEGF enhanced PCDH12 shedding, whereas PGE2 inhibited basal as well as VEGF-induced cleavage. In endothelial cells, histamine stimulates both Gq and Gs subunits through H1 and H2 receptors, respectively (33). Interestingly, stimulation of the Gq subunit, which activates the phospholipase C-PKC pathway, seems to be preponderant over Gs stimulation, because histamine significantly increased PCDH12 shedding.
Cadherin cleavage is an efficient means of remodeling cell-cell interaction. Indeed, prevention of PCDH12 cleavage by a metalloprotease inhibitor delayed cell migration in a cell monolayer wound assay, suggesting that the adhesive activity of PCDH12 is impaired by its high degradation rate in this model system. Interestingly, mouse placentas deficient in Pcdh12 exhibited an alteration in labyrinthine and intermediate layer segregation, which potentially reflects a modification of the capacity of glycogen cells to collectively migrate or aggregate. Similarly, PCDH12 could be involved in human microvillus formation or cytotrophoblast invasion of maternal uterine endometrium.
ADAM10 is expressed in syncytiotrophoblasts (37) and may thus directly regulate PCDH12 shedding within this cell layer. ADAM10 is also efficiently inhibited by tissue inhibitors of metalloproteinase 1 and 3 (38). This provides a potential external regulation mechanism as well. Tissue inhibitor of metalloproteinase 1 is highly expressed by maternal decidual cells and in the stromal compartment of placental villi (39). Tissue inhibitor of metalloproteinase 3 is expressed by invasive trophoblasts (40). Therefore, both inhibitors may regulate PCDH12 activity during critical phases of pregnancy. Importantly, ADAM10 expression is increased in syncytiotrophoblasts of PE placentas compared with normal placentas, which has been linked to the augmented release of soluble VEGFR1, a well established marker of PE (35, 37); therefore, similar to VEGFR1, the increase in seric levels of PCDH12 in patients with PE may be due to enhanced ADAM10 activity and thus PCDH12 shedding in the trophoblast layer directly exposed to the maternal bloodstream.
The extracellular domain of PCDH12 was detected in both human serum and urine. Therefore, PCDH12 extracellular domain seems to be relatively stable in human plasma and is eliminated, at least partially, through the urinary route. These findings lead to major issues that need to be further addressed: is the shed protein fragment biologically active, and is it involved in the pathogenesis of PE?
ADAM10 cleaves other members of the cadherin family, including N-, E-, and VE-cadherins as well as Pcdh γ B4 and C3 (41–44). Therefore, ADAM10-dependent shedding may be a general mechanism of irreversible intercellular junction disruption, as opposed to intracellular phosphorylation for example, which is only transient. The cytoplasmic domain of some proteins cleaved by γ-secretase, including Notch and ErbB4, have been shown to translocate to the nucleus where they activate gene transcription (28). In addition, the intracellular domain of Pcdhγ C3 tagged with the green fluorescent protein has been detected in the nucleus of transfected cells (45). In our hands, attempts to detect PCDH12 cytoplasmic domain in the nucleus were unsuccessful, even in the presence of the proteasome inhibitor lactacystin or the nucleus-sequestering drug leptomycin (data not shown). Although the possibility that the cytoplasmic domain has signaling functions cannot be excluded, these functions probably do not depend on translocation to the nucleus. Alternatively, as yet unidentified cytoplasmic partners of PCDH12 may be released after PCDH12 cleavage and may translocate to the nucleus. This type of mechanism has been demonstrated for β-catenin after N- or E-cadherin proteolysis (41, 42).
Our results provide novel insights into the cellular function of PCDH12 and its regulation. We have shown that PCDH12 controls cell migration, probably through increasing cell-cell adhesion. Its biological activity is highly regulated by ADAM10-dependent proteolytic cleavage, which is controlled by physiological agonists. The detection of PCDH12 extracellular domain in biological fluids provides evidence of in vivo shedding. Because of its high rate of expression in the placenta, it is tempting to speculate that PCDH12 in humans plays a role similar to that of murine Pcdh12 in placental development. Finally, we show for the first time that seric PCDH12 levels are increased in women with PE. The relevance of this finding in terms of clinical significance needs to be further investigated.
Supplementary Material
Acknowledgments
We thank Christophe Masselon and Magali Court from the Laboratoire d'Etude de la Dynamique des Protéomes for sharing their unpublished proteomic data on urine proteins. We further thank Prof. Andreas Ludwig for reagents and helpful discussion.
This work was supported by the CEA, INSERM, and Université Grenoble 1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- PCDH or Pcdh
- protocadherin
- CTF
- C-terminal fragment
- Fsk
- forskolin
- HUVEC
- human umbilical vein endothelial cell
- PE
- pre-eclampsia
- PGE2
- prostaglandin E2
- PMA
- phorbol 12-myristate 13-acetate
- Cter
- C-terminal
- EC
- endothelial cell(s).
REFERENCES
- 1. Wheelock M. J., Johnson K. R. (2003) Annu. Rev. Cell Dev. Biol. 19, 207–235 [DOI] [PubMed] [Google Scholar]
- 2. Junghans D., Haas I. G., Kemler R. (2005) Curr. Opin. Cell Biol. 17, 446–452 [DOI] [PubMed] [Google Scholar]
- 3. Morishita H., Yagi T. (2007) Curr. Opin. Cell Biol. 19, 584–592 [DOI] [PubMed] [Google Scholar]
- 4. Sano K., Tanihara H., Heimark R. L., Obata S., Davidson M., St John T., Taketani S., Suzuki S. (1993) EMBO J. 12, 2249–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu Q., Maniatis T. (1999) Cell 97, 779–790 [DOI] [PubMed] [Google Scholar]
- 6. Kuroda H., Inui M., Sugimoto K., Hayata T., Asashima M. (2002) Dev. Biol. 244, 267–277 [DOI] [PubMed] [Google Scholar]
- 7. Medina A., Swain R. K., Kuerner K. M., Steinbeisser H. (2004) EMBO J. 23, 3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aamar E., Dawid I. B. (2008) Dev. Biol. 318, 335–346 [DOI] [PubMed] [Google Scholar]
- 9. Bononi J., Cole A., Tewson P., Schumacher A., Bradley R. (2008) Mech. Dev. 125, 1033–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakao S., Platek A., Hirano S., Takeichi M. (2008) J. Cell Biol. 182, 395–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weiner J. A., Wang X., Tapia J. C., Sanes J. R. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X., Weiner J. A., Levi S., Craig A. M., Bradley A., Sanes J. R. (2002) Neuron 36, 843–854 [DOI] [PubMed] [Google Scholar]
- 13. Emond M. R., Jontes J. D. (2008) Dev. Biol. 321, 175–187 [DOI] [PubMed] [Google Scholar]
- 14. Okazaki N., Takahashi N., Kojima S., Masuho Y., Koga H. (2002) Carcinogenesis 23, 1139–1148 [DOI] [PubMed] [Google Scholar]
- 15. Yu J. S., Koujak S., Nagase S., Li C. M., Su T., Wang X., Keniry M., Memeo L., Rojtman A., Mansukhani M., Hibshoosh H., Tycko B., Parsons R. (2008) Oncogene 27, 4657–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahmed Z. M., Riazuddin S., Ahmad J., Bernstein S. L., Guo Y., Sabar M. F., Sieving P., Riazuddin S., Griffith A. J., Friedman T. B., Belyantseva I. A., Wilcox E. R. (2003) Hum. Mol. Genet. 12, 3215–3223 [DOI] [PubMed] [Google Scholar]
- 17. Reiners J., Märker T., Jürgens K., Reidel B., Wolfrum U. (2005) Mol. Vis. 11, 347–355 [PubMed] [Google Scholar]
- 18. Dibbens L. M., Tarpey P. S., Hynes K., Bayly M. A., Scheffer I. E., Smith R., Bomar J., Sutton E., Vandeleur L., Shoubridge C., Edkins S., Turner S. J., Stevens C., O'Meara S., Tofts C., Barthorpe S., Buck G., Cole J., Halliday K., Jones D., Lee R., Madison M., Mironenko T., Varian J., West S., Widaa S., Wray P., Teague J., Dicks E., Butler A., Menzies A., Jenkinson A., Shepherd R., Gusella J. F., Afawi Z., Mazarib A., Neufeld M. Y., Kivity S., Lev D., Lerman-Sagie T., Korczyn A. D., Derry C. P., Sutherland G. R., Friend K., Shaw M., Corbett M., Kim H. G., Geschwind D. H., Thomas P., Haan E., Ryan S., McKee S., Berkovic S. F., Futreal P. A., Stratton M. R., Mulley J. C., Gécz J. (2008) Nat. Genet. 40, 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morrow E. M., Yoo S. Y., Flavell S. W., Kim T. K., Lin Y., Hill R. S., Mukaddes N. M., Balkhy S., Gascon G., Hashmi A., Al-Saad S., Ware J., Joseph R. M., Greenblatt R., Gleason D., Ertelt J. A., Apse K. A., Bodell A., Partlow J. N., Barry B., Yao H., Markianos K., Ferland R. J., Greenberg M. E., Walsh C. A. (2008) Science 321, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Telo' P., Breviario F., Huber P., Panzeri C., Dejana E. (1998) J. Biol. Chem. 273, 17565–17572 [DOI] [PubMed] [Google Scholar]
- 21. Rampon C., Prandini M. H., Bouillot S., Pointu H., Tillet E., Frank R., Vernet M., Huber P. (2005) Exp. Cell Res. 302, 48–60 [DOI] [PubMed] [Google Scholar]
- 22. Bouillot S., Rampon C., Tillet E., Huber P. (2006) Placenta 27, 882–888 [DOI] [PubMed] [Google Scholar]
- 23. Rampon C., Bouillot S., Climescu-Haulica A., Prandini M. H., Cand F., Vandenbrouck Y., Huber P. (2008) Physiol. Genomics 34, 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cetin I., Huppertz B., Burton G., Cuckle H., Gonen R., Lapaire O., Mandia L., Nicolaides K., Redman C., Soothill P., Spencer K., Thilaganathan B., Williams D., Meiri H. (2011) Placenta 32, S4–16 [DOI] [PubMed] [Google Scholar]
- 25. Sibai B., Dekker G., Kupferminc M. (2005) Lancet 365, 785–799 [DOI] [PubMed] [Google Scholar]
- 26. Tillet E., Ruggiero F., Nishiyama A., Stallcup W. B. (1997) J. Biol. Chem. 272, 10769–10776 [DOI] [PubMed] [Google Scholar]
- 27. Wallez Y., Cand F., Cruzalegui F., Wernstedt C., Souchelnytskyi S., Vilgrain I., Huber P. (2007) Oncogene 26, 1067–1077 [DOI] [PubMed] [Google Scholar]
- 28. De Strooper B., Annaert W. (2010) Annu. Rev. Cell Dev. Biol. 26, 235–260 [DOI] [PubMed] [Google Scholar]
- 29. Huovila A. P., Turner A. J., Pelto-Huikko M., Kärkkäinen I., Ortiz R. M. (2005) Trends Biochem. Sci. 30, 413–422 [DOI] [PubMed] [Google Scholar]
- 30. Edwards D. R., Handsley M. M., Pennington C. J. (2008) Mol. Aspects Med. 29, 258–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hundhausen C., Misztela D., Berkhout T. A., Broadway N., Saftig P., Reiss K., Hartmann D., Fahrenholz F., Postina R., Matthews V., Kallen K. J., Rose-John S., Ludwig A. (2003) Blood 102, 1186–1195 [DOI] [PubMed] [Google Scholar]
- 32. Kojro E., Gimpl G., Lammich S., Marz W., Fahrenholz F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5815–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mehta D., Malik A. B. (2006) Physiol. Rev. 86, 279–367 [DOI] [PubMed] [Google Scholar]
- 34. Rao R., Redha R., Macias-Perez I., Su Y., Hao C., Zent R., Breyer M. D., Pozzi A. (2007) J. Biol. Chem. 282, 16959–16968 [DOI] [PubMed] [Google Scholar]
- 35. Maynard S. E., Min J. Y., Merchan J., Lim K. H., Li J., Mondal S., Libermann T. A., Morgan J. P., Sellke F. W., Stillman I. E., Epstein F. H., Sukhatme V. P., Karumanchi S. A. (2003) J. Clin. Invest. 111, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Venkatesha S., Toporsian M., Lam C., Hanai J., Mammoto T., Kim Y. M., Bdolah Y., Lim K. H., Yuan H. T., Libermann T. A., Stillman I. E., Roberts D., D'Amore P. A., Epstein F. H., Sellke F. W., Romero R., Sukhatme V. P., Letarte M., Karumanchi S. A. (2006) Nat. Med. 12, 642–649 [DOI] [PubMed] [Google Scholar]
- 37. Zhao S., Gu Y., Fan R., Groome L. J., Cooper D., Wang Y. (2010) Placenta 31, 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Amour A., Knight C. G., Webster A., Slocombe P. M., Stephens P. E., Knäuper V., Docherty A. J., Murphy G. (2000) FEBS Lett. 473, 275–279 [DOI] [PubMed] [Google Scholar]
- 39. Polette M., Nawrocki B., Pintiaux A., Massenat C., Maquoi E., Volders L., Schaaps J. P., Birembaut P., Foidart J. M. (1994) Lab. Invest. 71, 838–846 [PubMed] [Google Scholar]
- 40. Bass K. E., Li H., Hawkes S. P., Howard E., Bullen E., Vu T. K., McMaster M., Janatpour M., Fisher S. J. (1997) Dev. Genet. 21, 61–67 [DOI] [PubMed] [Google Scholar]
- 41. Maretzky T., Reiss K., Ludwig A., Buchholz J., Scholz F., Proksch E., de Strooper B., Hartmann D., Saftig P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9182–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reiss K., Maretzky T., Ludwig A., Tousseyn T., de Strooper B., Hartmann D., Saftig P. (2005) EMBO J. 24, 742–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reiss K., Maretzky T., Haas I. G., Schulte M., Ludwig A., Frank M., Saftig P. (2006) J. Biol. Chem. 281, 21735–21744 [DOI] [PubMed] [Google Scholar]
- 44. Schulz B., Pruessmeyer J., Maretzky T., Ludwig A., Blobel C. P., Saftig P., Reiss K. (2008) Circ. Res. 102, 1192–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haas I. G., Frank M., Véron N., Kemler R. (2005) J. Biol. Chem. 280, 9313–9319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.