Abstract
Background
Figs and fig-pollinating wasp species usually display a highly specific one-to-one association. However, more and more studies have revealed that the "one-to-one" rule has been broken. Co-pollinators have been reported, but we do not yet know how they evolve. They may evolve from insect speciation induced or facilitated by Wolbachia which can manipulate host reproduction and induce reproductive isolation. In addition, Wolbachia can affect host mitochondrial DNA evolution, because of the linkage between Wolbachia and associated mitochondrial haplotypes, and thus confound host phylogeny based on mtDNA. Previous research has shown that fig wasps have the highest incidence of Wolbachia infection in all insect taxa, and Wolbachia may have great influence on fig wasp biology. Therefore, we look forward to understanding the influence of Wolbachia on mitochondrial DNA evolution and speciation in fig wasps.
Results
We surveyed 76 pollinator wasp specimens from nine Ficus microcarpa trees each growing at a different location in Hainan and Fujian Provinces, China. We found that all wasps were morphologically identified as Eupristina verticillata, but diverged into three clades with 4.22-5.28% mtDNA divergence and 2.29-20.72% nuclear gene divergence. We also found very strong concordance between E. verticillata clades and Wolbachia infection status, and the predicted effects of Wolbachia on both mtDNA diversity and evolution by decreasing mitochondrial haplotypes.
Conclusions
Our study reveals that the pollinating wasp E. verticillata on F. microcarpa has diverged into three cryptic species, and Wolbachia may have a role in this divergence. The results also indicate that Wolbachia strains infecting E. verticillata have likely resulted in selective sweeps on host mitochondrial DNA.
Background
The system of figs and fig wasps is considered to be a classic example of coevolved mutualism. It is well known that in general each fig species has a unique pollinator, which is called the "one-to-one" rule. However, more and more examples of co-pollinators (two or more pollinating wasp species on a fig) have broken the "one-to-one" rule [1-10]. However, we do not yet know how they evolve. They might be distantly related species, suggesting host shifts, or sister species, suggesting speciation on the current host [11,12]. Host shifts might be more likely when a fig colonises a new habitat, or is near the edge of its geographic range, because the normal pollinator is rare or absent. In this scenario, the co-pollinators are usually not closely related species [2,5,8]. Alternatively, sister co-pollinators may evolve from a recent speciation event in the pollinator that is not accompanied by fig radiation [3,11,12]. However, the exact mode of speciation for co-pollinators has not been well understood yet.
Wolbachia bacteria are the most common intracellular bacteria in arthropods and nematodes, and can manipulate host reproduction in many ways [13]. Cytoplasmic incompatibility (CI), the most common effect on host reproduction, usually occurs between infected males and uninfected females (or females infected by a different incompatible Wolbachia strain), inducing progeny sterility or mortality [13]. This post-zygotic reproductive isolation can potentially cause or facilitate host speciation [14-18]. Fig wasp species have a very high incidence of Wolbachia infection [19-21], and previous work suggests that Wolbachia might have an influence on fig wasp speciation, because cryptic pollinator species have different Wolbachia infections [3]. So, Wolbachia may play a potential important role in co-pollinator speciation.
In addition, Wolbachia can influence host mitochondrial DNA evolution. Because Wolbachia and mitochondria are co-transmitted maternally, the spread of Wolbachia can result in the hitchhiking of mitochondrial haplotypes. One particular mitochondrial haplotype can sweep through a population, via hitchhiking, associated with the sweep of Wolbachia [22,23]. This affects mtDNA evolution by decreasing mitochondrial haplotype diversity and also sometimes the mtDNA divergence between species [15,22-27], and thus can confound phylogenies and barcodes based on host mtDNA [28,29]. Accordingly, we look forward to understanding the effect of Wolbachia on mtDNA evolution of fig wasps.
Ficus microcarpa Linn. is a functionally monoecious fig species. It is pollinated solely by Eupristina verticillata Waterston [30]. Here, we investigate the genetic variance and Wolbachia infection status of Eupristina verticillata in order to explore whether co-pollinators exist, the association between Wolbachia infection and co-pollinators speciation, and the effect of Wolbachia on host mtDNA evolution.
Results
Morphological study
We extracted DNA from the specimens non-destructively, so the specimens can be conserved as vouchers for morphological study. According to the keys of Bouček and Wiebes [30,31], all the specimens are morphologically identified as Eupristina verticillata.
Wolbachia infection
After initial screening for Wolbachia infection, we selected 76 pollinator specimens (35 infected, 41 uninfected) for further research. Only one specimen is infected by two Wolbachia strains, while the other 34 specimens are infected by a single strain (Table 1).
Table 1.
Summary of E. verticillata samples and their genetic characteristics.
| Locations | Wasp code | COI clades | ITS2 clades |
Wolbachia strains* |
|---|---|---|---|---|
| Haikou, Hainan | HK-EV-20 | 1 | Ⅰ | wEv1 |
| HK-EV-22 | 1 | Ⅰ | wEv1 | |
| HK-EV-27 | 1 | Ⅰ | wEv1 | |
| HK-EV-29 | 1 | Ⅰ | wEv1 | |
| HK-EV-30 | 1 | Ⅰ | wEv1 | |
| HK-EV-3 | 1 | Ⅰ | - | |
| HK-EV-7 | 1 | Ⅰ | - | |
| HK-EV-10 | 1 | Ⅰ | - | |
| HK-EV-16 | 1 | Ⅰ | - | |
| HK-EV-18 | 1 | Ⅰ | - | |
| Danzhou, Hainan | DZ-EV-1 | 2 | Ⅱ | wEv2 |
| DZ-EV-2 | 2 | Ⅱ | wEv2 | |
| DZ-EV-3 | 2 | Ⅱ | wEv2 | |
| DZ-EV-4 | 2 | Ⅱ | wEv2 | |
| DZ-EV-5 | 2 | Ⅱ | wEv2 | |
| DZ-EV-6 | 2 | Ⅱ | - | |
| DZ-EV-7 | 2 | Ⅱ | - | |
| DZ-EV-8 | 2 | Ⅱ | - | |
| DZ-EV-9 | 2 | Ⅱ | - | |
| DZ-EV-10 | 2 | Ⅱ | - | |
| Xidao, Hainan | XD-EV-2 | 2 | Ⅱ | wEv2 |
| XD-EV-5 | 1 | I | wEv1 | |
| XD-EV-7 | 2 | Ⅱ | wEv2 | |
| XD-EV-10 | 2 | Ⅱ | wEv2 | |
| XD-EV-13 | 2 | Ⅱ | wEv2 | |
| XD-EV-3 | 2 | Ⅱ | - | |
| XD-EV-6 | 2 | Ⅱ | - | |
| XD-EV-8 | 2 | Ⅱ | - | |
| XD-EV-9 | 2 | Ⅱ | - | |
| XD-EV-12 | 2 | Ⅱ | - | |
| Sanya, Hainan | SY-EV-22 | 1 | I | wEv1 |
| SY-EV-23 | 1 | I | wEv1 | |
| SY-EV-24 | 1 | I | wEv1 | |
| SY-EV-26 | 1 | I | wEv1 | |
| SY-EV-28 | 1 | I | wEv1 | |
| SY-EV-15 | 1 | I | - | |
| SY-EV-19 | 1 | I | - | |
| Lingshui, Hainan | LS-EV-16 | 1 | I | wEv1 |
| LS-EV-17 | 2 | II | wEv2 | |
| LS-EV-18 | 1 | I | wEv1 | |
| LS-EV-19 | 1 | I | wEv1 | |
| LS-EV-20 | 2 | II | wEv2 | |
| LS-EV-12 | 1 | I | - | |
| LS-EV-14 | 2 | II | - | |
| LS-EV-21 | 2 | II | - | |
| LS-EV-26 | 2 | II | - | |
| LS-EV-28 | 2 | II | - | |
| Wanning, Hainan | WN-EV-1 | 1 | I | wEv1 |
| WN-EV-5 | 1 | I | wEv3 | |
| WN-EV-7 | 1 | I | wEv1 | |
| WN-EV-8 | 1 | I | wEv1 | |
| WN-EV-9 | 1 | I | wEv1 | |
| WN-EV-2 | 1 | I | - | |
| WN-EV-3 | 1 | I | - | |
| WN-EV-4 | 1 | I | - | |
| WN-EV-11 | 1 | I | - | |
| Baisha, Hainan | BS-EV-4 | 2 | II | wEv2 |
| BS-EV-14 | 2 | II | wEv2 | |
| BS-EV-16 | 2 | II | wEv2 wEv3 | |
| BS-EV-17 | 2 | II | wEv2 | |
| BS-EV-19 | 2 | II | wEv2 | |
| BS-EV-5 | 2 | II | - | |
| BS-EV-8 | 2 | II | - | |
| BS-EV-10 | 1 | I | - | |
| BS-EV-15 | 2 | II | - | |
| BS-EV-18 | 2 | II | - | |
| Wenchang, Hainan | WC-EV-21 | 3 | Ⅲ | - |
| WC-EV-22 | 3 | Ⅲ | - | |
| WC-EV-23 | 3 | Ⅲ | - | |
| WC-EV-24 | 3 | Ⅲ | - | |
| WC-EV-25 | 3 | Ⅲ | - | |
| Wuping, Fujian | FW-EV-1 | 1 | Ⅰ | - |
| FW-EV-2 | 1 | Ⅰ | - | |
| FW-EV-3 | 1 | Ⅰ | - | |
| FW-EV-5 | 1 | Ⅰ | - | |
| FW-EV-9 | 1 | Ⅰ | - | |
* A line means no Wolbachia.
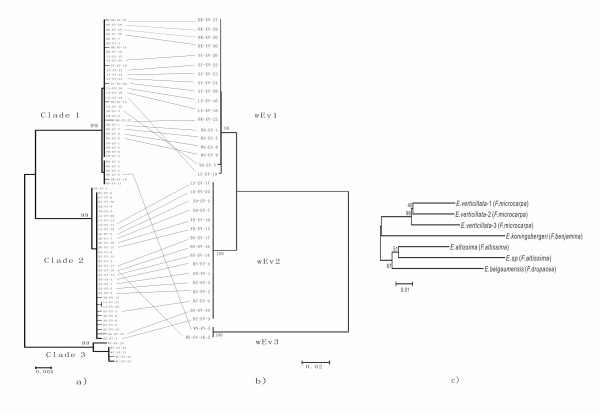
In view of the NJ-tree based on wsp sequences and the divergence among wsp haplotypes (Figure 1b), we recognise three Wolbachia strains (wEv1, wEv2 and wEv3). The divergence between wEv1 and wEv2 is much smaller (2.96%) than that between wEv1 and wEv3 (22.44%), and between wEv2 and wEv3 (23.20%).We then searched for similar wsp sequences on the NCBI database. The results show that wEv1 and wEv2 are 97-100% similar to the wsp sequences found in E. verticillata in Australia-Asia [20,21], whereas wEv3 is similar to a great number of wsp sequences found in many other insects, including other fig wasps, gall wasps, mosquitoes, bugs, fruit flies and so on.
Figure 1.
Phylogeny of Eupristina species and Correlated divergences of E. verticillata and Wolbachia strains. a) the tree based on E. verticillata COI gene. b) the tree based on Wolbachia wsp gene. c) phylogeny of Eupristina species based on COI gene.
Phylogeny and molecular variance
We obtain COI and ITS2 sequences for all 76 pollinator specimens. There are 24 unique COI haplotypes of 652bp in length, of which 68 nucleotide sites are polymorphic and 47 are parsimony informative. The NJ-tree based on COI sequences (Figure 1a) shows that all the specimens cluster into 3 distinct clades with high bootstrap values. We name them as Clade 1, Clade 2 and Clade 3. The mean COI genetic divergence within each clade is tiny (0.11%, 0.11%, 0.49% respectively), while the mean divergence among the clades is large (4.22%-5.28%) enough to suggest different species.
We also obtain 76 ITS2 sequences, whose length vary from 522 bp to 545 bp. The NJ-tree based on ITS2 sequences (Additional file 1) also shows three distinct clades (Clade I, Clade II and Clade III). The sequences within each clade are nearly or absolutely identical. The divergence among the clades, due mostly to length variance, is 2.29% between Clade I and II, 20.28% between Clade II and III, and 20.72% between Clade I and III. Remarkably, the NJ-trees based on COI and ITS2 sequences have very similar topologies, and the members which cluster together in COI clades also cluster together in ITS2 clades (Additional file 1).
Distribution of pollinators
A summary of E. verticillata samples and their genetic characteristics is shown in Table 1. All samples from Haikou, Sanya, Wanning, Wuping fall into Clade 1 and those from Danzhou and Wenchang belong to Clade 2 and Clade 3, respectively. For Xidao, Lingshui and Baisha, individuals of Clade 1 and Clade 2 coexist.
Mitochondrial DNA polymorphism and evolution pattern
We mapped the status of Wolbachia infection onto the E. verticillata clades based on COI sequences. This reveals very strong concordance between E. verticillata clades and Wolbachia infection status. Clade 1 and Clade 2 mainly harbour wEv1 and wEv2, respectively, while Clade 3 specimens are free from Wolbachia infection (Table 1). In order to examine the effect of Wolbachia on host mitochondrial DNA, we compared patterns of mitochondrial DNA polymorphism and evolution among the clades. The results are shown in Table 2.
Table 2.
Comparison of COI gene evolution of E. verticillata samples clades.
| n | h | Hd | S | Pi | D | Tajima's D | Fu and Li's D | Fu and Li's F | |
|---|---|---|---|---|---|---|---|---|---|
| Clade 1 | 37 | 9 | 0.535 | 9 | 0.00109 | 0.11% | -1.98912* | -3.55505** | -3.59153** |
| Clade 2 | 34 | 11 | 0.547 | 11 | 0.00108 | 0.11% | -2.31606*** | -3.75589** | -3.87463** |
| Clade 3 | 5 | 4 | 0.900 | 8 | 0.00491 | 0.49% | -1.17432 | -1.17432 | -1.22979 |
n = no. of fig wasps samples; h = no. of mitochondrial DNA haplotype; Hd = haplotype diversity; S = no. of segregating sites; Pi = nucleotide diversity; D = genetic divergence. *: p < 0.05;**: p < 0.02;***: p < 0.01.
The mean genetic divergence (D), haplotype diversity (Hd) and nucleotide diversity (Pi) among the three clades are significantly different. They are all much lower in Clade 1 and 2 than in Clade 3. In addition, we could reject neutral evolution for Clade 1 and 2 with neutrality values (Tajima's D, Fu and Li's D and Fu and Li's F) significantly less than zero, but not for Clade 3. In summary, we detect predicted effects of Wolbachia infection on both mtDNA diversity and evolution.
Discussion
Cryptic co-pollinators
In this research, data from both mitochondrial (COI) and nuclear (ITS2) markers indicate that E. verticillata wasps fall into three distinct species (clades). COI divergence among these clades is large (4.22%-5.28%) enough to suggest that these are different species. COI is the gene used for animal DNA barcoding and has the potential to facilitate both the identification of known species and the discovery of new ones [32]. Hebert et al. (2003) studied the ability of COI to distinguish 2238 species across 11 animal phyla, and the result revealed that more than 98% of species pairs showed greater than 2% sequence divergence [32]. In this study, the COI divergence is greater than 2%, so it suggests the existence of cryptic species within the morphologically defined E. verticillata. However, because Wolbachia can confound DNA barcoding based on mtDNA, we need nuclear markers to support the conclusion from the analysis of mtDNA. Nuclear marker ITS2 sequences, which are not affected by Wolbachia, also support the conclusion of cryptic species. ITS2 has been studied in other hymenopterans, and this nuclear DNA fragment shows high conservation within species and high divergence between congeneric species [33]. In our study, ITS2 sequences within each clade are nearly identical but diverge considerably among clades (2.29%, 20.28% and 20.72%). Therefore, according to the tree topologies and the divergence of COI and ITS2, we define the specimens in Clade 1, 2 and 3 as E. verticillata-1, E. verticillata-2 and E. verticillata-3.
How can cryptic co-pollinators occur?
As background mentioned, co-pollinators may occur as a result of host shift or speciation. In South China, F. microcarpa is widespread, so host shift event seems unlikely. In addition, survey of fig wasps in Hainan and other provinces in South China does not reveal that E. verticillata exists on other host figs (unpublished data). All the above information suggests that occurrence of co-pollinators on F. microcarpa is unlikely to be due to host shift.
Alternatively, co-pollinators may evolve from a recent speciation event in the pollinator that is not accompanied by fig radiation [3,11,12]. In this circumstance, the co-pollinators are likely to be sister or close related species [3,11,12]. Phylogenetic analysis indicates that the three species we found are not only cryptic species, but also closely related species (analysis combined with other Eupristina species, Figure 1c). Based on the commonly used mitochondrial DNA clock rate of 2.3% pairwise divergence/Myr [34], E. verticillata-1 diverged from E. verticillata-2 about 1.8 million years ago, and E. verticillata-3 diverged from E. verticillata-1 and E. verticillata-2 about 2.3 million years ago. Although E. verticillata-3 seems distant from the other two species particularly based on ITS2 sequences, in view of the phylogeny and identical morphology, we still expect that the three species share the same ancestor. Together with the evidence that in some locations (Xidao, Lingshui and Baisha), E. verticillata-1 and E. verticillata-2 occur on the same tree, we conclude that E. verticillata has diverged into three species on the same host. This pattern might be attributed to the comparatively longer generation time of figs compared to that of wasps [4,11].
New species often arise via geographic isolation, but the current geographic distributions of the three species do not suggest geographic isolation (Additional file 2). In some locations, E. verticillata-1 and E. verticillata-2 coexist. E. verticillata-3 is only found in Wenchang, but this phenomenon might only reflect low sampling to date. Additionally, speciation may also be driven by Wolbachia-induced reproductive incompatibility. Previous studies have indicated that Wolbachia can induce reproductive incompatibility, and then facilitate host speciation [14-18]. In this study, we note that the three species have different Wolbachia infection status. E. verticillata-1 and E. verticillata-2 are infected by closely related Wolbachia strains, wEv1 and wEv2, while E. verticillata-3 is uninfected. A blast search for similar wsp sequences on the NCBI database indicates that wEv1 and wEv2 are specifically related to E. verticillata. The strong correlation between phylogenetic trees of wasp species and Wolbachia strains suggests that Wolbachia may be involved in the divergence and speciation of hosts. In addition, we calculated the time when wEv1 and wEv2 diverged based on the wsp divergence rate (0.2% pairwise divergence/Myr) [35]. The divergence between wEv1 and wEv2 occurred about 15 MYA, earlier than the host wasp divergence. This makes it possible that wEv1 and wEv2 play a role in host speciation. Although our results show that wEv1 and wEv2 may be involved in host speciation, we can not demonstrate that Wolbachia cause reproductive isolation among these three host species. So, the exact mode of speciation for three E. verticillata still needs to be explored.
Effect of Wolbachia on host mitochondrial DNA polymorphism and evolution pattern
Many explanations have been proposed for lower mitochondrial DNA polymorphism and deviation from neutral evolution, including population expansion from a bottleneck [36,37], selective sweep [26] and selection against weakly deleterious mutations [38]. We analyse the population size change by DnaSP, and get no evidence for population expansion (Additional file 3) [39]. Therefore, we should consider the other two possibilities. It is notable that E. verticillata-1 and E. verticillata-2 are infected by Wolbachia and show lower mitochondrial DNA polymorphism than uninfected E. verticillata-3. The perfect concordance of mitochondrial DNA polymorphism and Wolbachia infection status suggests that Wolbachia-associated selective sweeps of the mitochondrion have occurred in E. verticillata-1 and E. verticillata-2. Decreased mitochondrial DNA polymorphism as a consequence of Wolbachia infection has also been reported in several other insects [15,22-27]. We also find that the COI genes of E. verticillata-1 and E. verticillata-2 deviate significantly from neutral evolution while this is not so for E. verticillata-3. Therefore, this is also likely due to an mtDNA sweep associated with the spread of Wolbachia, making the mtDNA undergo purifying selection. In addition, a selection against the weakly deleterious mutation may also induce lower mitochondrial DNA polymorphism and deviation from neutral evolution, and it needs to be explored further.
Conclusions
Our study reveals the molecular variance within E. verticillata, and indicates the presence of cryptic species. Further analyses suggest that the occurrence of cryptic species is attributable to successive speciation. A survey on the Wolbachia infection in E. verticillata points to a strong correlation between Wolbachia strains and the pollinator species, which may indicate an important role of Wolbachia in the formation of the cryptic species. Our study also reveals that Wolbachia strains infecting E. verticillata have likely resulted in selective sweeps on host mitochondrial DNA.
Methods
Specimen collection and identification
Ripe syconia were collected from nine F. microcarpa trees each growing at a different location in Hainan and Fujian provinces, China, between 2006 and 2008 (Table 1). Wasps from more than 200 syconia per tree were allowed to emerge and then stored in 95% ethanol at -20℃. All pollinator specimens were then examined under a Nikon SMZ80 microscope and identified following the keys of Bouček and Wiebes [30,31]. We then selected randomly 30 female pollinator specimens per tree for initial Wolbachia screening.
DNA extraction
Each specimen was washed in MILLI-Q water before total genomic DNA was extracted non-destructively from the whole specimen using Easypure genomic DNA Extraction kits (TransGen, Beijing, China). Extracted DNA solution was used for PCR amplification and then the specimens were restored in 95% ethanol as vouchers in Institute of Zoology, Chinese Academy of Sciences.
PCR amplification
The DNA qualities of extracts were first checked using COI PCR amplification (details below). The positives were then screened for Wolbachia infection by PCR, employing the primers wsp81F (5'-TGGTCCAATAAGTGATGAAGAAAC-3') and wsp691R (5'-AAAAATTAAACGCTACTCCA-3') [40], which amplify part of the Wolbachia surface protein gene (wsp). We also amplified the ftsZ and 16S rDNA regions using primers ftsZ F (5'-TACTGACTGTTGGAGTTGTAACTAACGCGT-3')-ftsZ R (5'-TGCCAGTTGCAAGAACAGAAACTCTAACTC-3') [41] and 16SwolF (5'-TTGTAGCCTGCTATGGTATAACT-3')-16SwolR (5'-GAATAGGTATGATTTTCATGT-3') [42] to verify Wolbachia infection status. After initial screening, we chose five infected individuals and five uninfected ones (or less than five if unavailable) from each sample location to amplify COI and ITS2 fragments.
We amplified the mitochondrial COI fragment using the barcode primer set of LCO1490 (5'-GGTCAACAAATCATAAAGATATTGG-3') and HCO2198 (5'-TAAACTTCAGGGTGACCAAAAAATCA-3') [43,44] with the following conditions: 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 45 s at 50°C, 1 min at 72°C, and a final elongation step of 10 min at 72°C. All PCR amplifications were performed using an Eppendorf Mastercycler gradient machine (Eppendorf) in 25 μl reaction volume: Tris-HCl 20 mM (pH 8.4), KCl 20 mM, (NH4)2SO4 10 mM, 5 pMol of each primer, 20 mM dNTPs, 1 μl of genomic DNA (~15 ng), and 2.5 unit of TransTaq polymerase (TransGen, Beijing, China).
5 μl of each PCR product was electrophoresed through 1% agarose gel to determine the size of fragment. Positive products were then purified with EasyPure PCR purification Kit (TransGen, Beijing). Because direct sequencing of the COI amplicon is difficult in Hymenoptera due to a poly-T region, the purified PCR products were cloned and 2-3 positive clones were sequenced with primer M13 on an ABI3730 capillary autosequencer.
The primers ITS2F (5'-ATTCCCGGACCACGCCTGGCTGA-3') and ITS2R (5'-TCCTCCGCTTATTGATATGC-3') [45] were used to amplify ITS2. The amplification was performed with the following conditions: 5 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C, and a final elongation step of 10 min at 72°C. The ITS2 products were purified and directly sequenced.
We also sequenced the wsp fragments directly from purified PCR products. If the direct sequencing failed (multiple peaks or frameshifts in electropherogram profiles may reveal the presence of more than one strain of Wolbachia), we cloned them and sequenced 5-6 positive clones.
Sequence analysis
The sequences were assembled and aligned in MEGA version 4.0 [46]. We then built neighbor-joining (NJ) tree for each gene region using Kimura 2-parameter (K2P) distance model and pairwise deletion in MEGA 4.0. Genetic distances between all specimens pairs were also calculated using the K2P model.
COI sequence polymorphism was calculated using DnaSP version 5.00.04 [47]. Polymorphism parameters estimated included nucleotide diversity (Pi), haplotype diversity (Hd), and the number of segregating sites (S). Neutrality tests were performed on the COI gene to determine if the frequency spectrum of polymorphisms conformed to the predictions of the neutral model of molecular evolution. Under selective neutrality, Tajima's D, Fu and Li's D and Fu and Li's F are all expected to equal zero. A value significantly less than zero indicates a higher-than-expected number of low-frequency variants, which can be caused by a recent selective sweep or population expansion or selection against weakly deleterious mutations [26,36-38], whereas a value significantly more than zero signifies low levels of both low and high frequency polymorphisms, indicating a decrease in population size or balancing selection [48].
All the sequences representing each COI and ITS2 haplotype and each Wolbachia strain have been deposited in GenBank [GenBank accession numbers HM582244-HM582279].
Authors' contributions
XJS and DWH conceived of the study and drafted the manuscript. XJS carried out the molecular genetic analyses. JHX participated in the design of the study and helped to draft the manuscript. JMC helped to draft the manuscript. GF participated in specimen identification. All authors read and approved the final manuscript.
Supplementary Material
E. verticillata NJ-trees. a) the tree based on COI gene. b) the tree based on ITS2.
Distribution of E. verticillata. In the pie chart, white circle represents E. verticillata-1, grey represents E. verticillata-2 and black represents E. verticillata-3.
Fit of equilibrium distributions for E. verticillata-1 population and E. verticillata-2 population. a) E. verticillata-1. b) E. verticillata-2. X axis: Pairwise Differences. Y axis: Frequency. The circles show the observed distribution of pairwise difference. The solid lines represent the expected equilibruim distributions. In equilibrium populations, the expected curves are free of waves. The observed curves with many peaks or resemblance to expected curves mean equilibrium population. On the contrary, unimodal curves represent population expansion [39].
Contributor Information
Xiao-Jing Sun, Email: sunxjx@gmail.com.
Jin-Hua Xiao, Email: xiaojh@ioz.ac.cn.
James M Cook, Email: james.cook@reading.ac.uk.
Gui Feng, Email: fenggui@gucas.ac.cn.
Da-Wei Huang, Email: huangdw@ioz.ac.cn.
Acknowledgements
This project was supported by National Natural Science Foundation of China (NSFC grant no. 31090253, 30900137), partially by Major Innovation Program of Chinese Academy of Sciences (KSCX2-YW-N-0807), by Program of Ministry of Science and Technology of the Republic of China (2006FY110500), by National Science Fund for Fostering Talents in Basic Research (Special subjects in animal taxonomy, NSFC- J0930004), and by a grant (No. O529YX5105) from the Key Laboratory of the Zoological Systematics and Evolution of the Chinese Academy of Sciences. We thank the anonymous reviewers for their valuable comments and suggestions.
References
- Rasplus JY. In: The Biodiversity of African Plants. van der Maesen LJ, van der Burgt XM, van Medenbach de Rooy JM, editor. Kluwer Academic Publishers; 1996. The one-to-one species specificity of the Ficus-Agaoninae mutualism: how casual? pp. 639–649. [Google Scholar]
- Ware AB, Compton SG. Breakdown of pollinator specificity in an African fig tree. Biotropica. 1992;24(4):544–549. doi: 10.2307/2389018. [DOI] [Google Scholar]
- Haine E, Martin J, Cook J. Deep mtDNA divergences indicate cryptic species in a fig-pollinating wasp. BMC Evol Biol. 2006;6(1):83. doi: 10.1186/1471-2148-6-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloud G, Carriere S, Kobbi M. Exceptions to the one: one relationship between African fig trees and their fig wasp pollinators: Possible evolutionary scenarios. J Biogeogr. 1996;23(4):513–520. doi: 10.1111/j.1365-2699.1996.tb00013.x. [DOI] [Google Scholar]
- Kerdelhue C, Le Clainche I, Rasplus JY. Molecular phylogeny of the Ceratosolen species pollinating Ficus of the subgenus Sycomorus sensu stricto: biogeographical history and origins of the species-specificity breakdown cases. Mol Phylogenet Evol. 1999;11(3):401–414. doi: 10.1006/mpev.1998.0590. [DOI] [PubMed] [Google Scholar]
- Machado CA, Robbins N, Gilbert MTP, Herre EA. Critical review of host specificity and its coevolutionary implications in the fig/fig-wasp mutualism. Proc Natl Acad Sci USA. 2005;102(suppl_1):6558–6565. doi: 10.1073/pnas.0501840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molbo D, Machado CA, Sevenster JG, Keller L, Herre EA. Cryptic species of fig-pollinating wasps: implications for the evolution of the fig-wasp mutualism, sex allocation, and precision of adaptation. Proc Natl Acad Sci USA. 2003;100(10):5867–5872. doi: 10.1073/pnas.0930903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton SG. A collapse of host specificity in some African fig wasps. South Afr J Sci. 1990;86(1):39–40. [Google Scholar]
- Peng YQ, Duan ZB, Yang DR, Rasplus JY. Co-occurrence of two Eupristina species on Ficus altissima in Xishuangbanna, SW China. Symbiosis. 2008;45(1-3):9–14. [Google Scholar]
- Lopez-Vaamonde C, Dixon DJ, Cook JM, Rasplus JY. Revision of the Australian species of Pleistodontes (Hymenoptera: Agaonidae) fig-pollinating wasps and their host-plant associations. Zool J Linn Soc. 2002;136(4):637–683. doi: 10.1046/j.1096-3642.2002.00040.x. [DOI] [Google Scholar]
- Cook JM, Rasplus JY. Mutualists with attitude: coevolving fig wasps and figs. Trends Ecol Evol. 2003;18(5):241–248. doi: 10.1016/S0169-5347(03)00062-4. [DOI] [Google Scholar]
- Cook JM, Segar ST. Speciation in fig wasps. Ecol Entomol. 2010;35:54–66. doi: 10.1111/j.1365-2311.2009.01148.x. [DOI] [Google Scholar]
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rew Micro. 2008;6(10):741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Werren JH. In: Endless Species and Speciation. Howard D, Berlocher S, editor. New York: Oxford University Press; 1997. Wolbachia and speciation. [Google Scholar]
- Shoemaker DD, Katju V, Jaenike J. Wolbachia and the evolution of reproductive isolation between Drosophilla recens and Drosophila subquinaria. Evolution. 1999;53(4):1157–1164. doi: 10.2307/2640819. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, O'Hara FP, Werren JH. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature. 2001;409(6821):707–710. doi: 10.1038/35055543. [DOI] [PubMed] [Google Scholar]
- Telschow A, Hammerstein P, Werren JH. The effect of Wolbachia versus genetic incompatibilities on reinforcement and speciation. Evolution. 2005;59(8):1607–1619. [PubMed] [Google Scholar]
- Telschow A, Flor M, Kobayashi Y, Hammerstein P, Werren JH. Wolbachia-Induced Unidirectional Cytoplasmic Incompatibility and Speciation: Mainland-Island Model. PLoS ONE. 2007;2(8):e701. doi: 10.1371/journal.pone.0000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haine ER, Cook JM. Convergent incidences of Wolbachia infection in fig wasp communities from two continents. Pro R Sco B. 2005;272(1561):421–429. doi: 10.1098/rspb.2004.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker DD, Machado CA, Molbo D, Werren JH, Windsor DM, Herre EA. The distribution of Wolbachia in fig wasps: correlations with host phylogeny, ecology and population structure. Pro R Soc Lond B. 2002;269(1506):2257–2267. doi: 10.1098/rspb.2002.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Cook JM, Xiao H, Hu HY, Niu LM, Huang DW. High incidences and similar patterns of Wolbachia infection in fig wasp communities from three different continents. Insect Science. 2010;17(2):101–111. doi: 10.1111/j.1744-7917.2009.01291.x. [DOI] [Google Scholar]
- Baudry E, Bartos J, Emerson K, Whitworth T, Werren JH. Wolbachia and genetic variability in the birdnest blowfly Protocalliphora sialia. Mol Eco. 2003;12(7):1843–1854. doi: 10.1046/j.1365-294X.2003.01855.x. [DOI] [PubMed] [Google Scholar]
- Kambhampati S, Rai KS, Verleye DM. Frequencies of mitochondrial DNA haplotypes in laboratory cage populations of the mosquito, Aedes albopictus. Genetics. 1992;132(1):205–209. doi: 10.1093/genetics/132.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard JW, Hatzidakis J, Karr TL, Kreitman M. Reduced variation in Drosophila simulans mitochondrial DNA. Genetics. 1996;144(4):1519–1528. doi: 10.1093/genetics/144.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon J, Cornel A, Scott T. Evolutionary history of a mosquito endosymbiont revealed through mitochondrial hitchhiking. Pro R Sco B. 2006;273(1594):1603–1611. doi: 10.1098/rspb.2006.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker DD, Dyer KA, Ahrens M, McAbee K, Jaenike J. Decreased diversity but increased substitution rate in host mtDNA as a consequence of Wolbachia endosymbiont infection. Genetics. 2004;168(4):2049–2058. doi: 10.1534/genetics.104.030890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM. Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics. 2003;164(1):5–12. doi: 10.1093/genetics/164.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst GD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Pro R Sco B. 2005;272(1572):1525–1534. doi: 10.1098/rspb.2005.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth TL, Dawson RD, Magalon H, Baudry E. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae) Pro R Sco B. 2007;274(1619):1731–1739. doi: 10.1098/rspb.2007.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebes JT. The Indo-Australian Agaoninae (pollinators of figs) Vol. 92. Amsterdam; New York: North-Holland; 1994. [Google Scholar]
- Bouček Z. Family Agaonidae. CAB, International, Wallingford, UK; 1988. Australasian Chalcidoidea (Hymenoptera): A Biosystematic Revision of Genera and Fourteen Families, with a Reclassification of Species; pp. 156–209. [Google Scholar]
- Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Lond B. 2003;270(Suppl 1):S96–99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YW, Zhou X, Feng G, Hu HY, NIu LM, Hebert PDN, Huang DW. COI and ITS2 sequences delimit species, reveal cryptic taxa and host specificity of fig-associated Sycophila (Hymenoptera, Eurytomidae) Mol Ecol Resour. 2010;10(1):31–40. doi: 10.1111/j.1755-0998.2009.02671.x. [DOI] [PubMed] [Google Scholar]
- Brower AV. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91(14):6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viljakainen L, Reuter M, Pamilo P. Wolbachia transmission dynamics in Formica wood ants. BMC Evol Biol. 2008;8(1):55. doi: 10.1186/1471-2148-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123(3):597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aris-Brosou S, Excoffier L. The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Mol Biol Evol. 1996;13(3):494–504. doi: 10.1093/oxfordjournals.molbev.a025610. [DOI] [PubMed] [Google Scholar]
- Seger J, Smith WA, Perry JJ, Hunn J, Kaliszewska ZA, Sala LL, Pozzi L, Rowntree VJ, Adler FR. Gene genealogies strongly distorted by weakly interfering mutations in constant environments. Genetics. 2010;184(2):529–545. doi: 10.1534/genetics.109.103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Bio Evol. 1992;9(3):552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Dobson SL, Bourtzis K, Braig HR, Jones BF, Zhou W, Rousset F, O'Neill SL. Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem Mol Biol. 1999;29(2):153. doi: 10.1016/S0965-1748(98)00119-2. [DOI] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9(4):393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- O'Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16S rRNA Phylogenetic Analysis of the Bacterial Endosymbionts Associated with Cytoplasmic Incompatibility in Insects. Proc Natl Acad Sci USA. 1992;89(7):2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotech. 1994;3(5):294–299. [PubMed] [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Pro R Soc Lond B. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bruns T, Lee S, Taylor J. PCR Protocols: A Guide to Methods and Applications. San Diego, Academic Press; 1990. Amplification and direct sequencing of fungal ribosomal genes for phylogenies; pp. 315–322. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Simonsen KL, Churchill GA, Aquadro CF. Properties of Statistical Tests of Neutrality for DNA Polymorphism Data. Genetics. 1995;141(1):413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
E. verticillata NJ-trees. a) the tree based on COI gene. b) the tree based on ITS2.
Distribution of E. verticillata. In the pie chart, white circle represents E. verticillata-1, grey represents E. verticillata-2 and black represents E. verticillata-3.
Fit of equilibrium distributions for E. verticillata-1 population and E. verticillata-2 population. a) E. verticillata-1. b) E. verticillata-2. X axis: Pairwise Differences. Y axis: Frequency. The circles show the observed distribution of pairwise difference. The solid lines represent the expected equilibruim distributions. In equilibrium populations, the expected curves are free of waves. The observed curves with many peaks or resemblance to expected curves mean equilibrium population. On the contrary, unimodal curves represent population expansion [39].