Abstract
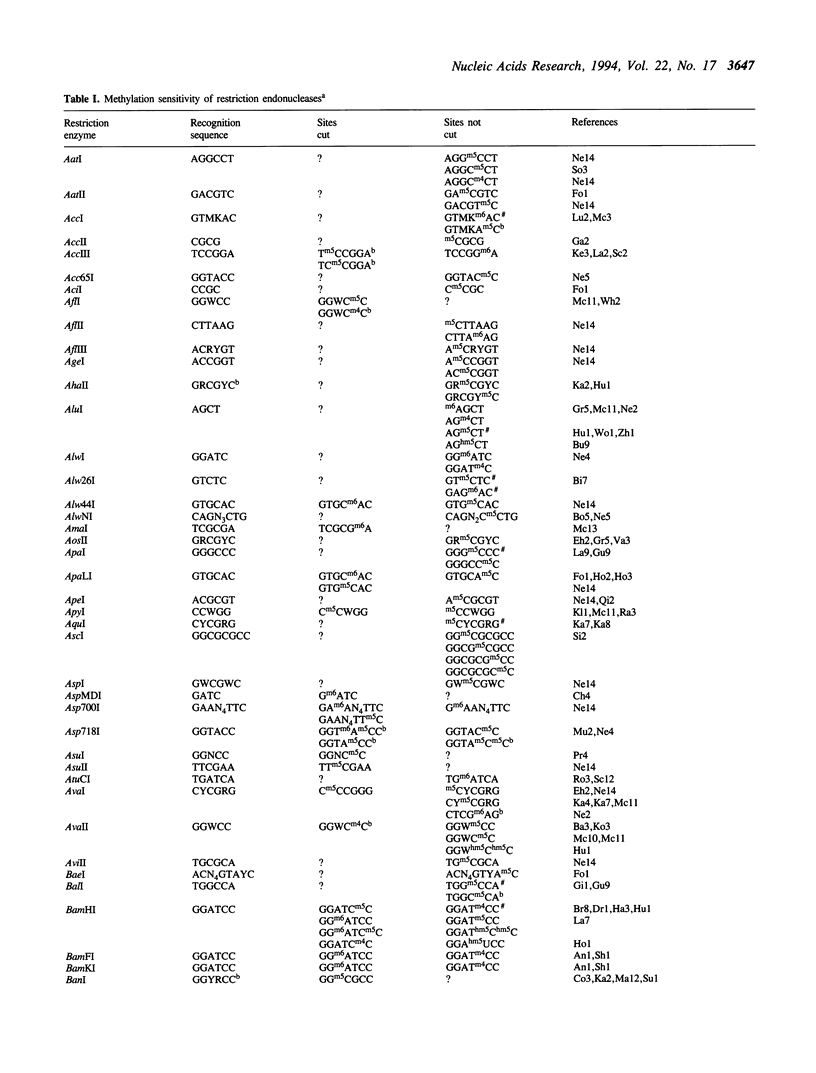
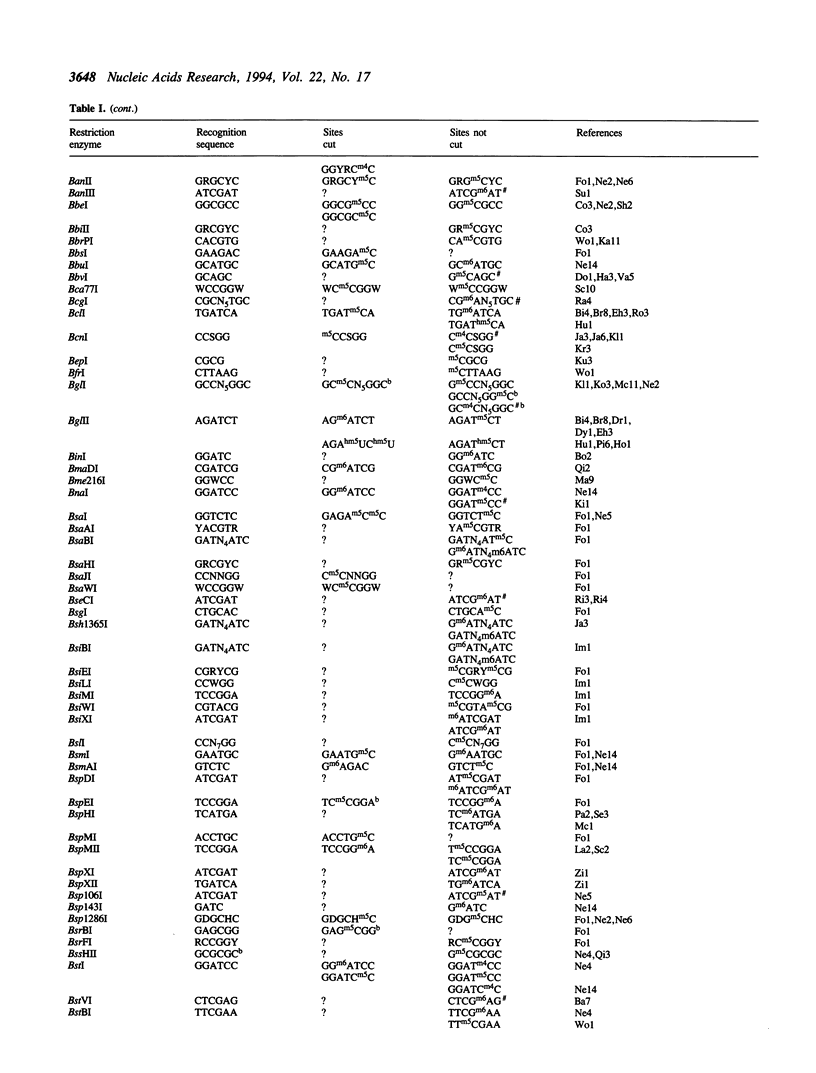
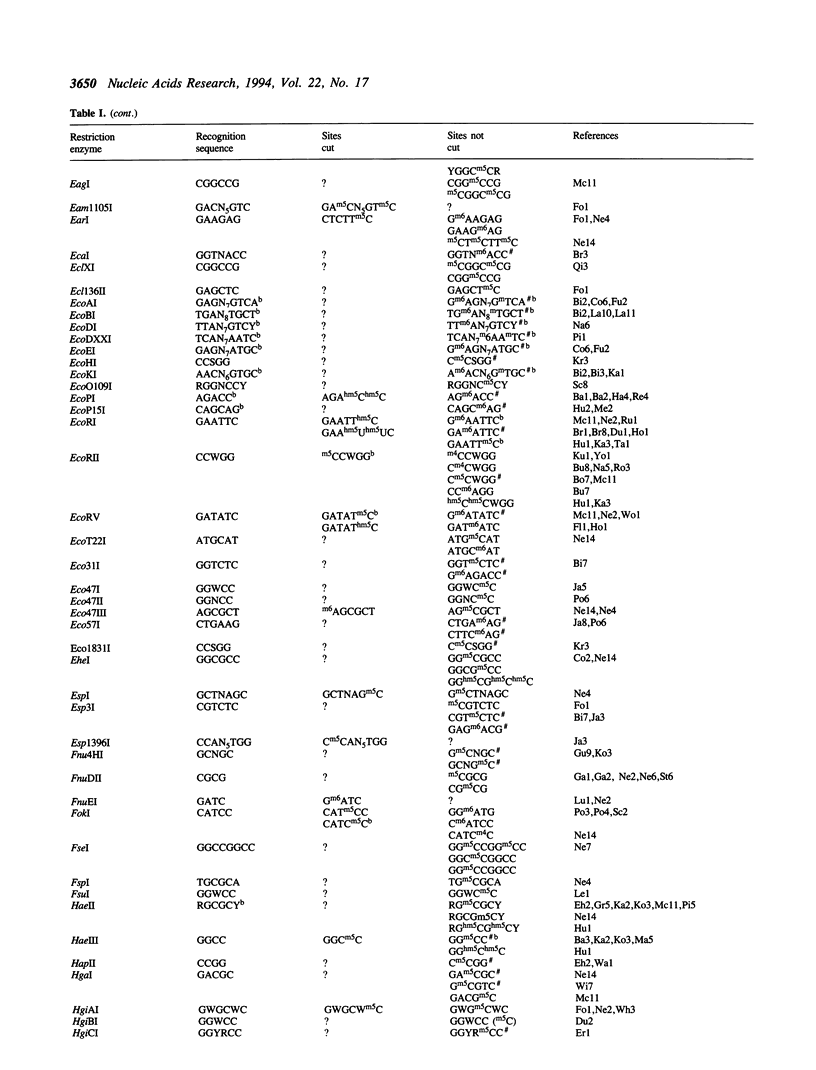
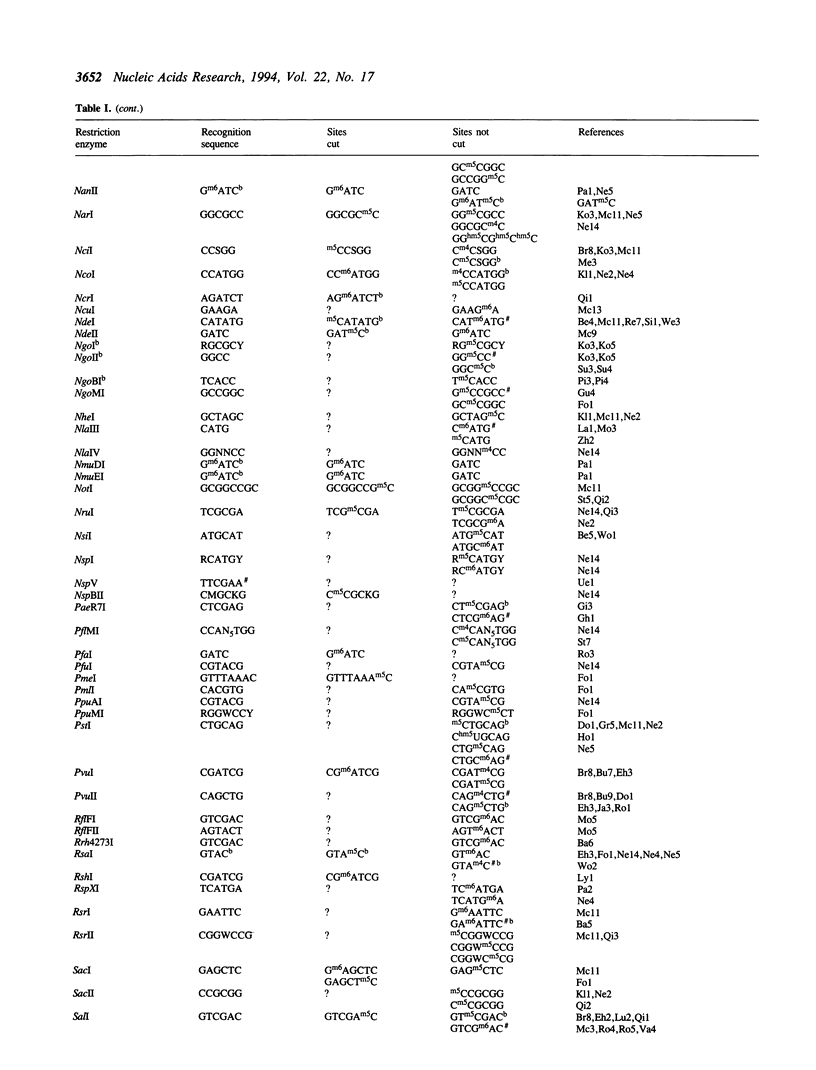
Restriction endonucleases have site-specific interactions with DNA that can often be inhibited by site-specific DNA methylation and other site-specific DNA modifications. However, such inhibition cannot generally be predicted. The empirically acquired data on these effects are tabulated for over 320 restriction endonucleases. In addition, a table of known site-specific DNA modification methyltransferases and their specificities is presented along with EMBL database accession numbers for cloned genes.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K. A cautionary note on the use of certain restriction endonucleases with methylated substrates. Gene. 1980 Oct;11(1-2):169–171. doi: 10.1016/0378-1119(80)90097-9. [DOI] [PubMed] [Google Scholar]
- Balganesh T. S., Reiners L., Lauster R., Noyer-Weidner M., Wilke K., Trautner T. A. Construction and use of chimeric SPR/phi 3T DNA methyltransferases in the definition of sequence recognizing enzyme regions. EMBO J. 1987 Nov;6(11):3543–3549. doi: 10.1002/j.1460-2075.1987.tb02681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas J. A., Ferretti J. J., Progulske-Fox A. Identification and sequence analysis of a methylase gene in Porphyromonas gingivalis. Nucleic Acids Res. 1991 Aug 11;19(15):4189–4192. doi: 10.1093/nar/19.15.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984 Nov;160(2):586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra R., Chiong M., González E., Vásquez C. A DNA-modification methylase from Bacillus stearothermophilus V. Biochem J. 1988 Oct 15;255(2):699–703. [PMC free article] [PubMed] [Google Scholar]
- Baryshev M. M., Bur'ianov Ia I., Kosykh V. G., Baev A. A. Vydelenie, ochistka i nekotorye svoistva restriktazy i metilazy BstN1 iz Bacillus stearothermophilus. Biokhimiia. 1989 Nov;54(11):1894–1903. [PubMed] [Google Scholar]
- Behrens B., Noyer-Weidner M., Pawlek B., Lauster R., Balganesh T. S., Trautner T. A. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987 Apr;6(4):1137–1142. doi: 10.1002/j.1460-2075.1987.tb04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hattar J., Jiricny J. Effect of cytosine methylation on the cleavage of oligonucleotide duplexes with restriction endonucleases HpaII and MspI. Nucleic Acids Res. 1988 May 11;16(9):4160–4160. doi: 10.1093/nar/16.9.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bingham A. H., Atkinson T., Sciaky D., Roberts R. J. A specific endonuclease from Bacillus caldolyticus. Nucleic Acids Res. 1978 Oct;5(10):3457–3467. doi: 10.1093/nar/5.10.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitinaité J. B., Klimasauskas S. J., Butkus V. V., Janulaitis A. A. Characterization of restriction-modification enzymes Cfr13 I from Citrobacter freundii RFL13. FEBS Lett. 1985 Mar 25;182(2):509–513. doi: 10.1016/0014-5793(85)80364-1. [DOI] [PubMed] [Google Scholar]
- Blumenthal R. M., Gregory S. A., Cooperider J. S. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J Bacteriol. 1985 Nov;164(2):501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar J. W., Zempsky W., Warder D., Bergson C., Ward D. C. Effect of nucleotide analogs on the cleavage of DNA by the restriction enzymes AluI, DdeI, HinfI, RsaI, and TaqI. J Biol Chem. 1983 Dec 25;258(24):15206–15213. [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Bougueleret L., Schwarzstein M., Tsugita A., Zabeau M. Characterization of the genes coding for the Eco RV restriction and modification system of Escherichia coli. Nucleic Acids Res. 1984 Apr 25;12(8):3659–3676. doi: 10.1093/nar/12.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A. C., Charles I. G., Keyte J. W., Brammar W. J. Isolation and computer-aided characterization of MmeI, a type II restriction endonuclease from Methylophilus methylotrophus. Nucleic Acids Res. 1986 Jul 11;14(13):5255–5274. doi: 10.1093/nar/14.13.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the cleavage by the EcoRI restriction endonuclease of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7270–7278. [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the methylation by the EcoRI modification methylase of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7279–7286. [PubMed] [Google Scholar]
- Brenner V., Venetianer P., Kiss A. Cloning and nucleotide sequence of the gene encoding the Ecal DNA methyltransferase. Nucleic Acids Res. 1990 Jan 25;18(2):355–359. doi: 10.1093/nar/18.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Hattman S. In vitro methylation of bacteriophage lambda DNA by wild type (dam+) and mutant (damh) forms of the phage T2 DNA adenine methylase. J Mol Biol. 1978 Dec 15;126(3):381–394. doi: 10.1016/0022-2836(78)90047-5. [DOI] [PubMed] [Google Scholar]
- Brooks J. E. Properties and uses of restriction endonucleases. Methods Enzymol. 1987;152:113–129. doi: 10.1016/0076-6879(87)52014-6. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Roberts R. J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982 Feb 11;10(3):913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhk H. J., Behrens B., Tailor R., Wilke K., Prada J. J., Günthert U., Noyer-Weidner M., Jentsch S., Trautner T. A. Restriction and modification in Bacillus subtilis: nucleotide sequence, functional organization and product of the DNA methyltransferase gene of bacteriophage SPR. Gene. 1984 Jul-Aug;29(1-2):51–61. doi: 10.1016/0378-1119(84)90165-3. [DOI] [PubMed] [Google Scholar]
- Bull L. N., Hewitt J. E., Cox D. R., Myers R. M. Sensitivity of HincII to CpG methylation. Nucleic Acids Res. 1993 Apr 25;21(8):2021–2021. doi: 10.1093/nar/21.8.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bur'ianov Ia I., Nesterenko V. F., Vagabova L. M. Prisutstvie dvukh DNK-tsitozin-metilaz v kletkakh Escherichia coli MRE 600. Dokl Akad Nauk SSSR. 1976;227(6):1472–1475. [PubMed] [Google Scholar]
- Bur'ianov Ia I., Zakharenko V. N., Baev A. A. Vydelenie, ochistka i nekotorye svoistva adeninovoi DNK-metilazy Eco dam. Dokl Akad Nauk SSSR. 1981;259(6):1492–1495. [PubMed] [Google Scholar]
- Burkhart J. G., Burkhart B. A., Sampson K., Malling H. V. Evidence for a previously undetected CpG methyl-directed restriction system in E. coli. Nucleic Acids Res. 1992 Aug 25;20(16):4368–4368. doi: 10.1093/nar/20.16.4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buryanov Y. I., Bogdarina I. G., Bayev A. A. Site specificity and chromatographic properties of E. coli K12 and EcoRIIDNA-cytosine methylases. FEBS Lett. 1978 Apr 15;88(2):251–254. doi: 10.1016/0014-5793(78)80186-0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., deBoer E., Wright S., Grosveld F. G., Flavell R. A. The sequence GGCmCGG is resistant to MspI cleavage. Nucleic Acids Res. 1983 Jun 11;11(11):3559–3569. doi: 10.1093/nar/11.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V., Klimasauskas S., Kersulyte D., Vaitkevicius D., Lebionka A., Janulaitis A. Investigation of restriction-modification enzymes from M. varians RFL19 with a new type of specificity toward modification of substrate. Nucleic Acids Res. 1985 Aug 26;13(16):5727–5746. doi: 10.1093/nar/13.16.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta M., Zacharias W., Nwankwo D., Wilson G. G., Wells R. D. Cloning, sequencing, in vivo promoter mapping, and expression in Escherichia coli of the gene for the HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4770–4777. [PubMed] [Google Scholar]
- Casjens S., Hayden M., Jackson E., Deans R. Additional restriction endonuclease cleavage sites on the bacteriophage P22 genome. J Virol. 1983 Feb;45(2):864–867. doi: 10.1128/jvi.45.2.864-867.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasegaran S., Lunnen K. D., Smith H. O., Wilson G. G. Cloning and sequencing the HinfI restriction and modification genes. Gene. 1988 Oct 30;70(2):387–392. doi: 10.1016/0378-1119(88)90210-7. [DOI] [PubMed] [Google Scholar]
- Chatterjee D. K., Hammond A. W., Blakesley R. W., Adams S. M., Gerard G. F. Genetic organization of the KpnI restriction--modification system. Nucleic Acids Res. 1991 Dec 11;19(23):6505–6509. doi: 10.1093/nar/19.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiura H. X., Kamiyama T., Hirano H., Futagami M., Watahiki M., Kobayashi K., Simidu U., Takagi J. Purification and characterization of AspMD1, an isoschizomer of Sau3AI, from a marine bacterium, Alcaligenes sp MD1. Nucleic Acids Res. 1992 Apr 25;20(8):1996–1996. doi: 10.1093/nar/20.8.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney C. A., Eykholt R. L., Bradbury E. M. Methylation is co-ordinated on the putative replication origins of Physarum ribosomal DNA. J Mol Biol. 1988 Dec 20;204(4):889–901. doi: 10.1016/0022-2836(88)90049-6. [DOI] [PubMed] [Google Scholar]
- Cooney C. A. The restriction enzyme EheI (GGC/GCC) is sensitive to CpG methylation. Nucleic Acids Res. 1990 Jun 25;18(12):3667–3667. doi: 10.1093/nar/18.12.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985 May 10;13(9):3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan G. M., Gann A. A., Murray N. E. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989 Jan 13;56(1):103–109. doi: 10.1016/0092-8674(89)90988-4. [DOI] [PubMed] [Google Scholar]
- Danaher R. J., Stein D. C. Expression of cloned restriction and modification genes, hjaIRM from Hyphomonas jannaschiana in Escherichia coli. Gene. 1990 Apr 30;89(1):129–132. doi: 10.1016/0378-1119(90)90215-d. [DOI] [PubMed] [Google Scholar]
- Daniel A. S., Fuller-Pace F. V., Legge D. M., Murray N. E. Distribution and diversity of hsd genes in Escherichia coli and other enteric bacteria. J Bacteriol. 1988 Apr;170(4):1775–1782. doi: 10.1128/jb.170.4.1775-1782.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R., van der Lelie D., Mercenier A., Daly C., Fitzgerald G. F. ScrFI restriction-modification system of Lactococcus lactis subsp. cremoris UC503: cloning and characterization of two ScrFI methylase genes. Appl Environ Microbiol. 1993 Mar;59(3):777–785. doi: 10.1128/aem.59.3.777-785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarev Skh, Prikhod'ko E. A., Prikhod'ko G. G., Krasnykh V. N. Vspl methylase belongs to m6A-gamma class of adenine methylases. Nucleic Acids Res. 1993 Apr 25;21(8):2015–2015. doi: 10.1093/nar/21.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingman D. W. Presence of N6-methyladenine in GATC sequences of Bacillus popilliae and Bacillus lentimorbus KLN2. J Bacteriol. 1990 Oct;172(10):6156–6159. doi: 10.1128/jb.172.10.6156-6159.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. P., Dobritsa S. V. DNA protection with the DNA methylase M . BbvI from Bacillus brevis var. GB against cleavage by the restriction endonucleases PstI and PvuII. Gene. 1980 Jul;10(2):105–112. doi: 10.1016/0378-1119(80)90128-6. [DOI] [PubMed] [Google Scholar]
- Doolittle M. M., Sirotkin K. Bacteriophage T2 and T4, dam+ and damh and Eco dam+ methylation: preference at different sites. Biochim Biophys Acta. 1988 Feb 28;949(2):240–246. doi: 10.1016/0167-4781(88)90088-7. [DOI] [PubMed] [Google Scholar]
- Dreiseikelmann B., Eichenlaub R., Wackernagel W. The effect of differential methylation by Escherichia coli of plasmid DNA and phage T7 and lambda DNA on the cleavage by restriction endonuclease MboI from Moraxella bovis. Biochim Biophys Acta. 1979 May 24;562(3):418–428. doi: 10.1016/0005-2787(79)90105-9. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Hedgpeth J., Boyer H. W., Goodman H. M. Physical identity of the SV40 deoxyribonucleic acid sequence recognized by the Eco RI restriction endonuclease and modification methylase. Biochemistry. 1974 Jan 29;13(3):503–512. doi: 10.1021/bi00700a016. [DOI] [PubMed] [Google Scholar]
- Dybvig K., Swinton D., Maniloff J., Hattman S. Cytosine methylation of the sequence GATC in a mycoplasma. J Bacteriol. 1982 Sep;151(3):1420–1424. doi: 10.1128/jb.151.3.1420-1424.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düsterhöft A., Erdmann D., Kröger M. Stepwise cloning and molecular characterization of the HgiDI restriction-modification system from Herpetosiphon giganteus Hpa2. Nucleic Acids Res. 1991 Mar 11;19(5):1049–1056. doi: 10.1093/nar/19.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wilson G. G., Kuo K. C., Gehrke C. W. N4-methylcytosine as a minor base in bacterial DNA. J Bacteriol. 1987 Mar;169(3):939–943. doi: 10.1128/jb.169.3.939-943.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann D., Horst G., Düsterhöft A., Kröger M. Stepwise cloning and genetic organization of the seemingly unclonable HgiCII restriction-modification system from Herpetosiphon giganteus strain Hpg9, using PCR technique. Gene. 1992 Aug 1;117(1):15–22. doi: 10.1016/0378-1119(92)90484-7. [DOI] [PubMed] [Google Scholar]
- Essani K., Goorha R., Granoff A. An animal virus-induced DNA methyltransferase. Gene. 1988 Dec 25;74(1):71–72. doi: 10.1016/0378-1119(88)90254-5. [DOI] [PubMed] [Google Scholar]
- Fehér Z., Kiss A., Venetianer P. Expression of a bacterial modification methylase gene in yeast. Nature. 1983 Mar 17;302(5905):266–268. doi: 10.1038/302266a0. [DOI] [PubMed] [Google Scholar]
- Ferrin L. J., Camerini-Otero R. D. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science. 1991 Dec 6;254(5037):1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- Fliess A., Wolfes H., Seela F., Pingoud A. Analysis of the recognition mechanism involved in the EcoRV catalyzed cleavage of DNA using modified oligodeoxynucleotides. Nucleic Acids Res. 1988 Dec 23;16(24):11781–11793. doi: 10.1093/nar/16.24.11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Bullas L. R., Delius H., Murray N. E. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Cowan G. M., Murray N. E. EcoA and EcoE: alternatives to the EcoK family of type I restriction and modification systems of Escherichia coli. J Mol Biol. 1985 Nov 5;186(1):65–75. doi: 10.1016/0022-2836(85)90257-8. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Murray N. E. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaido M. L., Prostko C. R., Strobl J. S. Isolation and characterization of BsuE methyltransferase, a CGCG specific DNA methyltransferase from Bacillus subtilis. J Biol Chem. 1988 Apr 5;263(10):4832–4836. [PubMed] [Google Scholar]
- Gaido M. L., Strobl J. S. Methylation sensitivity of the restriction enzymes FnuDII and AccII. Arch Microbiol. 1987 Jan;146(4):338–340. doi: 10.1007/BF00410932. [DOI] [PubMed] [Google Scholar]
- Gann A. A., Campbell A. J., Collins J. F., Coulson A. F., Murray N. E. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987 Jul;1(1):13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Garnett J., Halford S. E. Properties and subunit structure of EcoRV methyltransferase. Gene. 1988 Dec 25;74(1):73–76. doi: 10.1016/0378-1119(88)90255-7. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Ghosh S. S., Obermiller P. S., Kwoh T. J., Gingeras T. R. Analysis of substrate specificity of the PaeR7 endonuclease: effect of base methylation on the kinetics of cleavage. Nucleic Acids Res. 1990 Sep 11;18(17):5063–5068. doi: 10.1093/nar/18.17.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Brooks J. E. Cloned restriction/modification system from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1983 Jan;80(2):402–406. doi: 10.1073/pnas.80.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A., Hattman S., Pleger G. L. ( 6 N)methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J Cell Biol. 1973 Mar;56(3):697–701. doi: 10.1083/jcb.56.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen J. A., Vijg J. E. coli C: a convenient host strain for rescue of highly methylated DNA. Nucleic Acids Res. 1988 Oct 11;16(19):9343–9343. doi: 10.1093/nar/16.19.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Gupta M., Boyer H. W., Brown W. E., Rosenberg J. M. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J Biol Chem. 1981 Mar 10;256(5):2143–2153. [PubMed] [Google Scholar]
- Gruenbaum Y., Cedar H., Razin A. Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res. 1981 Jun 11;9(11):2509–2515. doi: 10.1093/nar/9.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler M., Braguglia D., Meyer J., Piekarowicz A., Bickle T. A. Recombination of constant and variable modules alters DNA sequence recognition by type IC restriction-modification enzymes. EMBO J. 1992 Jan;11(1):233–240. doi: 10.1002/j.1460-2075.1992.tb05046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J. S., Piekarowicz A., Chien R., Stein D. C. Cloning and linkage analysis of Neisseria gonorrhoeae DNA methyltransferases. J Bacteriol. 1992 Sep;174(17):5654–5660. doi: 10.1128/jb.174.17.5654-5660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Lauster R., Reiners L. Multispecific DNA methyltransferases from Bacillus subtilis phages. Properties of wild-type and various mutant enzymes with altered DNA affinity. Eur J Biochem. 1986 Sep 15;159(3):485–492. doi: 10.1111/j.1432-1033.1986.tb09912.x. [DOI] [PubMed] [Google Scholar]
- Günthert U., Trautner T. A. DNA methyltransferases of Bacillus subtilis and its bacteriophages. Curr Top Microbiol Immunol. 1984;108:11–22. doi: 10.1007/978-3-642-69370-0_2. [DOI] [PubMed] [Google Scholar]
- Hanish J., McClelland M. Enzymatic cleavage of a bacterial chromosome at a transposon-inserted rare site. Nucleic Acids Res. 1991 Feb 25;19(4):829–832. doi: 10.1093/nar/19.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanish J., McClelland M. Methylase-limited partial NotI cleavage for physical mapping of genomic DNA. Nucleic Acids Res. 1990 Jun 11;18(11):3287–3291. doi: 10.1093/nar/18.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Heidmann S., Seifert W., Kessler C., Domdey H. Cloning, characterization and heterologous expression of the SmaI restriction-modification system. Nucleic Acids Res. 1989 Dec 11;17(23):9783–9796. doi: 10.1093/nar/17.23.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Model P. Mutants of the EcoRI endonuclease with promiscuous substrate specificity implicate residues involved in substrate recognition. EMBO J. 1990 Oct;9(10):3369–3378. doi: 10.1002/j.1460-2075.1990.tb07538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentosh P., McCastlain J. C. Effects of 2-chloroadenine substitution in DNA on restriction endonuclease cleavage reactions. Nucleic Acids Res. 1991 Jun 11;19(11):3143–3148. doi: 10.1093/nar/19.11.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991 Jul;173(14):4363–4370. doi: 10.1128/jb.173.14.4363-4370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoet P. P., Coene M. M., Cocito C. G. Replication cycle of Bacillus subtilis hydroxymethyluracil-containing phages. Annu Rev Microbiol. 1992;46:95–116. doi: 10.1146/annurev.mi.46.100192.000523. [DOI] [PubMed] [Google Scholar]
- Hofer B., Kühlein B. The sensitivity of DNA cleavage by SnoI to methylation by M.EcoK. Nucleic Acids Res. 1989 Oct 11;17(19):8009–8009. doi: 10.1093/nar/17.19.8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. A., Card C., Benner J. S., Callahan H. L., Maunus R., Silber K., Wilson G., Brooks J. E. Cloning the DdeI restriction-modification system using a two-step method. Nucleic Acids Res. 1986 Oct 24;14(20):7939–7951. doi: 10.1093/nar/14.20.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. H., Farnet C. M., Ehrlich K. C., Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982 Mar 11;10(5):1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümbelin M., Suri B., Rao D. N., Hornby D. P., Eberle H., Pripfl T., Kenel S., Bickle T. A. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J Mol Biol. 1988 Mar 5;200(1):23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- Ikawa S., Shibata T., Ando T., Saito H. Genetic studies on site-specific endodeoxyribonucleases in Bacillus subtilis: multiple modification and restriction systems in transformants of Bacillus subtilis 168. Mol Gen Genet. 1980 Feb;177(3):359–368. doi: 10.1007/BF00271474. [DOI] [PubMed] [Google Scholar]
- Ito H., Shimato H., Sadaoka A., Kotani H., Kimizuka F., Kato I. Cloning and expression of the HpaI restriction-modification genes. Nucleic Acids Res. 1992 Feb 25;20(4):705–709. doi: 10.1093/nar/20.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs D., Brown N. L. Isolation and characterization of the M.EaeI modification methylase. Biochem J. 1986 Sep 1;238(2):613–615. doi: 10.1042/bj2380613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulaitis A., Klimasauskas S., Petrusyte M., Butkus V. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983 Sep 5;161(1):131–134. doi: 10.1016/0014-5793(83)80745-5. [DOI] [PubMed] [Google Scholar]
- Janulaitis A., Petrusyte M., Maneliene Z., Klimasauskas S., Butkus V. Purification and properties of the Eco57I restriction endonuclease and methylase--prototypes of a new class (type IV). Nucleic Acids Res. 1992 Nov 25;20(22):6043–6049. doi: 10.1093/nar/20.22.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulaitis A., Petrusyté M., Butkus V. Three sequence-specific endonucleases from Escherichia coli RFL47. FEBS Lett. 1983 Sep 19;161(2):213–216. doi: 10.1016/0014-5793(83)81010-2. [DOI] [PubMed] [Google Scholar]
- Janulaitis A., Vaisvila R., Timinskas A., Klimasauskas S., Butkus V. Cloning and sequence analysis of the genes coding for Eco57I type IV restriction-modification enzymes. Nucleic Acids Res. 1992 Nov 25;20(22):6051–6056. doi: 10.1093/nar/20.22.6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan N. C., Lautenberger J. A., Edgell M. H., Hutchison C. A., 3rd The nucleotide sequence recognized by the Escherichia coli K12 restriction and modification enzymes. J Mol Biol. 1979 May 15;130(2):191–209. doi: 10.1016/0022-2836(79)90426-1. [DOI] [PubMed] [Google Scholar]
- Kang S. C., Choi W. S., Yoo O. J. The effects of DNA methylation by Hha I methylase on the cleavage reactions by Hae II, Aha II and Ban I endonucleases. Biochem Biophys Res Commun. 1987 May 29;145(1):482–487. doi: 10.1016/0006-291x(87)91346-5. [DOI] [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman C., Tandeau de Marsac N., de Waard A. Isolation of a deoxycytidylate methyl transferase capable of protecting DNA uniquely against cleavage by endonuclease R.Aqu I (isoschizomer of Ava I). Nucleic Acids Res. 1986 Jul 11;14(13):5199–5205. doi: 10.1093/nar/14.13.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman C., de Waard A. Isolation and characterization of the modification methylase M . SinI. J Bacteriol. 1988 Jun;170(6):2533–2536. doi: 10.1128/jb.170.6.2533-2536.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami B., Sasaki A., Oka M., Maekawa Y. Nucleotide sequence of the gene coding for the BanIII DNA methyltransferase in Bacillus aneurinolyticus. Agric Biol Chem. 1990 Dec;54(12):3227–3233. [PubMed] [Google Scholar]
- Kelly S., Kaddurah-Daouk R., Smith H. O. Purification of the HhaII restriction endonuclease from an overproducer Escherichia coli clone. J Biol Chem. 1985 Dec 5;260(28):15339–15344. [PubMed] [Google Scholar]
- Kessler C., Neumaier P. S., Wolf W. Recognition sequences of restriction endonucleases and methylases--a review. Gene. 1985;33(1):1–102. doi: 10.1016/0378-1119(85)90119-2. [DOI] [PubMed] [Google Scholar]
- Kim E. L., Maliuta S. S. Purification of a DNA methyltransferase from Bacillus natto B3364. FEBS Lett. 1989 Sep 25;255(2):361–364. doi: 10.1016/0014-5793(89)81122-6. [DOI] [PubMed] [Google Scholar]
- Kiss A., Posfai G., Keller C. C., Venetianer P., Roberts R. J. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985 Sep 25;13(18):6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K., Suisha M., Kotani H., Yanase H., Kato N. Cloning and sequence analysis of the StsI restriction-modification gene: presence of homology to FokI restriction-modification enzymes. Nucleic Acids Res. 1992 Aug 25;20(16):4167–4172. doi: 10.1093/nar/20.16.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimashauskas S. I., Butkus V. V., Ianulaitis A. A. Spetsifichnost' deistviia DNK-metilaz BcnI, CfrI i Cfr10I. Mol Biol (Mosk) 1987 Jan-Feb;21(1):87–92. [PubMed] [Google Scholar]
- Kochanek S., Renz D., Doerfler W. Probing DNA-protein interactions in vitro with the CpG DNA methyltransferase. Nucleic Acids Res. 1993 May 25;21(10):2339–2342. doi: 10.1093/nar/21.10.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Kiss A., Venetianer P. Biochemical characterization of the restriction-modification system of Bacillus sphaericus. Eur J Biochem. 1978 Sep 1;89(2):523–529. doi: 10.1111/j.1432-1033.1978.tb12557.x. [DOI] [PubMed] [Google Scholar]
- Koob M., Burkiewicz A., Kur J., Szybalski W. RecA-AC: single-site cleavage of plasmids and chromosomes at any predetermined restriction site. Nucleic Acids Res. 1992 Nov 11;20(21):5831–5836. doi: 10.1093/nar/20.21.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch C., Hagblom P. In-vivo-modified gonococcal plasmid pJD1. A model system for analysis of restriction enzyme sensitivity to DNA modifications. Eur J Biochem. 1986 Dec 15;161(3):519–524. doi: 10.1111/j.1432-1033.1986.tb10473.x. [DOI] [PubMed] [Google Scholar]
- Korch C., Hagblom P., Normark S. Sequence-specific DNA modification in Neisseria gonorrhoeae. J Bacteriol. 1983 Sep;155(3):1324–1332. doi: 10.1128/jb.155.3.1324-1332.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch C., Hagblom P., Normark S. Type III 5-methylcytosine modification of DNA in Neisseria gonorrhoeae. J Bacteriol. 1985 Mar;161(3):1236–1237. doi: 10.1128/jb.161.3.1236-1237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosykh V. G., Buryanov Y. I., Bayev A. A. Molecular cloning of EcoRII endonuclease and methylase genes. Mol Gen Genet. 1980;178(3):717–718. doi: 10.1007/BF00337884. [DOI] [PubMed] [Google Scholar]
- Kosykh V. G., Glinskaite I. V., Bur'ianov Ia I., Baev A. A. Opredelenie lokalizatsii genov restriktazy i metilazy EcoRII na rekombinantnykh plazmidakh. Dokl Akad Nauk SSSR. 1982;265(3):727–730. [PubMed] [Google Scholar]
- Kosykh V. G., Solonin A. S., Buryanov YaI, Bayev A. A. Overproduction of the EcoRII endonuclease and methylase by Escherichia coli strains carrying recombinant plasmids constructed in vitro. Biochim Biophys Acta. 1981 Aug 27;655(1):102–106. doi: 10.1016/0005-2787(81)90072-1. [DOI] [PubMed] [Google Scholar]
- Kramarov V. M., Smolianinov V. V. DNK-metilaza iz Arthrobacter luteus zashchishchaet DNK ot deistviia saitspetsificheskoi éndonukleazy Alu I. Biokhimiia. 1981 Aug;46(8):1526–1529. [PubMed] [Google Scholar]
- Kravetz A. N., Zakharova M. V., Beletskaya I. V., Sineva E. V., Denjmuchametov M. M., Petrov S. I., Glatman L. I., Solonin A. S. The cleavage sites and localization of genes encoding the restriction endonucleases Eco1831I and EcoHI. Gene. 1993 Jul 15;129(1):153–154. doi: 10.1016/0378-1119(93)90712-c. [DOI] [PubMed] [Google Scholar]
- Kretz P. L., Reid C. H., Greener A., Short J. M. Effect of lambda packaging extract mcr restriction activity on DNA cloning. Nucleic Acids Res. 1989 Jul 11;17(13):5409–5409. doi: 10.1093/nar/17.13.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubareva E. A., Gromova E. S., Romanova E. A., Oretskaia T. S., Shabarova Z. A. Osobennosti rasshchepleniia éndonukleazami restriktsii MvaI i EcoRII substratov, modifitsirovannykh po aminogruppam geterotsiklicheskikh osnovanii. Bioorg Khim. 1990 Apr;16(4):501–506. [PubMed] [Google Scholar]
- Kubareva E. A., Pein C. D., Gromova E. S., Kuznezova S. A., Tashlitzki V. N., Cech D., Shabarova Z. A. The role of modifications in oligonucleotides in sequence recognition by MvaI restriction endonuclease. Eur J Biochem. 1988 Aug 15;175(3):615–618. doi: 10.1111/j.1432-1033.1988.tb14236.x. [DOI] [PubMed] [Google Scholar]
- Labbé D., Höltke H. J., Lau P. C. Cloning and characterization of two tandemly arranged DNA methyltransferase genes of Neisseria lactamica: an adenine-specific M.NlaIII and a cytosine-type methylase. Mol Gen Genet. 1990 Oct;224(1):101–110. doi: 10.1007/BF00259456. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Mannarelli B. M., Springhorn S. S., Greenberg B. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell. 1986 Sep 26;46(7):993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Springhorn S. S. Cloning in Streptococcus pneumoniae of the gene for DpnII DNA methylase. J Bacteriol. 1984 Mar;157(3):934–936. doi: 10.1128/jb.157.3.934-936.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Landry D., Looney M. C., Feehery G. R., Slatko B. E., Jack W. E., Schildkraut I., Wilson G. G. M.FokI methylates adenine in both strands of its asymmetric recognition sequence. Gene. 1989 Apr 15;77(1):1–10. doi: 10.1016/0378-1119(89)90353-3. [DOI] [PubMed] [Google Scholar]
- Lange C., Noyer-Weidner M., Trautner T. A., Weiner M., Zahler S. A. M.H2I, a multispecific 5C-DNA methyltransferase encoded by Bacillus amyloliquefaciens phage H2. Gene. 1991 Apr;100:213–218. doi: 10.1016/0378-1119(91)90369-m. [DOI] [PubMed] [Google Scholar]
- Larimer F. W. Cleavage by ApaI is inhibited by overlapping dcm methylation. Nucleic Acids Res. 1987 Nov 11;15(21):9087–9087. doi: 10.1093/nar/15.21.9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Kan N. C., Lackey D., Linn S., Edgell M. H., Hutchison C. A., 3rd Recognition site of Escherichia coli B restriction enzyme on phi XsB1 and simian virus 40 DNAs: an interrupted sequence. Proc Natl Acad Sci U S A. 1978 May;75(5):2271–2275. doi: 10.1073/pnas.75.5.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. I. Purification, subunit structure, and catalytic properties of the modification methylase. J Biol Chem. 1972 Oct 10;247(19):6176–6182. [PubMed] [Google Scholar]
- Lee S. F., Forsberg C. W., Gibbins A. M. Type II DNA restriction-modification system and an endonuclease from the ruminal bacterium Fibrobacter succinogenes S85. J Bacteriol. 1992 Aug;174(16):5275–5283. doi: 10.1128/jb.174.16.5275-5283.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. K., Tu J. Ssp RF1, a novel class-II restriction endonuclease from Synechococcus RF-1 recognizing 5'TT/CGAA-3'. Nucleic Acids Res. 1991 Sep 11;19(17):4770–4770. doi: 10.1093/nar/19.17.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodwick D., Ross H. N., Harris J. E., Almond J. W., Grant W. D. dam methylation in the archaebacteria. J Gen Microbiol. 1986 Nov;132(11):3055–3059. doi: 10.1099/00221287-132-11-3055. [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Daniel A. S., Braymer H. D., Murray N. E. Organization and sequence of the hsd genes of Escherichia coli K-12. J Mol Biol. 1987 Nov 20;198(2):159–170. doi: 10.1016/0022-2836(87)90303-2. [DOI] [PubMed] [Google Scholar]
- Looney M. C., Moran L. S., Jack W. E., Feehery G. R., Benner J. S., Slatko B. E., Wilson G. G. Nucleotide sequence of the FokI restriction-modification system: separate strand-specificity domains in the methyltransferase. Gene. 1989 Aug 15;80(2):193–208. doi: 10.1016/0378-1119(89)90284-9. [DOI] [PubMed] [Google Scholar]
- Lui A. C., McBride B. C., Vovis G. F., Smith M. Site specific endonuclease from Fusobacterium nucleatum. Nucleic Acids Res. 1979 Jan;6(1):1–15. doi: 10.1093/nar/6.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnen K. D., Barsomian J. M., Camp R. R., Card C. O., Chen S. Z., Croft R., Looney M. C., Meda M. M., Moran L. S., Nwankwo D. O. Cloning type-II restriction and modification genes. Gene. 1988 Dec 25;74(1):25–32. doi: 10.1016/0378-1119(88)90242-9. [DOI] [PubMed] [Google Scholar]
- Lynn S. P., Cohen L. K., Gardner J. F., Kaplan S. Characterization of a site-specific restriction endonuclease from Rhodopseudomonas sphaeroides. J Bacteriol. 1979 May;138(2):505–509. doi: 10.1128/jb.138.2.505-509.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil D. J. Characterization of a unique methyl-specific restriction system in Streptomyces avermitilis. J Bacteriol. 1988 Dec;170(12):5607–5612. doi: 10.1128/jb.170.12.5607-5612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Mosig G. Regulation of a new bacteriophage T4 gene, 69, that spans an origin of DNA replication. EMBO J. 1984 Dec 1;3(12):2863–2871. doi: 10.1002/j.1460-2075.1984.tb02221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa Y., Yasukawa H., Kawakami B. Cloning and nucleotide sequences of the BanI restriction-modification genes in Bacillus aneurinolyticus. J Biochem. 1990 Apr;107(4):645–649. doi: 10.1093/oxfordjournals.jbchem.a123101. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Wold B., Dervan P. B. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989 Aug 18;245(4919):725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- Mannarelli B. M., Balganesh T. S., Greenberg B., Springhorn S. S., Lacks S. A. Nucleotide sequence of the Dpn II DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4468–4472. doi: 10.1073/pnas.82.13.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko N. I., Kramarov V. M., Pachkunov D. M. Isolation and some properties of the site-specific endonuclease and methylase Bme2161 from Bacillus megaterium 216. Eur J Biochem. 1987 Jun 15;165(3):565–570. doi: 10.1111/j.1432-1033.1987.tb11477.x. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- McClelland M., Kessler L. G., Bittner M. Site-specific cleavage of DNA at 8- and 10-base-pair sequences. Proc Natl Acad Sci U S A. 1984 Feb;81(4):983–987. doi: 10.1073/pnas.81.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M., Cantor C. R. Purification of Mbo II methylase (GAAGmA) from Moraxella bovis: site specific cleavage of DNA at nine and ten base pair sequences. Nucleic Acids Res. 1985 Oct 25;13(20):7171–7182. doi: 10.1093/nar/13.20.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M. Enhancement of the apparent cleavage specificities of restriction endonucleases: applications to megabase mapping of chromosomes. Gene Amplif Anal. 1987;5:257–282. [PubMed] [Google Scholar]
- McClelland M., Nelson M. The effect of site specific methylation on restriction endonuclease digestion. Nucleic Acids Res. 1985;13 (Suppl):r201–r207. doi: 10.1093/nar/13.suppl.r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The effect of sequence specific DNA methylation on restriction endonuclease cleavage. Nucleic Acids Res. 1981 Nov 25;9(22):5859–5866. doi: 10.1093/nar/9.22.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The effect of site specific methylation on restriction endonuclease cleavage (update). Nucleic Acids Res. 1983 Jan 11;11(1):r169–r173. doi: 10.1093/nar/11.1.235-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan R. R., Ulrich E., Bird A. P. Restriction endonuclease NciI is not blocked by CpG methylation. Nucleic Acids Res. 1993 Nov 25;21(23):5517–5518. doi: 10.1093/nar/21.23.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner Z., Hattman S. Molecular cloning, sequencing, and mapping of the bacteriophage T2 dam gene. J Bacteriol. 1988 Nov;170(11):5177–5184. doi: 10.1128/jb.170.11.5177-5184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P., Rubin R. A. Role of the 2-amino group of deoxyguanosine in sequence recognition by EcoRI restriction and modification enzymes. J Biol Chem. 1977 Oct 25;252(20):7273–7278. [PubMed] [Google Scholar]
- Molloy P. L., Watt F. Effect of cytosine methylation on cutting by the restriction enzyme MaeII. Nucleic Acids Res. 1988 Mar 25;16(5):2335–2335. doi: 10.1093/nar/16.5.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison M., Mackie R. I., White B. A. Partial characterization of a DNA restriction endonuclease from Ruminococcus flavefaciens FD-1 and its inhibition by site-specific adenine methylation. Appl Environ Microbiol. 1992 Jan;58(1):66–69. doi: 10.1128/aem.58.1.66-69.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Ozawa O., Kanematsu T., Yamada Y. Characterization of restriction endonuclease CbiI, an isoschizomer of AsuII, from Clostridium bifermentans strain B-4. Nucleic Acids Res. 1991 Jun 25;19(12):3458–3458. doi: 10.1093/nar/19.12.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural R. J. Cleavage by the restriction endonuclease Asp718, an isoschizomer of KpnI, is sensitive to Escherichia coli Dcm methylation. Nucleic Acids Res. 1987 Nov 11;15(21):9085–9085. doi: 10.1093/nar/15.21.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja V., Shepherd J. C., Pripfl T., Bickle T. A. Two type I restriction enzymes from Salmonella species. Purification and DNA recognition sequences. J Mol Biol. 1985 Apr 20;182(4):579–587. doi: 10.1016/0022-2836(85)90243-8. [DOI] [PubMed] [Google Scholar]
- Nagaraja V., Stieger M., Nager C., Hadi S. M., Bickle T. A. The nucleotide sequence recognised by the Escherichia coli D type I restriction and modification enzyme. Nucleic Acids Res. 1985 Jan 25;13(2):389–399. doi: 10.1093/nar/13.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narva K. E., Wendell D. L., Skrdla M. P., Van Etten J. L. Molecular cloning and characterization of the gene encoding the DNA methyltransferase, M.CviBIII, from Chlorella virus NC-1A. Nucleic Acids Res. 1987 Dec 10;15(23):9807–9823. doi: 10.1093/nar/15.23.9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. M., Miceli S. M., Lechevalier M. P., Roberts R. J. FseI, a new type II restriction endonuclease that recognizes the octanucleotide sequence 5' GGCCGGCC 3'. Nucleic Acids Res. 1990 Apr 25;18(8):2061–2064. doi: 10.1093/nar/18.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., Christ C., Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984 Jul 11;12(13):5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Effect of site-specific methylation on DNA modification methyltransferases and restriction endonucleases. Nucleic Acids Res. 1989;17 (Suppl):r389–r415. doi: 10.1093/nar/17.suppl.r389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., McClelland M. Use of DNA methyltransferase/endonuclease enzyme combinations for megabase mapping of chromosomes. Methods Enzymol. 1992;216:279–303. doi: 10.1016/0076-6879(92)16027-h. [DOI] [PubMed] [Google Scholar]
- Nelson M., Raschke E., McClelland M. Effect of site-specific methylation on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1993 Jul 1;21(13):3139–3154. doi: 10.1093/nar/21.13.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P. S., Papas T. S., Schweinfest C. W. Restriction endonuclease cleavage of 5-methyl-deoxycytosine hemimethylated DNA at high enzyme-to-substrate ratios. Nucleic Acids Res. 1993 Feb 11;21(3):681–686. doi: 10.1093/nar/21.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterenko V. F., Bur'ianov Ia I., Baev A. A. Determinirovanie DNK-tsitozin-metilazy I (M.Eco MRE 600 I) plazmidoi Col A. Dokl Akad Nauk SSSR. 1980;250(5):1265–1267. [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- Newman P. C., Williams D. M., Cosstick R., Seela F., Connolly B. A. Interaction of the EcoRV restriction endonuclease with the deoxyadenosine and thymidine bases in its recognition hexamer d(GATATC). Biochemistry. 1990 Oct 23;29(42):9902–9910. doi: 10.1021/bi00494a021. [DOI] [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Anikeicheva N. V., Debov S. S. Determination of the recognition sites of cytosine DNA-methylases from Escherichia coli SK. Nucleic Acids Res. 1979 Sep 25;7(2):517–528. doi: 10.1093/nar/7.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Sharkova E. V., Suchkov S. V., Somodi P., Földes I., Debov S. S. Sequence specificity of isolated DNA-adenine methylases from Mycobacterium smegmatis (butyricum) and Shigella sonnei 47 cells. Biochem Int. 1985 Mar;10(3):405–413. [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Suchkov S. V., Kartashova I. M., Debov S. S. Sequence specificity of isolated DNA-cytosine methylases from Shigella sonnei 47 cells. Biochem Int. 1984 Dec;9(6):771–781. [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Kupsch J., Bergbauer M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: structural relatedness and gene expression. Gene. 1985;35(1-2):143–150. doi: 10.1016/0378-1119(85)90166-0. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Pawlek B., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: DNA methylation potential of the related bacteriophages Z, SPR, SP beta, phi 3T, and rho 11. J Virol. 1983 May;46(2):446–453. doi: 10.1128/jvi.46.2.446-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyer-Weidner M., Pawlek B., Jentsch S., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: gene coding for a BsuR-specific modification methyltransferase in the temperate bacteriophage phi 3T. J Virol. 1981 Jun;38(3):1077–1080. doi: 10.1128/jvi.38.3.1077-1080.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur I., Szyf M., Razin A., Glaser G., Rottem S., Razin S. Procaryotic and eucaryotic traits of DNA methylation in spiroplasmas (mycoplasmas). J Bacteriol. 1985 Oct;164(1):19–24. doi: 10.1128/jb.164.1.19-24.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwankwo D., Wilson G. Cloning of two type II methylase genes that recognise asymmetric nucleotide sequences: FokI and HgaI. Mol Gen Genet. 1987 Oct;209(3):570–574. doi: 10.1007/BF00331164. [DOI] [PubMed] [Google Scholar]
- Nölling J., de Vos W. M. Characterization of the archaeal, plasmid-encoded type II restriction-modification system MthTI from Methanobacterium thermoformicicum THF: homology to the bacterial NgoPII system from Neisseria gonorrhoeae. J Bacteriol. 1992 Sep;174(17):5719–5726. doi: 10.1128/jb.174.17.5719-5726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nölling J., van Eeden F. J., Eggen R. I., de Vos W. M. Modular organization of related Archaeal plasmids encoding different restriction-modification systems in Methanobacterium thermoformicicum. Nucleic Acids Res. 1992 Dec 25;20(24):6501–6507. doi: 10.1093/nar/20.24.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktavcovcá B., Godány A., Pristas P., Stevcíková B., Farkasovská J. Isolation and characterization of the modification methylase M.SauLPI from Streptomyces aureofaciens B-96. Nucleic Acids Res. 1993 Oct 11;21(20):4843–4843. doi: 10.1093/nar/21.20.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Ueda T. Synthesis of decadeoxyribonucleotides containing N6-methyladenine, N4-methylcytosine, and 5-methylcytosine: recognition and cleavage by restriction endonucleases (nucleosides and nucleotides part 74). Nucleic Acids Res. 1987 Jan 12;15(1):219–232. doi: 10.1093/nar/15.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J., Taylor I., Dutta C. F., Kneale G., Firman K. High-level expression of the cloned genes encoding the subunits of and intact DNA methyltransferase, M.EcoR124. Gene. 1992 Mar 1;112(1):21–27. doi: 10.1016/0378-1119(92)90298-4. [DOI] [PubMed] [Google Scholar]
- Pech M., Streeck R. E., Zachau H. G. Patchwork structure of a bovine satellite DNA. Cell. 1979 Nov;18(3):883–893. doi: 10.1016/0092-8674(79)90140-5. [DOI] [PubMed] [Google Scholar]
- Piekarowicz A., Yuan R., Stein D. C. Identification of a new restriction endonuclease, R.NgoBI, from Neisseria gonorrhoeae. Nucleic Acids Res. 1988 Oct 25;16(20):9868–9868. doi: 10.1093/nar/16.20.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarowicz A., Yuan R., Stein D. C. Purification and characterization of DNA methyltransferases from Neisseria gonorrhoeae. Nucleic Acids Res. 1988 Jul 11;16(13):5957–5972. doi: 10.1093/nar/16.13.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Two restriction endonucleases from Bacillus globiggi. Nucleic Acids Res. 1976 Jul;3(7):1747–1760. doi: 10.1093/nar/3.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C., Shepherd J. C., Bickle T. A. DNA recognition by a new family of type I restriction enzymes: a unique relationship between two different DNA specificities. EMBO J. 1987 May;6(5):1493–1497. doi: 10.1002/j.1460-2075.1987.tb02391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Baldauf F., Erdei S., Pósfai J., Venetianer P., Kiss A. Structure of the gene coding for the sequence-specific DNA-methyltransferase of the B. subtilis phage SPR. Nucleic Acids Res. 1984 Dec 11;12(23):9039–9049. doi: 10.1093/nar/12.23.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Kiss A., Erdei S., Pósfai J., Venetianer P. Structure of the Bacillus sphaericus R modification methylase gene. J Mol Biol. 1983 Nov 5;170(3):597–610. doi: 10.1016/s0022-2836(83)80123-5. [DOI] [PubMed] [Google Scholar]
- Pósfai G., Szybalski W. A simple method for locating methylated bases in DNA using class-IIS restriction enzymes. Gene. 1988 Dec 25;74(1):179–181. doi: 10.1016/0378-1119(88)90280-6. [DOI] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint A., Cedar H. In vitro methylation of DNA with Hpa II methylase. Nucleic Acids Res. 1981 Feb 11;9(3):633–646. doi: 10.1093/nar/9.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc Natl Acad Sci U S A. 1986 Dec;83(23):9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Urieli S., Pollack Y., Gruenbaum Y., Glaser G. Studies on the biological role of dna methylation; IV. Mode of methylation of DNA in E. coli cells. Nucleic Acids Res. 1980 Apr 25;8(8):1783–1792. doi: 10.1093/nar/8.8.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Pein C. D., Butkus V., Krüger D. H. An improved method for the detection of Dcm methylation in DNA molecules. Gene. 1990 Oct 30;95(1):161–162. doi: 10.1016/0378-1119(90)90429-u. [DOI] [PubMed] [Google Scholar]
- Richards D. F., Linnett P. E., Oultram J. D., Young M. Restriction endonucleases in Clostridium pasteurianum ATCC 6013 and C. thermohydrosulfuricum DSM 568. J Gen Microbiol. 1988 Dec;134(12):3151–3157. doi: 10.1099/00221287-134-12-3151. [DOI] [PubMed] [Google Scholar]
- Rina M., Dialektakis D., Clark D., Pagomenou M., Bouriotis V. Isolation and identification of restriction endonuclease BseCI. Nucleic Acids Res. 1992 Apr 11;20(7):1807–1807. doi: 10.1093/nar/20.7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1988;16 (Suppl):r271–r313. doi: 10.1093/nar/16.suppl.r271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodicio M. R., Chater K. F. The SalI (SalGI) restriction-modification system of Streptomyces albus G. Gene. 1988 Dec 25;74(1):39–42. doi: 10.1016/0378-1119(88)90246-6. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. II. Partial recognition site base sequences. J Mol Biol. 1973 Dec 25;81(4):445–459. doi: 10.1016/0022-2836(73)90516-0. [DOI] [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Sato S., Nakazawa K., Shinomiya T. A DNA methylase from Thermus thermophilus HB8. J Biochem. 1980 Sep;88(3):737–747. doi: 10.1093/oxfordjournals.jbchem.a133026. [DOI] [PubMed] [Google Scholar]
- Scherzer E., Auer B., Schweiger M. Identification, purification, and characterization of Escherichia coli virus T1 DNA methyltransferase. J Biol Chem. 1987 Nov 5;262(31):15225–15231. [PubMed] [Google Scholar]
- Schlagman S. L., Hattman S. The bacteriophage T2 and T4 DNA-[N6-adenine] methyltransferase (Dam) sequence specificities are not identical. Nucleic Acids Res. 1989 Nov 25;17(22):9101–9112. doi: 10.1093/nar/17.22.9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagman S. L., Miner Z., Fehér Z., Hattman S. The DNA [adenine-N6]methyltransferase (Dam) of bacteriophage T4. Gene. 1988 Dec 20;73(2):517–530. doi: 10.1016/0378-1119(88)90516-1. [DOI] [PubMed] [Google Scholar]
- Schlagman S., Hattman S., May M. S., Berger L. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R. Eco RII). J Bacteriol. 1976 May;126(2):990–996. doi: 10.1128/jb.126.2.990-996.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Scherzer E., Auer B., de Groot E. J., Schweiger M. Primary structure of a DNA (N6-adenine)-methyltransferase from Escherichia coli virus T1. DNA sequence, genomic organization, and comparative analysis. J Biol Chem. 1990 Apr 15;265(11):6086–6091. [PubMed] [Google Scholar]
- Schoner B., Kelly S., Smith H. O. The nucleotide sequence of the HhaII restriction and modification genes from Haemophilus haemolyticus. Gene. 1983 Oct;24(2-3):227–236. doi: 10.1016/0378-1119(83)90083-5. [DOI] [PubMed] [Google Scholar]
- Seeber S., Kessler C., Götz F. Cloning, expression and characterization of the Sau3AI restriction and modification genes in Staphylococcus carnosus TM300. Gene. 1990 Sep 28;94(1):37–43. doi: 10.1016/0378-1119(90)90465-4. [DOI] [PubMed] [Google Scholar]
- Seela F., Röling A. 7-Deazapurine containing DNA: efficiency of c7GdTP, c7AdTP and c7IdTP incorporation during PCR-amplification and protection from endodeoxyribonuclease hydrolysis. Nucleic Acids Res. 1992 Jan 11;20(1):55–61. doi: 10.1093/nar/20.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Garrett P. W., Haack K. R., Jensen B. C., Schabtach E. DNA methylation and control of genome organization in Neurospora crassa. Gene. 1988 Dec 25;74(1):109–111. doi: 10.1016/0378-1119(88)90264-8. [DOI] [PubMed] [Google Scholar]
- Servos S., Silva C., Dougan G., Charles I. G. The commercially available restriction enzyme BspHI is blocked by overlapping methylation. Nucleic Acids Res. 1991 Jan 11;19(1):183–183. doi: 10.1093/nar/19.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields S. L., Burbank D. E., Grabherr R., van Etten J. L. Cloning and sequencing the cytosine methyltransferase gene M. CviJI from Chlorella virus IL-3A. Virology. 1990 May;176(1):16–24. doi: 10.1016/0042-6822(90)90225-g. [DOI] [PubMed] [Google Scholar]
- Silber K. R., Polisson C., Rees P. A., Benner J. S. Cloning, purification and characterization of the M.NdeI methyltransferase from Neisseria denitrificans. Gene. 1988 Dec 25;74(1):43–44. doi: 10.1016/0378-1119(88)90247-8. [DOI] [PubMed] [Google Scholar]
- Sladek T. L., Nowak J. A., Maniloff J. Mycoplasma restriction: identification of a new type of restriction specificity for DNA containing 5-methylcytosine. J Bacteriol. 1986 Jan;165(1):219–225. doi: 10.1128/jb.165.1.219-225.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko B. E., Croft R., Moran L. S., Wilson G. G. Cloning and analysis of the HaeIII and HaeII methyltransferase genes. Gene. 1988 Dec 25;74(1):45–50. doi: 10.1016/0378-1119(88)90248-x. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Annau T. M., Chandrasegaran S. Finding sequence motifs in groups of functionally related proteins. Proc Natl Acad Sci U S A. 1990 Jan;87(2):826–830. doi: 10.1073/pnas.87.2.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. D., Longo M., Gerard G. F., Chatterjee D. K. Cloning and characterization of genes for the PvuI restriction and modification system. Nucleic Acids Res. 1992 Nov 11;20(21):5743–5747. doi: 10.1093/nar/20.21.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Bhagwat A. S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987 Jan 12;15(1):313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stakenas P. S., Zaretskaia N. M., Manelene Z. P., Mauritsas M. M., Butkus V. V., Ianulaitis A. A. Mikoplazmennaia sistema restriktsii--modifikatsii MunI i ee vozmozhnaia rol' v protsessakh patogeneza. Mol Biol (Mosk) 1992 May-Jun;26(3):546–557. [PubMed] [Google Scholar]
- Stefan C., Xia Y. N., Van Etten J. L. Molecular cloning and characterization of the gene encoding the adenine methyltransferase M.CviRI from Chlorella virus XZ-6E. Nucleic Acids Res. 1991 Jan 25;19(2):307–311. doi: 10.1093/nar/19.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein D. C., Chien R., Seifert H. S. Construction of a Neisseria gonorrhoeae MS11 derivative deficient in NgoMI restriction and modification. J Bacteriol. 1992 Aug;174(15):4899–4906. doi: 10.1128/jb.174.15.4899-4906.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel H. M., Schmitz G. G., Kaluza K., Jarsch M., Kessler C. MamI, a novel class-II restriction endonuclease from Microbacterium ammoniaphilum recognizing 5'-GATNN decreases NNATC-3'. Gene. 1990 Jul 2;91(1):95–100. doi: 10.1016/0378-1119(90)90167-p. [DOI] [PubMed] [Google Scholar]
- Strobl J. S., Thompson E. B. Methylation of either cytosine in the recognition sequence CGCG inhibits ThaI cleavage of DNA. Nucleic Acids Res. 1984 Nov 12;12(21):8073–8083. doi: 10.1093/nar/12.21.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki H., Maekawa Y., Kanazawa S., Takanami M. New restriction endonucleases from Acetobacter aceti and Bacillus aneurinolyticus. Nucleic Acids Res. 1982 Oct 11;10(19):5747–5752. doi: 10.1093/nar/10.19.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. M., Saunders J. R. Sequence analysis of the NgoPII methyltransferase gene from Neisseria gonorrhoeae P9: homologies with other enzymes recognizing the sequence 5'-GGCC-3'. Nucleic Acids Res. 1988 May 25;16(10):4369–4387. doi: 10.1093/nar/16.10.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilák L., Venetianer P., Kiss A. Cloning and nucleotide sequence of the genes coding for the Sau96I restriction and modification enzymes. Nucleic Acids Res. 1990 Aug 25;18(16):4659–4664. doi: 10.1093/nar/18.16.4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznyter L. A., Slatko B., Moran L., O'Donnell K. H., Brooks J. E. Nucleotide sequence of the DdeI restriction-modification system and characterization of the methylase protein. Nucleic Acids Res. 1987 Oct 26;15(20):8249–8266. doi: 10.1093/nar/15.20.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao T., Walter J., Brennan K. J., Cotterman M. M., Blumenthal R. M. Sequence, internal homology and high-level expression of the gene for a DNA-(cytosine N4)-methyltransferase, M.Pvu II. Nucleic Acids Res. 1989 Jun 12;17(11):4161–4175. doi: 10.1093/nar/17.11.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasseron-de Jong J. G., Aker J., Giphart-Gassler M. The ability of the restriction endonuclease EcoRI to digest hemi-methylated versus fully cytosine-methylated DNA of the herpes tk promoter region. Gene. 1988 Dec 25;74(1):147–149. doi: 10.1016/0378-1119(88)90272-7. [DOI] [PubMed] [Google Scholar]
- Taylor J. D., Badcoe I. G., Clarke A. R., Halford S. E. EcoRV restriction endonuclease binds all DNA sequences with equal affinity. Biochemistry. 1991 Sep 10;30(36):8743–8753. doi: 10.1021/bi00100a005. [DOI] [PubMed] [Google Scholar]
- Theriault G., Roy P. H., Howard K. A., Benner J. S., Brooks J. E., Waters A. F., Gingeras T. R. Nucleotide sequence of the PaeR7 restriction/modification system and partial characterization of its protein products. Nucleic Acids Res. 1985 Dec 9;13(23):8441–8461. doi: 10.1093/nar/13.23.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault G., Roy P. H. Cloning of Pseudomonas plasmid pMG7 and its restriction-modification system in Escherichia coli. Gene. 1982 Oct;19(3):355–359. doi: 10.1016/0378-1119(82)90026-9. [DOI] [PubMed] [Google Scholar]
- Tran-Betcke A., Behrens B., Noyer-Weidner M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: comparison of their nucleotide sequences. Gene. 1986;42(1):89–96. doi: 10.1016/0378-1119(86)90153-8. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Balganesh T. S., Pawlek B. Chimeric multispecific DNA methyltransferases with novel combinations of target recognition. Nucleic Acids Res. 1988 Jul 25;16(14A):6649–6658. doi: 10.1093/nar/16.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Günthert U., Canosi U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol Gen Genet. 1980;180(2):361–367. doi: 10.1007/BF00425849. [DOI] [PubMed] [Google Scholar]
- Ueno T., Ito H., Kimizuka F., Kotani H., Nakajima K. Gene structure and expression of the MboI restriction--modification system. Nucleic Acids Res. 1993 May 25;21(10):2309–2313. doi: 10.1093/nar/21.10.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T., Ito H., Kotani H., Kimizuka F., Nakajima K. Cloning and expression of the NspV restriction-modification genes of Nostoc sp. strain PCC7524. Nucleic Acids Res. 1993 Aug 11;21(16):3899–3899. doi: 10.1093/nar/21.16.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Razin A. Sequence and substrate specificity of isolated DNA methylases from Escherichia coli C. J Bacteriol. 1983 Jan;153(1):274–280. doi: 10.1128/jb.153.1.274-280.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cott E. M., Wilson G. G. Cloning the FnuDI, NaeI, NcoI and XbaI restriction-modification systems. Gene. 1988 Dec 25;74(1):55–59. doi: 10.1016/0378-1119(88)90251-x. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Dobritsa A. P. On the nature of the cytosine-methylated sequence in DNA of Bacillus brevis var. G.-B. Biochim Biophys Acta. 1975 Sep 12;407(1):61–72. doi: 10.1016/0005-2787(75)90023-4. [DOI] [PubMed] [Google Scholar]
- Venetianer P., Kiss A. The restriction-modification enzymes of Bacillus sphaericus R. Gene Amplif Anal. 1981;1:209–215. [PubMed] [Google Scholar]
- Vinogradova M. N., Gromova E. S., Uporova T. M., Nikol'skaia I. I., Shabarova Z. A. Endonukleaza restriktsii SsoII: vzaimodeistvie s modifitsirovannymi substratami. Dokl Akad Nauk SSSR. 1987;295(3):732–736. [PubMed] [Google Scholar]
- Vovis G. F., Lacks S. Complementary action of restriction enzymes endo R-DpnI and Endo R-DpnII on bacteriophage f1 DNA. J Mol Biol. 1977 Sep 25;115(3):525–538. doi: 10.1016/0022-2836(77)90169-3. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Y., Hartley J. L., Donelson J. E., Walder J. A. Cloning and expression of the Pst I restriction-modification system in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1503–1507. doi: 10.1073/pnas.78.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Y., Hartley J. L., Donelson J. E., Walder J. A. Cloning of the Pst I restriction-modification system. Gene Amplif Anal. 1981;1:217–227. [PubMed] [Google Scholar]
- Walder R. Y., Langtimm C. J., Chatterjee R., Walder J. A. Cloning of the MspI modification enzyme. The site of modification and its effects on cleavage by MspI and HpaII. J Biol Chem. 1983 Jan 25;258(2):1235–1241. [PubMed] [Google Scholar]
- Walder R. Y., Walder J. A., Donelson J. E. The organization and complete nucleotide sequence of the PstI restriction-modification system. J Biol Chem. 1984 Jun 25;259(12):8015–8026. [PubMed] [Google Scholar]
- Walter J., Noyer-Weidner M., Trautner T. A. The amino acid sequence of the CCGG recognizing DNA methyltransferase M.BsuFI: implications for the analysis of sequence recognition by cytosine DNA methyltransferases. EMBO J. 1990 Apr;9(4):1007–1013. doi: 10.1002/j.1460-2075.1990.tb08203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. Y., Zhang X. Y., Khan R., Zhou Y. W., Huang L. H., Ehrlich M. Methylated DNA-binding protein from human placenta recognizes specific methylated sites on several prokaryotic DNAs. Nucleic Acids Res. 1986 Dec 22;14(24):9843–9860. doi: 10.1093/nar/14.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M. D., McClelland M. Enzymatic cleavage of a bacterial genome at a 10-base-pair recognition site. Proc Natl Acad Sci U S A. 1989 Jan;86(1):51–55. doi: 10.1073/pnas.86.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead P. R., Jacobs D., Brown N. L. Restriction endonucleases from Herpetosiphon giganteus: an example of the evolution of DNA recognition specificity? Nucleic Acids Res. 1986 Sep 11;14(17):7031–7045. doi: 10.1093/nar/14.17.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiatr C. L., Witmer H. J. Selective protection of 5' ... GGCC ... 3' and 5' ... GCNGC ... 3' sequences by the hypermodified oxopyrimidine in Bacillus subtilis bacteriophage SP10 DNA. J Virol. 1984 Oct;52(1):47–54. doi: 10.1128/jvi.52.1.47-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Levy D., Perucho M. The somatic replication of DNA methylation. Cell. 1981 Apr;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Wilke K., Rauhut E., Noyer-Weidner M., Lauster R., Pawlek B., Behrens B., Trautner T. A. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988 Aug;7(8):2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G. Cloned restriction-modification systems--a review. Gene. 1988 Dec 25;74(1):281–289. doi: 10.1016/0378-1119(88)90304-6. [DOI] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., D'Arcy A., Blöcker H., Frank R., van Boom J. H. Crystallization of complexes of EcoRV endonuclease with cognate and non-cognate DNA fragments. J Mol Biol. 1991 Jan 20;217(2):235–238. doi: 10.1016/0022-2836(91)90536-f. [DOI] [PubMed] [Google Scholar]
- Withers B. E., Ambroso L. A., Dunbar J. C. Structure and evolution of the XcyI restriction-modification system. Nucleic Acids Res. 1992 Dec 11;20(23):6267–6273. doi: 10.1093/nar/20.23.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. K., McClelland M. PCR with 5-methyl-dCTP replacing dCTP. Nucleic Acids Res. 1991 Mar 11;19(5):1081–1085. doi: 10.1093/nar/19.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock D. M., Crowther P. J., Diver W. P., Graham M., Bateman C., Baker D. J., Smith S. S. RglB facilitated cloning of highly methylated eukaryotic DNA: the human L1 transposon, plant DNA, and DNA methylated in vitro with human DNA methyltransferase. Nucleic Acids Res. 1988 May 25;16(10):4465–4482. doi: 10.1093/nar/16.10.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. High level expression and purification of HhaI methyltransferase. Nucleic Acids Res. 1988 Jan 25;16(2):703–717. doi: 10.1093/nar/16.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Burbank D. E., Uher L., Rabussay D., Van Etten J. L. IL-3A virus infection of a Chlorella-like green alga induces a DNA restriction endonuclease with novel sequence specificity. Nucleic Acids Res. 1987 Aug 11;15(15):6075–6090. doi: 10.1093/nar/15.15.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Burbank D. E., Uher L., Rabussay D., Van Etten J. L. Restriction endonuclease activity induced by PBCV-1 virus infection of a Chlorella-like green alga. Mol Cell Biol. 1986 May;6(5):1430–1439. doi: 10.1128/mcb.6.5.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Morgan R., Schildkraut I., Van Etten J. L. A site-specific single strand endonuclease activity induced by NYs-1 virus infection of a Chlorella-like green alga. Nucleic Acids Res. 1988 Oct 25;16(20):9477–9487. doi: 10.1093/nar/16.20.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Burbank D. E., Van Etten J. L. Restriction endonuclease activity induced by NC-1A virus infection of a Chlorella-like green alga. Nucleic Acids Res. 1986 Aug 11;14(15):6017–6030. doi: 10.1093/nar/14.15.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. L., Kapfer W., Walter J., Trautner T. A. BsuBI--an isospecific restriction and modification system of PstI: characterization of the BsuBI genes and enzymes. Nucleic Acids Res. 1992 Dec 25;20(24):6517–6523. doi: 10.1093/nar/20.24.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolov A. A., Vinogradova M. N., Gromova E. S., Rosenthal A., Cech D., Veiko V. P., Metelev V. G., Kosykh V. G., Buryanov Y. I., Bayev A. A. Interaction of EcoRII restriction and modification enzymes with synthetic DNA fragments. VI. The binding and cleavage of substrates containing nucleotide analogs. Nucleic Acids Res. 1985 Dec 20;13(24):8983–8998. doi: 10.1093/nar/13.24.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo O. J., Agarwal K. L. Isolation and characterization of two proteins possessing Hpa II methylase activity. J Biol Chem. 1980 Jul 10;255(13):6445–6449. [PubMed] [Google Scholar]
- Yoshimori R., Roulland-Dussoix D., Boyer H. W. R factor-controlled restriction and modification of deoxyribonucleic acid: restriction mutants. J Bacteriol. 1972 Dec;112(3):1275–1279. doi: 10.1128/jb.112.3.1275-1279.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Hammer S. M., Hirsch M. S., Mulder C. Methylation of the viral genome in an in vitro model of herpes simplex virus latency. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2207–2210. doi: 10.1073/pnas.79.7.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Kilpatrick M. W., Wells R. D. HhaI methylase and restriction endonuclease as probes for B to Z DNA conformational changes in d(GCGC) sequences. Nucleic Acids Res. 1984 Oct 25;12(20):7677–7692. doi: 10.1093/nar/12.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Tao T., Wilson G. G., Blumenthal R. M. The M.AluI DNA-(cytosine C5)-methyltransferase has an unusually large, partially dispensable, variable region. Nucleic Acids Res. 1993 Feb 25;21(4):905–911. doi: 10.1093/nar/21.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Nelson M., Nietfeldt J. W., Burbank D. E., Van Etten J. L. Characterization of Chlorella virus PBCV-1 CviAII restriction and modification system. Nucleic Acids Res. 1992 Oct 25;20(20):5351–5356. doi: 10.1093/nar/20.20.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieger M., Patillon M., Roizes G., Lerouge T., Dupret D., Jeltsch J. M. Two restriction endonucleases from Bacillus sphaericus: BspXI and BspXII. Nucleic Acids Res. 1987 May 11;15(9):3919–3919. doi: 10.1093/nar/15.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Campa A. G., Kale P., Springhorn S. S., Lacks S. A. Proteins encoded by the DpnII restriction gene cassette. Two methylases and an endonuclease. J Mol Biol. 1987 Aug 5;196(3):457–469. doi: 10.1016/0022-2836(87)90024-6. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]


