Abstract
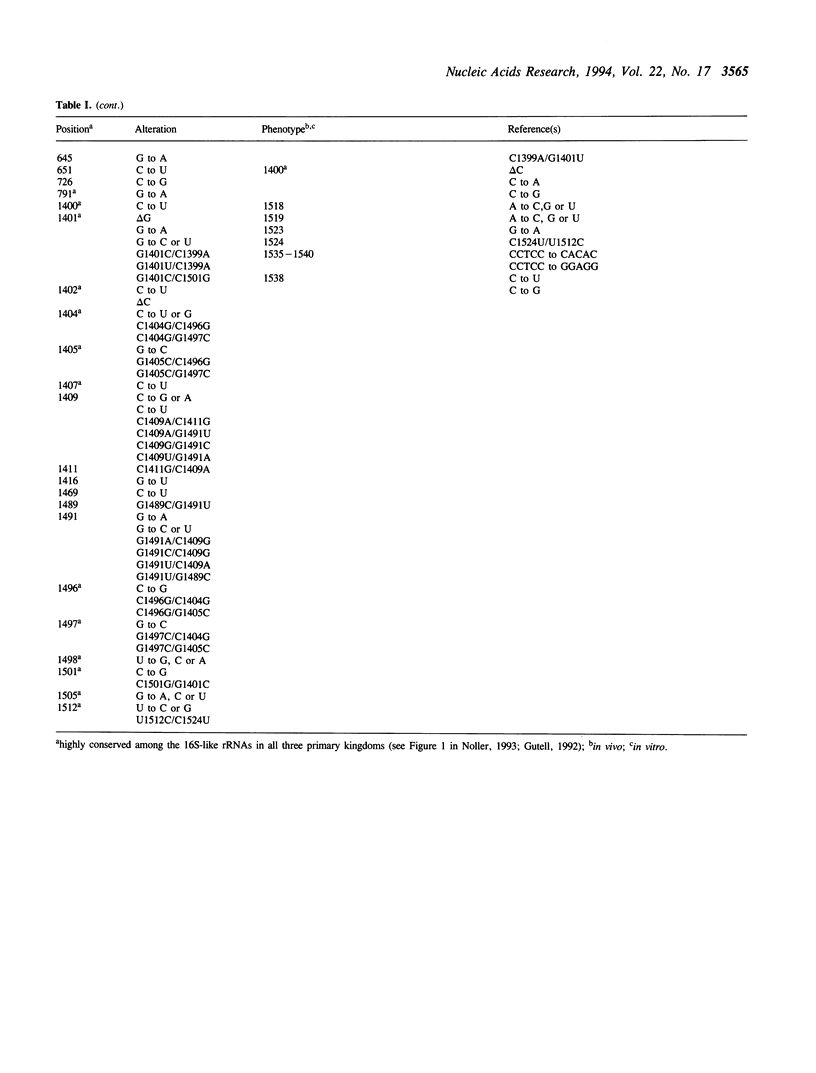
The 16S ribosomal RNA mutation database (16SMDB), provides a list of mutated positions in 16S ribosomal RNA from Escherichia coli and the identity of each alteration. Information provided for each mutation includes: (1) a brief description of the phenotype(s) associated with each mutation, (2) whether a mutant phenotype has been detected by in vivo or in vitro methods, and (3) relevant literature citations. The database is available via ftp.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brink M. F., Verbeet M. P., de Boer H. A. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993 Oct;12(10):3987–3996. doi: 10.1002/j.1460-2075.1993.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammel C. S., Noller H. F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993 Apr;7(4):660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- Pinard R., Payant C., Melançon P., Brakier-Gingras L. The 5' proximal helix of 16S rRNA is involved in the binding of streptomycin to the ribosome. FASEB J. 1993 Jan;7(1):173–176. doi: 10.1096/fasebj.7.1.7678560. [DOI] [PubMed] [Google Scholar]
- Triman K., Becker E., Dammel C., Katz J., Mori H., Douthwaite S., Yapijakis C., Yoast S., Noller H. F. Isolation of temperature-sensitive mutants of 16 S rRNA in Escherichia coli. J Mol Biol. 1989 Oct 20;209(4):645–653. doi: 10.1016/0022-2836(89)92000-7. [DOI] [PubMed] [Google Scholar]


