Abstract
We have studied the role of extended protein DNA contacts and DNA topology on the ability of Escherichia coli RNA polymerase to form open complexes at several related promoters. The -35 region of several Escherichia coli promoters do not have homology with the consensus sequence, but still drive activator independent transcription initiation. This is due to the presence of a TG motif upstream from the -10 hexamer creating an 'extended -10' promoter. We have previously shown that two 'extended -10' promoters, galP1 and pBla, can form open complexes at lower temperatures than the galP1 derivative, galPcon6, which has a consensus -35 hexamer. Here we report further investigations into the mechanism of open complex formation by RNA polymerase, in particular the thermal energy requirement. A single base pair change in galPcon6 creating an 'extended -10' sequence, results in a 20 degrees C reduction in the temperature requirement for open complex formation. The DNA topology has also been shown to effect the thermal energy requirement for strand separation. Promoters carried on supercoiled plasmids form open complexes at lower temperatures than when present on linear DNA templates. We have also shown that in vivo, RNA polymerase can form open complexes at lower temperatures than those observed for linear templates in vitro, but requires slightly higher temperatures than supercoiled templates in vitro, however the promoter hierachy remains the same.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belyaeva T., Griffiths L., Minchin S., Cole J., Busby S. The Escherichia coli cysG promoter belongs to the 'extended -10' class of bacterial promoters. Biochem J. 1993 Dec 15;296(Pt 3):851–857. doi: 10.1042/bj2960851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Gralla J. D. All three elements of the lac ps promoter mediate its transcriptional response to DNA supercoiling. J Mol Biol. 1987 May 5;195(1):89–97. doi: 10.1016/0022-2836(87)90329-9. [DOI] [PubMed] [Google Scholar]
- Busby S., Spassky A., Chan B. RNA polymerase makes important contacts upstream from base pair -49 at the Escherichia coli galactose operon P1 promoter. Gene. 1987;53(2-3):145–152. doi: 10.1016/0378-1119(87)90002-3. [DOI] [PubMed] [Google Scholar]
- Chan B., Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989 Dec 14;84(2):227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- Chan B., Minchin S., Busby S. Unwinding of duplex DNA during transcription initiation at the Escherichia coli galactose operon overlapping promoters. FEBS Lett. 1990 Jul 2;267(1):46–50. doi: 10.1016/0014-5793(90)80284-p. [DOI] [PubMed] [Google Scholar]
- Chan B., Spassky A., Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus -35 region sequences. Biochem J. 1990 Aug 15;270(1):141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein E., Drlica K. Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4046–4050. doi: 10.1073/pnas.81.13.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
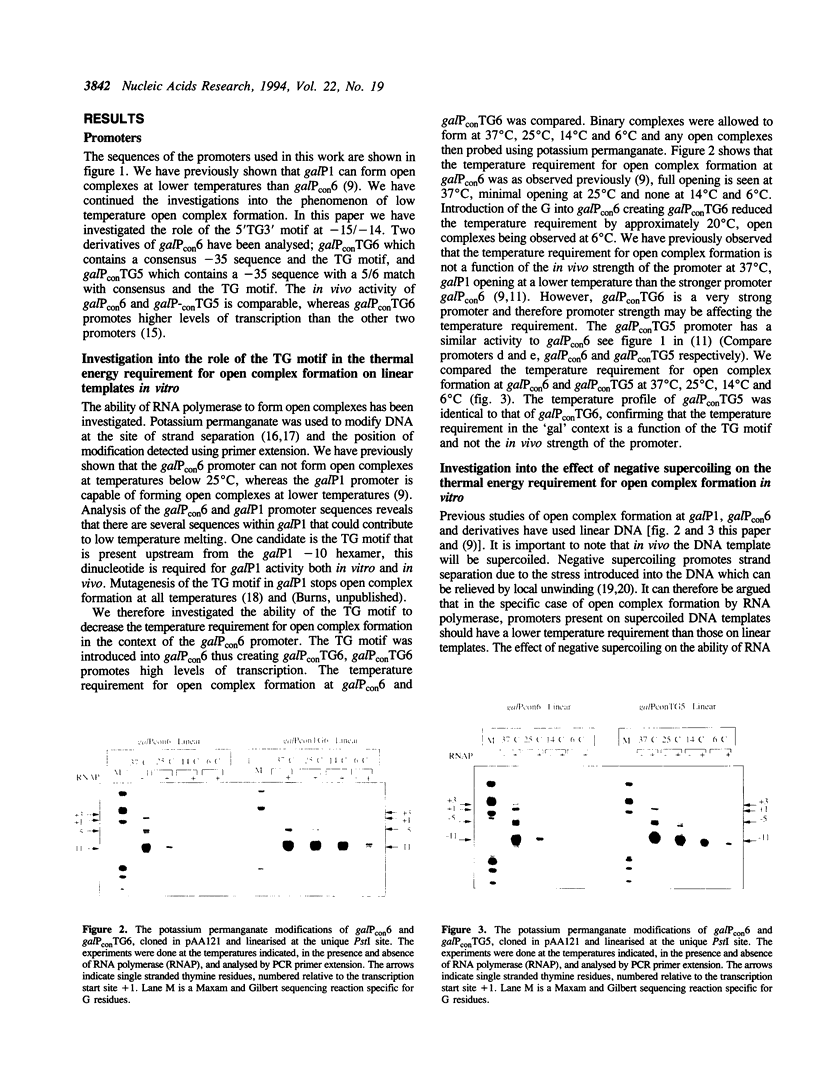
- Grimes E., Busby S., Minchin S. Different thermal energy requirement for open complex formation by Escherichia coli RNA polymerase at two related promoters. Nucleic Acids Res. 1991 Nov 25;19(22):6113–6118. doi: 10.1093/nar/19.22.6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987 May 5;262(13):6389–6395. [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Minchin S., Busby S. Location of close contacts between Escherichia coli RNA polymerase and guanine residues at promoters either with or without consensus -35 region sequences. Biochem J. 1993 Feb 1;289(Pt 3):771–775. doi: 10.1042/bj2890771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnambalam S., Webster C., Bingham A., Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal -35 region sequences. J Biol Chem. 1986 Dec 5;261(34):16043–16048. [PubMed] [Google Scholar]
- Roe J. H., Burgess R. R., Record M. T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985 Aug 5;184(3):441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993 Nov 26;262(5138):1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Footprinting protein-DNA complexes in vivo. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Suh W. C., Ross W., Record M. T., Jr Two open complexes and a requirement for Mg2+ to open the lambda PR transcription start site. Science. 1993 Jan 15;259(5093):358–361. doi: 10.1126/science.8420002. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Variation of the average rotation angle of the DNA helix and the superhelical turns of covalently closed cyclic lambda DNA. J Mol Biol. 1969 Jul 14;43(1):25–39. doi: 10.1016/0022-2836(69)90076-x. [DOI] [PubMed] [Google Scholar]
- Wood D. C., Lebowitz J. Effect of supercoiling on the abortive initiation kinetics of the RNA-I promoter of ColE1 plasmid DNA. J Biol Chem. 1984 Sep 25;259(18):11184–11187. [PubMed] [Google Scholar]