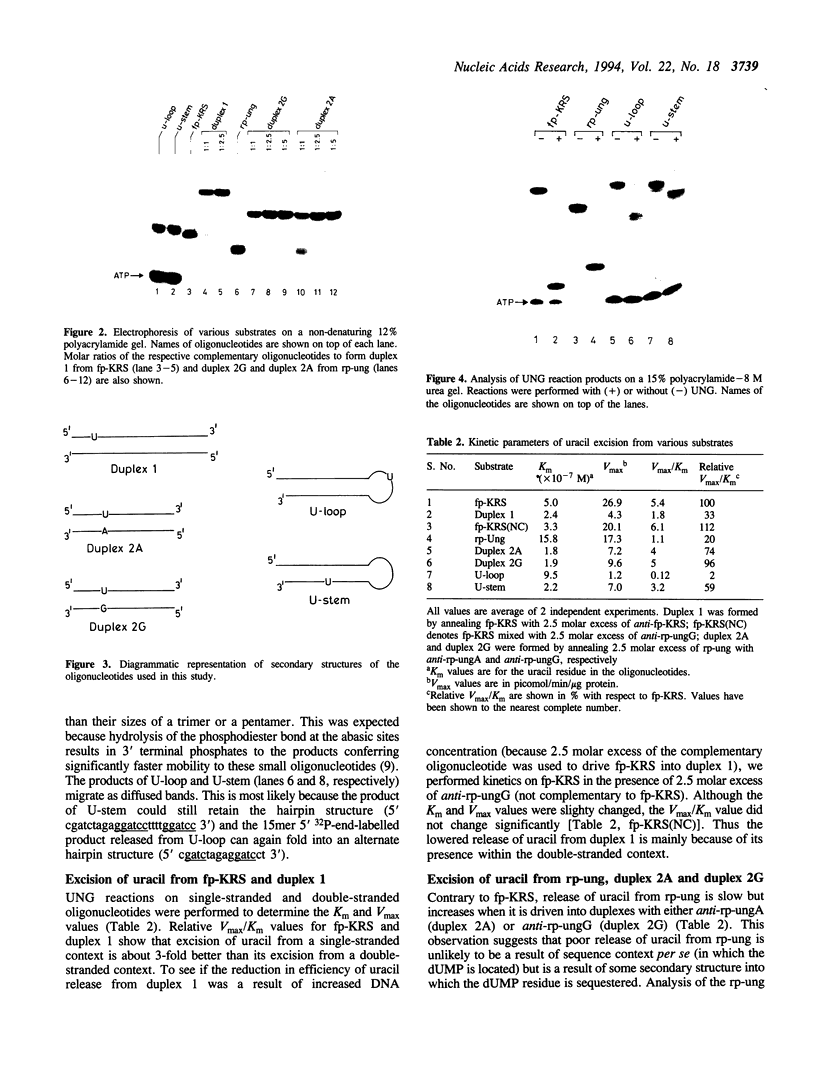
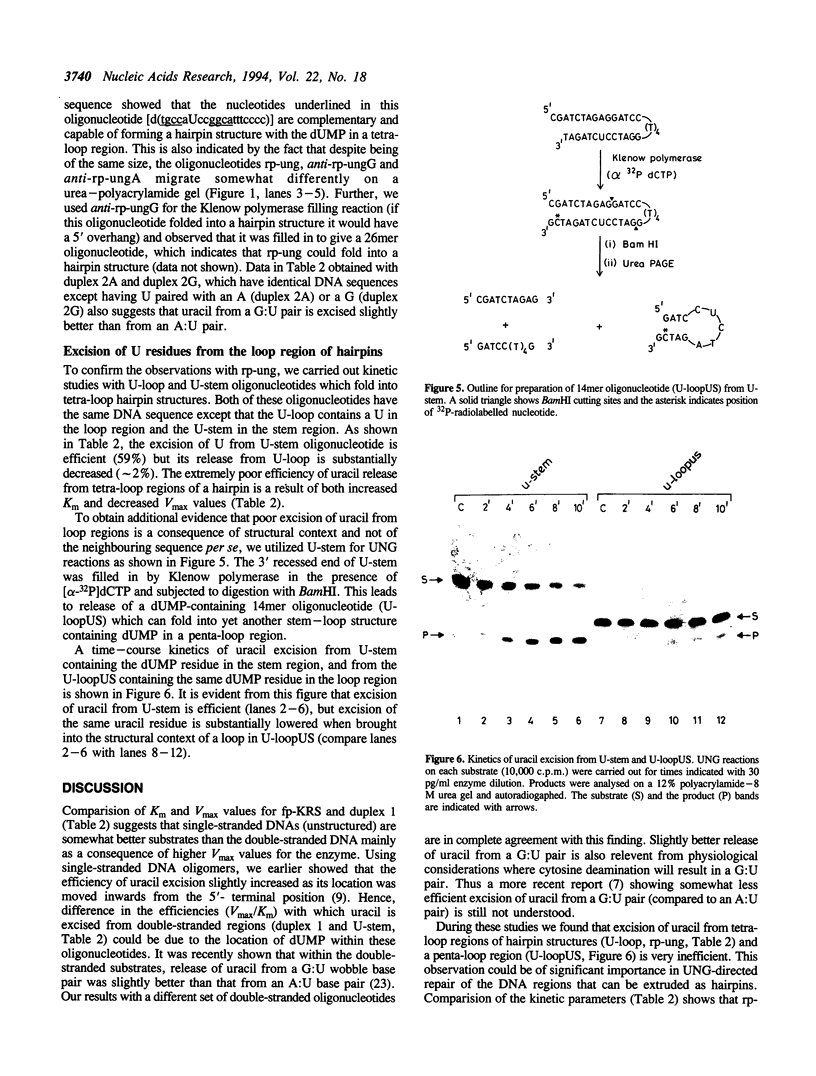
Abstract
Kinetic parameters for uracil DNA glycosylase (E. coli)-catalysed excision of uracil from DNA oligomers containing dUMP in different structural contexts were determined. Our results show that single-stranded oligonucleotides (unstructured) are used as somewhat better substrates than the double-stranded oligonucleotides. This is mainly because of the favourable Vmax value of the enzyme for single-stranded substrates. More interestingly, however, we found that uracil release from loop regions of DNA hairpins is extremely inefficient. The poor efficiency with which uracil is excised from loop regions is a result of both increased Km and lowered Vmax values. This observation may have significant implications in uracil DNA glycosylase-directed repair of DNA segments that can be extruded as hairpins. In addition, these studies are useful in designing oligonucleotides for various applications in DNA research where the use of uracil DNA glycosylase is sought.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball J. K., Desselberger U. The use of uracil-N-glycosylase in the preparation of PCR products for direct sequencing. Nucleic Acids Res. 1992 Jun 25;20(12):3255–3255. doi: 10.1093/nar/20.12.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Craig A. G., Nizetic D., Lehrach H. Labelling oligonucleotides to high specific activity (I). Nucleic Acids Res. 1989 Jun 26;17(12):4605–4610. doi: 10.1093/nar/17.12.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOWD J. E., RIGGS D. S. A COMPARISON OF ESTIMATES OF MICHAELIS-MENTEN KINETIC CONSTANTS FROM VARIOUS LINEAR TRANSFORMATIONS. J Biol Chem. 1965 Feb;240:863–869. [PubMed] [Google Scholar]
- Delort A. M., Duplaa A. M., Molko D., Teoule R., Leblanc J. P., Laval J. Excision of uracil residues in DNA: mechanism of action of Escherichia coli and Micrococcus luteus uracil-DNA glycosylases. Nucleic Acids Res. 1985 Jan 25;13(2):319–335. doi: 10.1093/nar/13.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devchand P. R., McGhee J. D., van de Sande J. H. Uracil-DNA glycosylase as a probe for protein--DNA interactions. Nucleic Acids Res. 1993 Jul 25;21(15):3437–3443. doi: 10.1093/nar/21.15.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftedal I., Guddal P. H., Slupphaug G., Volden G., Krokan H. E. Consensus sequences for good and poor removal of uracil from double stranded DNA by uracil-DNA glycosylase. Nucleic Acids Res. 1993 May 11;21(9):2095–2101. doi: 10.1093/nar/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann M. W., Pon R. T., van de Sande J. H. A general method for the purification of synthetic oligodeoxyribonucleotides containing strong secondary structure by reversed-phase high-performance liquid chromatography on PRP-1 resin. Anal Biochem. 1987 Sep;165(2):399–405. doi: 10.1016/0003-2697(87)90288-0. [DOI] [PubMed] [Google Scholar]
- Hirao I., Kawai G., Yoshizawa S., Nishimura Y., Ishido Y., Watanabe K., Miura K. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 1994 Feb 25;22(4):576–582. doi: 10.1093/nar/22.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan H., Wittwer C. U. Uracil DNa-glycosylase from HeLa cells: general properties, substrate specificity and effect of uracil analogs. Nucleic Acids Res. 1981 Jun 11;9(11):2599–2613. doi: 10.1093/nar/9.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc J. P., Laval J. Comparison at the molecular level of uracil-DNA glycosylases from different origins. Biochimie. 1982 Aug-Sep;64(8-9):735–738. doi: 10.1016/s0300-9084(82)80120-x. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem. 1977 May 25;252(10):3286–3294. [PubMed] [Google Scholar]
- Longo M. C., Berninger M. S., Hartley J. L. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990 Sep 1;93(1):125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. Uracil interference, a rapid and general method for defining protein-DNA interactions involving the 5-methyl group of thymines: the GCN4-DNA complex. Nucleic Acids Res. 1992 Feb 25;20(4):771–775. doi: 10.1093/nar/20.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashtchian A., Buchman G. W., Schuster D. M., Berninger M. S. Uracil DNA glycosylase-mediated cloning of polymerase chain reaction-amplified DNA: application to genomic and cDNA cloning. Anal Biochem. 1992 Oct;206(1):91–97. doi: 10.1016/s0003-2697(05)80015-6. [DOI] [PubMed] [Google Scholar]
- Sakumi K., Sekiguchi M. Structures and functions of DNA glycosylases. Mutat Res. 1990 Sep-Nov;236(2-3):161–172. doi: 10.1016/0921-8777(90)90003-n. [DOI] [PubMed] [Google Scholar]
- Smith C., Day P. J., Walker M. R. Generation of cohesive ends on PCR products by UDG-mediated excision of dU, and application for cloning into restriction digest-linearized vectors. PCR Methods Appl. 1993 May;2(4):328–332. doi: 10.1101/gr.2.4.328. [DOI] [PubMed] [Google Scholar]
- Talpaert-Borlé M., Campagnari F., Creissen D. M. Properties of purified uracil-DNA glycosylase from calf thymus. An in vitro study using synthetic DNA-like substrates. J Biol Chem. 1982 Feb 10;257(3):1208–1214. [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Varshney U., van de Sande J. H. Specificities and kinetics of uracil excision from uracil-containing DNA oligomers by Escherichia coli uracil DNA glycosylase. Biochemistry. 1991 Apr 23;30(16):4055–4061. doi: 10.1021/bi00230a033. [DOI] [PubMed] [Google Scholar]
- Verri A., Mazzarello P., Spadari S., Focher F. Uracil-DNA glycosylases preferentially excise mispaired uracil. Biochem J. 1992 Nov 1;287(Pt 3):1007–1010. doi: 10.1042/bj2871007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Vogel H. J. Two-dimensional NMR and restrained molecular dynamics studies of the hairpin d(T8C4A8): detection of an extraloop cytosine. Biochemistry. 1993 Jan 19;32(2):637–645. doi: 10.1021/bi00053a032. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., Ramsing N. B., Germann M. W., Elhorst W., Kalisch B. W., von Kitzing E., Pon R. T., Clegg R. C., Jovin T. M. Parallel stranded DNA. Science. 1988 Jul 29;241(4865):551–557. doi: 10.1126/science.3399890. [DOI] [PubMed] [Google Scholar]