Abstract
Multiple MAP kinase pathways share components yet initiate distinct biological processes. Signaling fidelity can be maintained by scaffold proteins and restriction of signaling complexes to discreet subcellular locations. For example, the yeast MAP kinase scaffold Ste5 binds to phospholipids produced at the plasma membrane and promotes selective MAP kinase activation. Here we show that Pik1, a phosphatidylinositol 4-kinase that localizes primarily to the Golgi, also regulates MAP kinase specificity but does so independently of Ste5. Pik1 is required for full activation of the MAP kinases Fus3 and Hog1 and represses activation of Kss1. Further, we show by genetic epistasis analysis that Pik1 likely regulates Ste11 and Ste50, components shared by all three MAP kinase pathways, through their interaction with the scaffold protein Opy2. These findings reveal a new regulator of signaling specificity functioning at endomembranes rather than at the plasma membrane.
Keywords: MAP kinases (MAPKs), Phosphatidylinositol, Phosphatidylinositol Signaling, Phosphatidylinositol Kinase, Signal Transduction, Yeast, Fus3, Hog1, Kss1, Pheromone
Introduction
Cells growing in complex environments are exposed to multiple chemical and physical stimuli. Many external stimuli activate MAP kinase pathways to elicit intracellular biological processes. In some cases, a single stimulus will activate multiple MAP kinases, yet signaling specificity is maintained (1). How cells regulate the activation of different MAP kinase pathways to invoke the appropriate biological response is not well understood.
The yeast Saccharomyces cerevisiae provides a versatile model for understanding the coordinated regulation of multiple MAP kinases. In yeast, three well characterized MAP kinase pathways respond to different external stimuli to initiate distinct and sometimes mutually exclusive biological processes (Fig. 1A) (2). First, mating pheromones activate a pathway that induces cell cycle arrest, polarized cell expansion, and the fusion of haploid a- and α-type cells to form an a/α diploid. This process is mediated by a heterotrimeric G protein and a protein kinase cascade comprised of Ste20, Ste11, Ste7, and the MAP kinase Fus3 (3). Second, nutrient deprivation results in the activation of the same kinase components, with the exception of the MAP kinase Kss1 (4, 5), and induces filamentous growth as well as increased adherence and invasion into the substratum (6). Third, osmotic stress activates the MAP kinase Hog1 in the high osmolarity glycerol (HOG)2 response pathway (7) and induces glycerol production to counterbalance osmotic pressure and enable cell survival (8, 9).
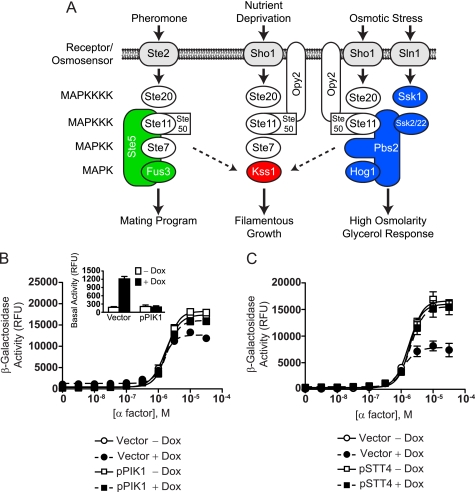
FIGURE 1.
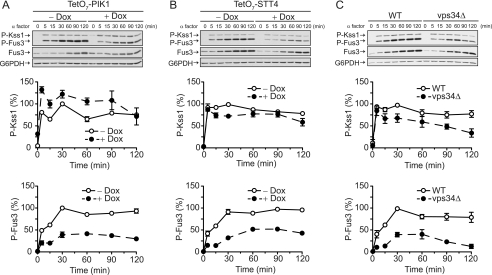
Pik1 is required for proper pheromone signaling. A, Three yeast MAP kinase pathways respond to different stimuli and regulate distinct biological processes, yet all three pathways share signaling components. The MAP kinase kinase kinase (MAPKKK) Ste11 and the adapter Ste50 regulate all three pathways. Some pathway components are not shown for the sake of clarity. MAPKKKK, MAP kinase kinase kinase kinase. B, TetO7-PIK1 cells were transformed with the FUS1-lacZ transcription reporter and pRS315-PIK1 (pPIK1) or pRS315 (Vector) and treated with α factor pheromone at the indicated concentration for 90 min. β-galactosidase activity was measured spectrofluorometrically. The inset shows activity in cells not stimulated with α factor. Dox, doxycycline; RFU, relative fluorescence units. C, TetO7-STT4 cells were transformed with the FUS1-lacZ reporter and pRS315-STT4 (pSTT4) or empty vector and treated with α factor pheromone. Results are the mean ± S.E. for three individual experiments each performed in triplicate.
Despite profound differences in stimulus and response, different MAP kinase pathways will often share signaling components. For example, the MAP kinase kinase kinase Ste11 and its adapter protein Ste50 are shared by the mating, filamentous growth, and HOG pathways (10–12). Ste7 is shared by the mating and filamentous pathways (2). Yet remarkably little pathway cross-talk is observed. In the pheromone response pathway, MAP kinase specificity is maintained by the scaffold protein Ste5, which binds Ste11, Ste7, and Fus3 (13). Upon pheromone stimulation, Ste5 translocates to the plasma membrane by binding to G protein βγ subunits (14) and facilitates signal propagation through Fus3 but not Kss1 or Hog1 (15). In the HOG pathway, specificity is maintained by another scaffold protein, Pbs2, which also functions as the kinase that activates Hog1. However, despite the existence of these scaffolds, Kss1 is still partially activated in response to pheromone (16, 17) and osmotic stress (18). Therefore, scaffolds that associate with Fus3 or Hog1 are not sufficient to prevent activation of Kss1. Additional mechanisms are likely to be required to maintain proper balance between Fus3, Hog1, and Kss1 activation.
Previous reports demonstrated a role for phospholipids in maintaining signaling fidelity. For example, deletion of the PtdIns 3-kinase Vps34 alters Fus3 activation but not Kss1 (19), indicating that phosphorylated phosphoinositides may play an important role in maintaining MAP kinase specificity. Ste5 is likewise required for Fus3 activation. Ste5 binds PtdIns-4-P and PtdIns-4,5-P2 in vitro (20, 21), and Ste5 translocation to the plasma membrane in vivo requires the PtdIns 4-kinase Stt4 and the PtdIns-4-P 5-kinase Mss4 (21, 22). Although both Stt4 and Mss4 localize primarily to the plasma membrane (23, 24), Vps34 localizes to endosomes (25, 26). Together these findings show that phospholipids play an important role in maintaining MAP kinase specificity and that these phospholipids do not necessarily originate at the plasma membrane.
In a recent screen of essential genes we identified regulators of pheromone signaling, including Stt4, as well as a second PtdIns 4-kinase, Pik1 (27). As noted above, Stt4 promotes the activation of Fus3. Here we show that Pik1 regulates the activity of three different MAP kinases in yeast. Although Pik1 enhances Fus3 and Hog1 activation, it inhibits Kss1 activation. We demonstrate further that Pik1 regulates MAP kinase signaling through a mechanism distinct from that of Stt4. Although Stt4 acts by promoting Ste5 translocation to the plasma membrane (22), Pik1 exerts its effects through Ste11 and the adapter protein Ste50 and requires the scaffold protein Opy2. These findings reveal a novel role for PtdIns-4-P at endomembranes in maintaining specificity across multiple MAP kinase pathways.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Growth Conditions
Standard procedures for the growth, maintenance, and transformation of yeast and bacteria and for the manipulation of DNA were used throughout. Cells were grown in selective medium containing 2% (w/v) dextrose or galactose to induce gene expression. Yeast strains used are listed in supplemental Table S1. Plasmids used are listed in supplemental Table S2. Plasmid pRS313 GAL-STE5-CTM was created by SacI and ApaI digestion of pGS5-CTM (14) and ligation into the corresponding sites of pRS313.
The yeast TetO7 strains (28) were grown in selective medium to an A600 nm of ∼0.8, reinoculated at 1:80 into medium containing doxycycline at a final concentration of 10 μg/ml, and grown to an A600 nm of ∼0.8. To activate the pheromone pathway, α factor pheromone was added at a final concentration of 3 μm for 30 min unless noted otherwise. To induce osmotic stress, KCl was added to a final concentration of 0.5 m for 10 min unless noted otherwise. Time courses were halted by the addition of trichloroacetic acid at a final concentration of 5%.
Cell Extracts and Immunoblotting
Protein extracts were produced by glass bead lysis in trichloroacetic acid as described previously (29). Protein extracts were resolved by 12.5% SDS-PAGE and Coomassie-stained or subjected to immunoblotting with phospho-p44/42 MAPK antibodies (9101L, Cell Signaling Technology) at 1:500, Fus3 antibodies (sc-6773, Santa Cruz Biotechnology, Inc.) at 1:500, phospho-p38 MAPK antibodies (9211L, Cell Signaling Technology) at 1:500, Hog1 antibodies (sc-6815, Santa Cruz Biotechnology) at 1:500, and glucose-6-phosphate dehydrogenase (G6PDH) antibodies (A9521, Sigma-Aldrich) at 1:50,000. Immunoreactive species were visualized by chemiluminescent detection (PerkinElmer Life Sciences) of horseradish peroxidase-conjugated antibodies (170-5047 and 170-5046, Bio-Rad). Protein concentration was determined by detergent-compatible protein assay (500-0112, Bio-Rad). Band intensity was quantified by scanning densitometry using Image J (National Institutes of Health). Phospho-Fus3 and phospho-Kss1 values were normalized to G6PDH loading control, and phospho-Hog1 values were normalized to total Hog1.
Transcriptional Reporter Assay
FUS1-LacZ levels were measured 90 min after treatment with α factor pheromone using a β-galactosidase assay and fluorescein di-β-d-galactopyranoside as described previously (30).
Microscopy
Cells were visualized with differential interference contrast and fluorescence microscopy using an Olympus Fluoview 1000 confocal microscope equipped with a 488-nm laser (blue argon for GFP). Images were analyzed using ImageJ (National Institutes of Health).
RESULTS
Pik1 Is Required for Proper Pheromone Signaling
In a recent screen to identify essential genes required for proper pheromone signaling, we identified two PtdIns 4-kinases, Stt4 and Pik1. Although Stt4 and Pik1 have the same enzymatic activity, they are both essential, suggesting they have non-redundant functions in vivo (31). Furthermore, Stt4 and Pik1 localize to different parts of the cell: Stt4 at the plasma membrane (23) and Pik1 at the Golgi and nucleus (32). Given that depleting the cell of either STT4 or PIK1 diminishes pheromone signaling (27), we reasoned that they might likewise have non-redundant functions in the pheromone response pathway. Stt4 has a known role in promoting Ste5 translocation to the plasma membrane (22). Here we investigate the signaling properties of Pik1.
Previous research on Pik1 and Stt4 was conducted using temperature-sensitive (ts) alleles. The use of ts alleles requires growth at suboptimal temperatures and introduces destabilizing mutations that could alter enzyme function or protein-protein interactions. Growth at high temperatures can impair MAP kinase activity independent of any gene mutations. For example, Garrenton et al. (22) reported an ∼50% reduction in Fus3 activation in wild-type cells grown at 37 °C versus 26 °C (see Fig. 5 in 22). Thus, the use of higher growth temperatures could obscure small differences resulting from partial loss of function ts alleles.
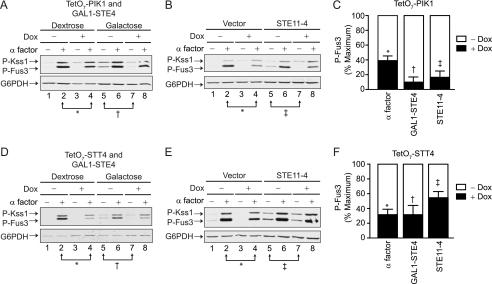
FIGURE 5.
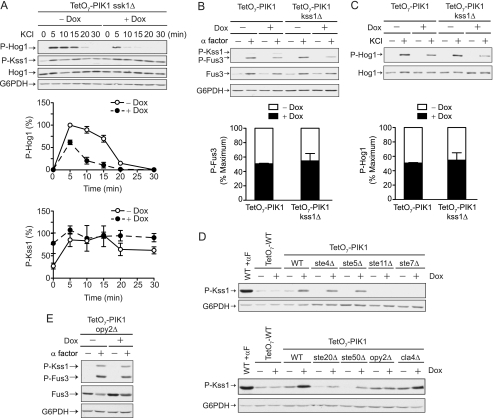
Pik1 acts via Ste11 or a pathway component downstream of Ste11. A, TetO7-PIK1 cells were transformed with pRS315-GAL1-STE4 and grown in selective medium containing dextrose or galactose to induce Ste4 (Gβ) protein expression. Cells were treated with 10 μg/ml doxycycline for 15 h and 3 μm α factor pheromone for 30 min, as indicated. Cell lysates were resolved by 12.5% SDS-PAGE and immunoblotting with phospho-p44/42 antibodies (P-Kss1 and P-Fus3) and G6PDH antibodies as a loading control. B, TetO7-PIK1 cells were transformed with pRS425 (Vector) or pRS425-STE11–4. C, P-Fus3 levels from A and B were quantified by scanning densitometry and analyzed with ImageJ software. Results are the mean ± S.E. for three individual experiments. D, TetO7-STT4 cells treated as in A. E, TetO7-STT4 cells treated as in B. F, P-Fus3 levels from D and E were quantified as in C.
To best determine the contribution of Pik1 in pheromone signaling and to verify a role for Stt4, we used strains where the native promoter was replaced with a doxycycline-repressible (TetO7) promoter. Cells were grown in the presence or absence of doxycycline to repress gene expression. Pathway activation was measured using a highly specific pheromone-inducible promoter (from FUS1) fused to the β-galactosidase gene. As a control we established that knockdown of PIK1 or treatment with doxycycline alone had no effect on overall protein expression (supplemental Fig. S1). As shown in Fig. 1, knockdown expression of TetO7-PIK1 or TetO7-STT4 results in dampened transcriptional output upon pheromone stimulation. Furthermore, knockdown of PIK1 results in constitutive activation in the absence of pheromone (27). Thus, knockdown of PIK1 paradoxically yields both a dampened maximum response and increased basal activity. To confirm the integrity of these strains, we expressed single-copy plasmids containing the wild-type gene in the corresponding TetO7 strain. For both PIK1 (Fig. 1B) and STT4 (C), introduction of the absent gene restored normal pheromone responses.
To determine whether depletion of PIK1 results in a subsequent loss of intracellular PtdIns 4-P, we visualized PtdIns 4-P in vivo using three well characterized GFP-tagged biosensors. First, the pleckstrin homology (PH) domain from phospholipase Cδ (PHPLCδ-GFP) binds specifically to PtdIns-4,5-P2 at the plasma membrane and has been used to monitor plasma membrane pools of both PtdIns 4-P and PtdIns-4,5-P2 (33). Second, the PH domain from FAPP1 (PHFAPP1-GFP) binds specifically to PtdIns 4-P at the Golgi (33). Third, the conserved region 2 domain from bovine lactadherin (C2lact-GFP) binds phosphatidylserine, an abundant component of all membranes (34), and serves as a reference control. Knockdown of PIK1 resulted in a partial loss of Golgi staining of PHFAPP1-GFP but no change in localization of PHPLCδ-GFP or C2lact-GFP (supplemental Fig. S2A). In contrast, knockdown of STT4 resulted in a partial loss of plasma membrane staining of PHPLCδ-GFP but no change in localization of PHFAPP1-GFP or C2lact-GFP (supplemental Fig. S2B) (23). Thus, partial knockdown of either PIK1 or STT4 is sufficient to observe a dampened pheromone response, underscoring the importance of PtdIns 4-P in maintaining proper pheromone signaling.
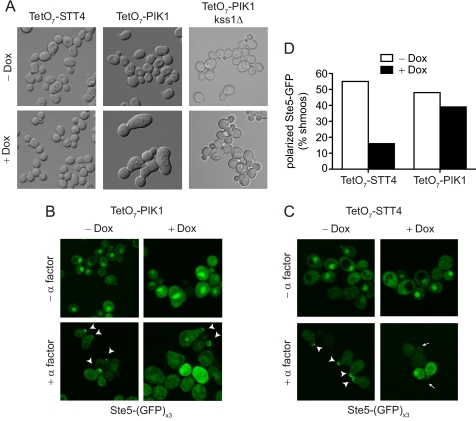
Loss of Pik1 Induces Elongated Growth
Cells exposed to pheromone undergo cell cycle arrest and form mating projections in preparation for mating (“shmoo” morphology). At low doses of the pheromone, cells continue to divide in a bipolar fashion and elongate along pheromone gradients in the direction of a potential mating partner (chemotropic growth) (35–38). When elongated cells encounter a sufficiently high level of pheromone, they undergo cell cycle arrest and form shmoos. Because knockdown of PIK1 results in constitutive induction of pheromone-responsive genes (see Fig. 1, B and inset), we considered whether these cells also exhibit an altered morphology. Differential interference contrast microscopy revealed that PIK1 knockdown results in large and elongated cells, even in the absence of pheromone (Fig. 2A). The observed cellular elongation was most similar to that of cells exposed to low doses of pheromone (35–38) and is consistent with our observation that PIK1 knockdown results in a small but significant increase in basal activation of the pheromone pathway. Considering that the MAPK Kss1 induces chemotropic growth at low doses of pheromone, we asked whether the elongated growth phenotype was dependent on Kss1. We found that deletion of KSS1 from the TetO7-PIK1 strain restored normal cellular morphology, suggesting that PIK1 knockdown leads to constitutive activation of Kss1 (Fig. 2A). In contrast, knockdown of STT4 had no effect on cell morphology, suggesting further that Pik1 and Stt4 regulate signaling in fundamentally different ways.
FIGURE 2.
Pik1 alters cell morphology but does not alter Ste5 localization. A, differential interference contrast image of TetO7-STT4, TetO7-PIK1, and TetO7-PIK1 kss1Δ cells treated with 10 μg/ml doxycycline for 15 h where indicated (+ Dox). B, GFP fluorescence of TetO7-PIK1 cells expressing pRS316-Ste5-(GFP)x3. Cells were treated with doxycycline for 15 h and 3 μm α factor pheromone for 90 min. Arrow heads indicate Ste5-GFP localized to shmoo tips. C, TetO7-STT4 cells treated as in B. Arrows indicate the absence of Ste5-GFP at shmoo tips. D, quantitation of the percentage of shmoos with polarized Ste5-GFP from B and C (n > 50 shmoos).
Pik1 Regulates Pheromone Signaling Independently of Ste5
Prior to pheromone stimulation, the MAP kinase scaffold Ste5 is localized diffusely in the nucleus and cytoplasm. After pheromone stimulation, Ste5 translocates to the plasma membrane. In an stt4ts strain, however, Ste5 is no longer localized to the plasma membrane, most likely because of diminished synthesis of PtdIns 4-P or PtdIns-4,5-P2 (14, 21, 22). Accordingly, Stt4 (like Ste5) is required for full activation of Fus3. Pik1 also generates PtdIns-4-P and is required for Fus3 activity, yet Pik1 is absent from the plasma membrane. Thus, we investigated whether Pik1 affects Ste5 localization in some other way. To this end, we expressed STE5-(GFP)x3 in a TetO7-PIK1 strain and treated cells with and without α factor pheromone. Consistent with previous data from ts strains, knockdown of PIK1 had no effect on Ste5-GFP localization (Fig. 2, B and D), whereas knockdown of STT4 resulted in a marked loss of Ste5-GFP from the shmoo tip (Fig. 2, C and D). These results indicate that Ste5 translocation to the plasma membrane requires Stt4 but is largely unaffected by Pik1.
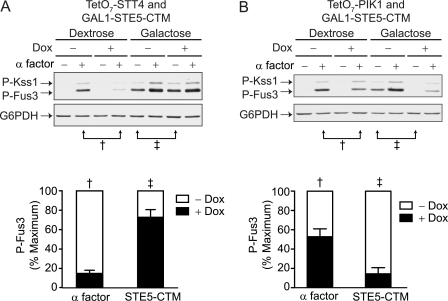
In addition to binding phospholipids, Ste5 binds several signaling components, including Gβγ (14) and all three kinases in the MAP kinase cascade: Ste11, Ste7, and Fus3 (39, 40). Although available evidence indicates that Stt4 helps to recruit this scaffolded complex to the plasma membrane, the differences between Stt4 and Pik1 suggest that Pik1 might activate the complex without contributing to Ste5-membrane association. To test this model, we tethered Ste5 to the plasma membrane using a C-terminal transmembrane (CTM) fusion protein and expressed it under the control of a galactose-inducible promoter. Ste5-CTM results in constitutive association of the MAP kinase cascade with upstream activators and therefore results in constitutive activation of Fus3. Ste5-CTM is localized only at the plasma membrane and therefore should not interact with Pik1 or PtdIns 4-P at the Golgi or nucleus (14). We then monitored Fus3 activation using an antibody that recognizes the dually phosphorylated and fully activated form of the kinase. As shown in Fig. 3, Ste5-CTM strongly activates Fus3 (14). As expected, Fus3 activity was largely unaffected by the loss of STT4 (Fig. 3A, lane 5 versus lane 7). In contrast, this response was substantially diminished by the loss of PIK1 (Fig. 3B, lane 5 versus lane 7). Thus, Stt4 and Pik1 regulate MAP kinase signaling by distinct mechanisms. Whereas expression of Ste5-CTM bypasses a need for PtdIns-4-P at the plasma membrane, these cells remain sensitive to changes in PtdIns-4-P by Pik1 within the cell.
FIGURE 3.
Pik1 regulates pheromone signaling independently of Ste5. A, TetO7-STT4 cells were transformed with pRS313-GAL1-STE5-CTM and grown in selective medium containing dextrose or galactose to induce Ste5-CTM protein expression. Cells were treated with 10 μg/ml doxycycline (Dox) for 15 h and 3 μm α factor pheromone for 30 min, as indicated. Cell lysates were resolved by 12.5% SDS-PAGE and immunoblotting with phospho-p44/42 antibodies (P-Kss1 and P-Fus3) and G6PDH antibodies as a loading control. Phosphorylated and activated Fus3 (P-Fus3) was quantified by scanning densitometry and analyzed with ImageJ software. Results were normalized to P-Fus3 levels of untreated samples. B, TetO7-PIK1 cells treated as in A. Results are the mean ± S.E. for three individual experiments.
Pik1 Is Required for Full Fus3 Activation and Inhibits Kss1 Activation
Although necessary for Fus3 signaling, Ste5 actually slows the rate of Fus3 activation (41). A second MAP kinase, Kss1, is also activated by pheromones but is not scaffolded and is activated comparatively quickly (35). To determine whether STT4 or PIK1 affects the kinetics of activation, we monitored Fus3 and Kss1 at multiple time points following pheromone treatment. Although knockdown in either case reduced the magnitude of Fus3 phosphorylation, the dynamics of activation were largely unchanged: Fus3 activation remained slow whereas Kss1 activation remained fast (Fig. 4, A and B). There were, however, notable differences in the behavior of Kss1. In contrast to STT4, loss of PIK1 resulted in marked elevation of Kss1 activity, particularly in the absence of pheromone stimulation (Fig. 4A). The 22–51% increase in Kss1 activation is particularly striking when compared with the 28–61% reduction in Fus3 activation. Thus, knockdown of PIK1 results in the constitutive activation of Kss1, which subsequently drives elongated growth (Fig. 2A). Treatment of cells harboring the TetO7 promoter attached to a non-expressible genetic element (TetO7-WT) with doxycycline had no effect on activation of Fus3 or Kss1, indicating that doxycycline alone has no effect on pathway activation (supplemental Fig. S3A).
FIGURE 4.
Pik1 is required for full Fus3 activation and represses basal Kss1 activation. A, TetO7-PIK1 cells were treated with 10 μg/ml doxycycline for 15 h and 3 μm α factor pheromone for the times indicated. Cell lysates were resolved by 12.5% SDS-PAGE and immunoblotting with phospho-p44/42 antibodies (P-Fus3 and P-Kss1), Fus3 antibodies, or G6PDH antibodies as a loading control. Note that pheromone stimulation induces FUS3 but not KSS1 expression. Bands were quantified by scanning densitometry and analyzed with ImageJ software. Results are the mean ± S.E. for three individual experiments. B, TetO7-STT4 cells treated as in A. C, BY4741 (WT) and isogenic vps34Δ cells treated as in A, except that no doxycycline was added to the cultures.
As an additional control, we monitored MAP kinase activity in cells lacking Vps34, a PtdIns 3-kinase required for full activation of Fus3 (19) (Fig. 4C). Again, the dynamics of activation were largely unaltered (Fig. 4C), even as overall Fus3 activity was diminished by 29–66%. The reduction we observed here is comparable with that reported by Slessareva et al. (19) but somewhat greater than the ∼20% difference reported by Garrenton et al. (22). These data reveal that Pik1 differentially regulates MAP kinase activation and is required to maintain MAP kinase specificity. Although Pik1 is required for stimulus-dependent activation of Fus3, it is also required to limit the activation of Kss1.
Pik1 Functions at the Level of Ste11
Given that Pik1 activates Fus3 while inhibiting Kss1, we hypothesized that Pik1 must regulate a pathway component upstream of both kinases. To better define which component is targeted by Pik1, we took a genetic epistasis approach. Using constitutively active mutants, we stimulated the pathway at multiple points, bypassing the need for pheromones and the pheromone receptor. First we overexpressed the G protein βγ subunits (STE4Gβ expressed using a galactose-inducible promoter). Because Gpa1 cannot sequester excess Gβγ (42–44), the overproduced Ste4Gβ is free to activate effectors even in the absence of any stimulus. As shown in Fig. 5, knockdown of PIK1 dampened Gβγ-mediated activation of Fus3 (Fig. 5, A and C). Next, we overexpressed the constitutively active STE11–4 mutant (45). STE11 encodes the kinase that phosphorylates Ste7, which in turn phosphorylates and activates Fus3 and Kss1. Once again knockdown of PIK1 dampened STE11–4-mediated activation of Fus3 (Fig. 5, B and C). Knockdown of STT4 likewise dampened STE11–4-mediated activation of Fus3 (Fig. 5, E and F). Thus, Pik1 promotes signaling upon activation by pheromone, the G protein, the kinase scaffold, and the protein kinase Ste11.
Pik1 Regulates the HOG Pathway
The data presented above indicate that Pik1 regulates the pheromone pathway and that Pik1 acts on or downstream of Ste11. We have largely excluded Ste5 as a target for Pik1 regulation, leaving three likely targets: Ste11, its binding partner Ste50, or its direct substrate Ste7 (Fig. 1A). To further distinguish between these candidate targets, we examined Pik1 regulation of the HOG pathway (10–12). The pheromone and HOG pathways share the use of Ste11 and Ste50 but not Ste7. We measured activation of Hog1 and Kss1 in response to the addition of 0.5 m KCl. Knockdown of PIK1 resulted in diminished Hog1 activity (supplemental Fig. S4). Furthermore, we again observed constitutive activation of Kss1 as well as an overall increase in Kss1 activation in response to salt stress.
The HOG pathway has two signaling branches that converge to activate Hog1. Our candidate target proteins, Ste11 and Ste50, only function in the Sho1 branch. To determine which branch of the HOG pathway is targeted by Pik1, we deleted SSK1 (eliminates signaling via the Sln1 branch, see Fig. 1A) and measured the activation of Hog1 in response to osmotic stress. Once again we observed diminished Hog1 activity and elevated Kss1 activity (Fig. 6A). These data indicate that Pik1 regulates the Sho1 branch of the HOG pathway.
FIGURE 6.
Pik1 acts via Ste11. A, TetO7-PIK1 ssk1Δ cells were treated with doxycycline (Dox) for 15 h and 0.5 m KCl for the times indicated and analyzed by immunoblotting with phospho-p38 (P-Hog1) antibodies, phospho-p44/42 (P-Kss1) antibodies, Hog1 antibodies, or G6PDH antibodies as a loading control. B, TetO7-PIK1 and TetO7-PIK1 kss1Δ cells were treated with doxycycline for 15 h and 3 μm α factor pheromone for 30 min, as indicated. Immunoblots were analyzed with phospho-p44/42, Fus3, and G6PDH antibodies. C, TetO7-PIK1 and TetO7-PIK1 kss1Δ cells were treated with doxycycline for 15 h and 0.5 Μ KCl for 10 min. Immunoblots were analyzed with phospho-p38 and G6PDH antibodies. D, Wild-type, TetO7-PIK1, and isogenic cells carrying ste4Δ, ste5Δ, ste11Δ, ste7Δ, ste20Δ, ste50Δ, opy2Δ, or cla4Δ mutations were treated with doxycycline for 15 h and α factor (αF) for 30 min and analyzed with phospho-p44/42 antibodies (P-Kss1). E, TetO7-PIK1 opy2Δ cells were treated as in B. All bands were quantified by scanning densitometry and analyzed with ImageJ software. Results are the mean ± S.E. for three individual experiments.
Recent reports indicate that Kss1 activates a Hog1-specific phosphatase Ptp2 (46). To determine whether high basal activation of Kss1 was in any way responsible for the diminished Hog1 response, we deleted KSS1 from the TetO7-PIK1 strain. As shown in Fig. 6, the loss of KSS1 did not affect the ability of Pik1 to regulate activation of either Fus3 (Fig. 6B) or Hog1 (C).
The available data indicate that Pik1 and Stt4 act in different ways to promote Fus3 signaling. Pik1 acts via Ste11, whereas Stt4 acts via Ste5. However, Pik1 also acts to limit Kss1 signaling. To further establish the site of Pik1 action, we monitored Kss1 activity in the absence of several signaling components that lie upstream of Kss1. In accordance with the model, we found that constitutive activation of Kss1 was dependent on Ste11 and Ste7 but not on Ste4 or Ste5 (Fig. 6D). Constitutive activation of Kss1 was also partially dependent on Ste50 and the MAP kinase kinase kinase kinase (MAPKKKK) Ste20, but not on another MAPKKKK Cla4.
Furthermore, we found that constitutive activation of Kss1 was dependent on the protein Opy2 (Fig. 6D), which binds Ste50 and recruits Ste50 and Ste11 to the plasma membrane (47). These data are consistent with our hypothesis that Pik1 regulates signaling at the level of Ste11 and indicate that Pik1 may regulate Ste11 via its interaction with Ste50 and Opy2. According to this model, deletion of OPY2 should restore normal pathway activity. As expected, deletion of OPY2 in the TetO7-PIK1 cells restored normal Fus3 and Kss1 activation in response to pheromones (Fig. 6E) and restored normal cell morphology (supplemental Fig. S5). These data indicate that Pik1 regulates MAPK signaling through Opy2 and Ste11.
DISCUSSION
Signal transduction systems will often share core signaling components yet maintain specificity and avoid pathway cross-talk. In yeast, three proteins have been found to preferentially regulate Fus3 and not Kss1. First, the scaffold protein Ste5 binds Fus3 and is required for Fus3 catalytic activity. Ste5 is not required by other MAP kinases and thus helps to differentiate pheromone signaling from other signaling systems (15, 39). Second, the PtdIns 4-kinase Stt4 promotes activation of Fus3 and does so by helping to recruit Ste5 to the plasma membrane (20–22). Like Ste5, Stt4 is required by Fus3 but not by other MAP kinases. Third, the PtdIns 3-kinase Vps34 promotes activation of Fus3, again in preference to Kss1 (19). Although functionally similar to Stt4 (see Fig. 4), Vps34 is expressed at endosomes rather than at the plasma membrane (25). Here we have investigated the function of another PtdIns kinase, Pik1. Like Stt4, Pik1 generates PtdIns 4-P and selectively regulates MAP kinase activity. Like Vps34, Pik1 is an endomembrane protein. Thus, Pik1 joins a small but growing number of factors that promote MAP kinase signaling specificity. Unlike any of the previously characterized regulators, however, Pik1 activates two MAP kinases (Fus3 and Hog1) while simultaneously inhibiting a third, competing MAP kinase (Kss1).
Although much has been learned, important questions remain for the future. Currently, there is little evidence to suggest the enzymatic activity of any PtdIns kinase (in yeast) is dynamically regulated. Garrenton et al. (22) showed that pheromone treatment does not change total cellular PtdIns-3-P or PtdIns-4-P levels. Thus, it is likely that activation by PtdIns-4-P occurs in cooperation with another, as yet unidentified, signaling event.
Another question is how the location of Pik1 at endomembranes contributes to its unique function in signaling. Activation of signal transduction pathways usually requires the assembly of signaling components at the plasma membrane. In pheromone signaling, several mechanisms are required to recruit components to activated transmembrane receptors. The heterotrimeric G protein subunits Gα and Gγ (48, 49) and the small G protein Cdc42 (50) are covalently modified with lipid moieties that anchor them to the plasma membrane. The scaffold Ste5 translocates from the cytoplasm to the plasma membrane by binding to the Gβγ dimer (14) as well as to Stt4-derived PtdIns-4-P and PtdIns-4,5-P2 (20, 21). Additionally, the PAK-family kinase Ste20 and the closely related kinase Cla4 translocate to the plasma membrane by binding both PtdIns-4,5-P2 and Cdc42 (51, 52). Therefore, spatial restriction of signaling components to areas near activated receptors may help prevent aberrant activation of parallel pathways. By analogy, Pik1 may function to redirect a signaling protein from its normal location at the plasma membrane to another location within the cell.
Although localization studies can be informative, a more pressing (and difficult) question is the direct target of Pik1 and PtdIns-4-P. Our epistasis analysis reveals that Pik1 likely regulates Ste11 and Ste50. Ste50 is required for Ste11 catalytic activity, and both are shared among the three MAP kinase pathways (11, 47, 53–56). This makes Ste50 or Ste11 an ideal target for coordinated regulation of all three pathways. Furthermore, Ste11 and Ste50 translocate from the cytoplasm to puncta after exposure to osmotic stress (10). Although the puncta were not identified by colocalization with known organelle markers, it might be useful to determine whether they coincide with the distribution of Pik1 or PtdIns-4-P. However, we consider it unlikely that either Ste11 or Ste50 bind directly to PtdIns-4-P. Neither protein contains a typical phospholipid-binding motif such as a PH or Phox homology domain (57). However, the BLAST CDD database does predict a low confidence Bin/Amphiphysin/Rvs domain in Ste50 (E value = 0.19). Bin/Amphiphysin/Rvs domains are dimerization, lipid-binding, and curvature-sensing modules found on many proteins involved in protein trafficking (58). It is possible that this putative Bin/Amphiphysin/Rvs domain is responsible for osmotic stress-induced localization of Ste11 and Ste50 at puncta. Alternatively, Ste11 and Ste50 could interact with a third protein containing a known lipid-binding domain. This model is particularly attractive because Ste50 binds to the transmembrane protein Opy2 (47). Opy2, in turn, functions as a scaffold protein that recruits Ste50 and Ste11 to the plasma membrane where they initiate signaling in the HOG and FG pathways (46, 47). Thus, Opy2 serves a function analogous to Ste5 in the pheromone pathway (14). Given the parallels between Opy2 and Ste5, it is possible that they are also regulated in a similar manner.
Recent investigations have revealed that MAPK function can be controlled by the association and dissociation of Ste50 and Opy2 (59). Specifically, feedback phosphorylation of Ste50 by Fus3, Kss1, or Hog1 disrupts Ste50-Opy2 binding and inhibits the HOG pathway. Here, we show that knockdown of Pik1 likewise inhibits Hog1 activation. Conversely, knockdown of Pik1 leads to constitutive Kss1 activation and accompanying changes in cell morphology. Moreover, deletion of OPY2 restores normal MAPK signaling in the face of PIK1 depletion. Taken together, these data indicate that Opy2 could inhibit mating by sequestering Ste50-Ste11. In support of this hypothesis, Edwards et al. (60) reported that overexpression of Opy2 inhibits the pheromone response. However, the exact role of Pik1 and PtdIns-4-P in regulating Opy2, Ste50, and Ste11 will require further investigation.
Finally, our data show that Pik1 positively regulates Fus3 but negatively affects Kss1. Fus3 is known to down-regulate Kss1 (17). Although we propose that Pik1 affects a shared upstream component that results in the differential regulation of both Fus3 and Kss1, it is possible that Pik1 regulates Fus3 directly but Kss1 indirectly. We consider this unlikely, however, because loss of STT4 or VPS34 dampens Fus3 activation without a concomitant increase in Kss1 activation. Thus, simply dampening Fus3 does not result in a constitutively active Kss1. We also considered a previous suggestion that Pik1 is needed for efficient mRNA export and protein synthesis (22). Under conditions where MAP kinase activity is severely affected, however, we observed no changes in the expression of control proteins, including alcohol dehydrogenase (PADH1-RFP), glucose 6-phosphate dehydrogenase, or Hog1 (27). We also observed no change in global protein expression as determined by Coomassie-staining of whole cell lysates.
In conclusion, we show that Pik1 and PtdIns-4-P promote the activation of Fus3 and Hog1 while repressing activation of Kss1. By acting on all three MAP kinases, Pik1 appears well positioned to coordinate cellular responses in the face of competing signals. Together with previous demonstrations of signal regulation by a PtdIns 3-kinase at the endosome, there is growing evidence for signal coordination by endomembrane-associated second messengers (19, 61–64). Ste11 is homologous to human MEKK3, and Ste50 is similar to human OSM (osmosensing scaffold for MEKK3) (65). Given this conservation across species, our findings are likely to translate to human MAP kinase pathways as well.
Supplementary Material
Acknowledgments
We thank Nan Hao and Michal Nagiec for critical reading of this manuscript; M. Winters and P. Pryciak, C. Stefan and S. Emr, and T. Yeung and S. Grinstein for their generous gift of plasmids; and P. Pryciak for communicating unpublished results.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM059167 and GM080739.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- HOG
- high osmolarity glycerol
- PtdIns
- phosphatidylinositol
- G6PDH
- glucose-6-phosphate dehydrogenase
- ts
- temperature-sensitive
- TetO7
- tetracycline-repressible promoter
- PH
- pleckstrin homology
- C2
- conserved region 2
- CTM
- C-terminal transmembrane domain.
REFERENCES
- 1. Turjanski A. G., Vaqué J. P., Gutkind J. S. (2007) Oncogene 26, 3240–3253 [DOI] [PubMed] [Google Scholar]
- 2. Chen R. E., Thorner J. (2007) Biochim. Biophys. Acta 1773, 1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dohlman H. G., Thorner J. W. (2001) Annu. Rev. Biochem. 70, 703–754 [DOI] [PubMed] [Google Scholar]
- 4. Roberts R. L., Fink G. R. (1994) Genes Dev. 8, 2974–2985 [DOI] [PubMed] [Google Scholar]
- 5. Liu H., Styles C. A., Fink G. R. (1993) Science 262, 1741–1744 [DOI] [PubMed] [Google Scholar]
- 6. Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. (1992) Cell 68, 1077–1090 [DOI] [PubMed] [Google Scholar]
- 7. Brewster J. L., de Valoir T., Dwyer N. D., Winter E., Gustin M. C. (1993) Science 259, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 8. Hohmann S. (2002) Microbiol. Mol. Biol. Rev. 66, 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O'Rourke S. M., Herskowitz I., O'Shea E. K. (2002) Trends Genet. 18, 405–412 [DOI] [PubMed] [Google Scholar]
- 10. Posas F., Witten E. A., Saito H. (1998) Mol. Cell. Biol. 18, 5788–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rad M. R., Xu G., Hollenberg C. P. (1992) Mol. Gen. Genet. 236, 145–154 [DOI] [PubMed] [Google Scholar]
- 12. Ramezani Rad M., Jansen G., Bühring F., Hollenberg C. P. (1998) Mol. Gen. Genet. 259, 29–38 [DOI] [PubMed] [Google Scholar]
- 13. Elion E. A. (2001) J. Cell Sci. 114, 3967–3978 [DOI] [PubMed] [Google Scholar]
- 14. Pryciak P. M., Huntress F. A. (1998) Genes Dev. 12, 2684–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Drogen F., Stucke V. M., Jorritsma G., Peter M. (2001) Nat. Cell Biol. 3, 1051–1059 [DOI] [PubMed] [Google Scholar]
- 16. Gartner A., Nasmyth K., Ammerer G. (1992) Genes Dev. 6, 1280–1292 [DOI] [PubMed] [Google Scholar]
- 17. Sabbagh W., Jr., Flatauer L. J., Bardwell A. J., Bardwell L. (2001) Mol. Cell 8, 683–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hao N., Zeng Y., Elston T. C., Dohlman H. G. (2008) J. Biol. Chem. 283, 33798–33802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Slessareva J. E., Routt S. M., Temple B., Bankaitis V. A., Dohlman H. G. (2006) Cell 126, 191–203 [DOI] [PubMed] [Google Scholar]
- 20. Garrenton L. S., Young S. L., Thorner J. (2006) Genes Dev. 20, 1946–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winters M. J., Lamson R. E., Nakanishi H., Neiman A. M., Pryciak P. M. (2005) Mol. Cell 20, 21–32 [DOI] [PubMed] [Google Scholar]
- 22. Garrenton L. S., Stefan C. J., McMurray M. A., Emr S. D., Thorner J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 11805–11810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Audhya A., Emr S. D. (2002) Dev. Cell 2, 593–605 [DOI] [PubMed] [Google Scholar]
- 24. Homma K., Terui S., Minemura M., Qadota H., Anraku Y., Kanaho Y., Ohya Y. (1998) J. Biol. Chem. 273, 15779–15786 [DOI] [PubMed] [Google Scholar]
- 25. Herman P. K., Emr S. D. (1990) Mol. Cell. Biol. 10, 6742–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stack J. H., DeWald D. B., Takegawa K., Emr S. D. (1995) J. Cell Biol. 129, 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cappell S. D., Baker R., Skowyra D., Dohlman H. G. (2010) Mol. Cell 38, 746–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mnaimneh S., Davierwala A. P., Haynes J., Moffat J., Peng W. T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., Trochesset M., Morse D., Krogan N. J., Hiley S. L., Li Z., Morris Q., Grigull J., Mitsakakis N., Roberts C. J., Greenblatt J. F., Boone C., Kaiser C. A., Andrews B. J., Hughes T. R. (2004) Cell 118, 31–44 [DOI] [PubMed] [Google Scholar]
- 29. Lee M. J., Dohlman H. G. (2008) Curr. Biol. 18, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffman G. A., Garrison T. R., Dohlman H. G. (2002) Methods Enzymol. 344, 617–631 [DOI] [PubMed] [Google Scholar]
- 31. Audhya A., Foti M., Emr S. D. (2000) Mol. Biol. Cell 11, 2673–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Strahl T., Hama H., DeWald D. B., Thorner J. (2005) J. Cell Biol. 171, 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stefan C. J., Audhya A., Emr S. D. (2002) Mol. Biol. Cell 13, 542–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yeung T., Gilbert G. E., Shi J., Silvius J., Kapus A., Grinstein S. (2008) Science 319, 210–213 [DOI] [PubMed] [Google Scholar]
- 35. Hao N., Nayak S., Behar M., Shanks R. H., Nagiec M. J., Errede B., Hasty J., Elston T. C., Dohlman H. G. (2008) Mol. Cell 30, 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dorer R., Pryciak P. M., Hartwell L. H. (1995) J. Cell Biol. 131, 845–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Erdman S., Snyder M. (2001) Genetics 159, 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Paliwal S., Iglesias P. A., Campbell K., Hilioti Z., Groisman A., Levchenko A. (2007) Nature 446, 46–51 [DOI] [PubMed] [Google Scholar]
- 39. Choi K. Y., Satterberg B., Lyons D. M., Elion E. A. (1994) Cell 78, 499–512 [DOI] [PubMed] [Google Scholar]
- 40. Marcus S., Polverino A., Barr M., Wigler M. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 7762–7766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhattacharyya R. P., Reményi A., Good M. C., Bashor C. J., Falick A. M., Lim W. A. (2006) Science 311, 822–826 [DOI] [PubMed] [Google Scholar]
- 42. Cole G. M., Stone D. E., Reed S. I. (1990) Mol. Cell. Biol. 10, 510–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nomoto S., Nakayama N., Arai K., Matsumoto K. (1990) EMBO J. 9, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Whiteway M., Hougan L., Thomas D. Y. (1990) Mol. Cell. Biol. 10, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stevenson B. J., Rhodes N., Errede B., Sprague G. F., Jr. (1992) Genes Dev. 6, 1293–1304 [DOI] [PubMed] [Google Scholar]
- 46. Yang H. Y., Tatebayashi K., Yamamoto K., Saito H. (2009) EMBO J. 28, 1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu C., Jansen G., Zhang J., Thomas D. Y., Whiteway M. (2006) Genes Dev. 20, 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stone D. E., Cole G. M., de Barros Lopes M., Goebl M., Reed S. I. (1991) Genes Dev. 5, 1969–1981 [DOI] [PubMed] [Google Scholar]
- 49. Hirschman J. E., Jenness D. D. (1999) Mol. Cell. Biol. 19, 7705–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Johnson D. I. (1999) Microbiol. Mol. Biol. Rev. 63, 54–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takahashi S., Pryciak P. M. (2007) Mol. Biol. Cell 18, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wild A. C., Yu J. W., Lemmon M. A., Blumer K. J. (2004) J. Biol. Chem. 279, 17101–17110 [DOI] [PubMed] [Google Scholar]
- 53. Xu G., Jansen G., Thomas D. Y., Hollenberg C. P., Ramezani Rad M. (1996) Mol. Microbiol. 20, 773–783 [DOI] [PubMed] [Google Scholar]
- 54. Wu C., Leberer E., Thomas D. Y., Whiteway M. (1999) Mol. Biol. Cell 10, 2425–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Truckses D. M., Bloomekatz J. E., Thorner J. (2006) Mol. Cell. Biol. 26, 912–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tatebayashi K., Yamamoto K., Tanaka K., Tomida T., Maruoka T., Kasukawa E., Saito H. (2006) EMBO J. 25, 3033–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lemmon M. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 58. Habermann B. (2004) EMBO Rep. 5, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamamoto K., Tatebayashi K., Tanaka K., Saito H. (2010) Mol. Cell 40, 87–98 [DOI] [PubMed] [Google Scholar]
- 60. Edwards M. C., Liegeois N., Horecka J., DePinho R. A., Sprague G. F., Jr., Tyers M., Elledge S. J. (1997) Genetics 147, 1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Calebiro D., Nikolaev V. O., Gagliani M. C., de Filippis T., Dees C., Tacchetti C., Persani L., Lohse M. J. (2009) PLoS Biol. 7, e1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Galperin E., Sorkin A. (2008) Traffic 9, 1776–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sorkin A., von Zastrow M. (2009) Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zoncu R., Perera R. M., Balkin D. M., Pirruccello M., Toomre D., De Camilli P. (2009) Cell 136, 1110–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uhlik M. T., Abell A. N., Johnson N. L., Sun W., Cuevas B. D., Lobel-Rice K. E., Horne E. A., Dell'Acqua M. L., Johnson G. L. (2003) Nat. Cell Biol. 5, 1104–1110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.