Abstract
Purpose
To determine the frequency of TET2 mutations, their associations with clinical and molecular characteristics and outcome, and the associated gene- and microRNA-expression signatures in patients with primary cytogenetically normal acute myeloid leukemia (CN-AML).
Patients and Methods
Four-hundred twenty-seven patients with CN-AML were analyzed for TET2 mutations by polymerase chain reaction and direct sequencing and for established prognostic gene mutations. Gene- and microRNA-expression profiles were derived using microarrays.
Results
TET2 mutations, found in 23% of patients, were associated with older age (P < .001) and higher pretreatment WBC (P = .04) compared with wild-type TET2 (TET2-wt). In the European LeukemiaNet (ELN) favorable-risk group (patients with CN-AML who have mutated CEBPA and/or mutated NPM1 without FLT3 internal tandem duplication [FLT3-ITD]), TET2-mutated patients had shorter event-free survival (EFS; P < .001) because of a lower complete remission (CR) rate (P = .007), and shorter disease-free survival (DFS; P = .003), and also had shorter overall survival (P = .001) compared with TET2-wt patients. TET2 mutations were not associated with outcomes in the ELN intermediate-I–risk group (CN-AML with wild-type CEBPA and wild-type NPM1 and/or FLT3-ITD). In multivariable models, TET2 mutations were associated with shorter EFS (P = .004), lower CR rate (P = .03), and shorter DFS (P = .05) only among favorable-risk CN-AML patients. We identified a TET2 mutation-associated gene-expression signature in favorable-risk but not in intermediate-I–risk patients and found distinct mutation-associated microRNA signatures in both ELN groups.
Conclusion
TET2 mutations improve the ELN molecular-risk classification in primary CN-AML because of their adverse prognostic impact in an otherwise favorable-risk patient subset. Our data suggest that these patients may be candidates for alternative therapies.
INTRODUCTION
The tet oncogene family member 2 (TET2) gene is located at chromosome band 4q24. Mutations in TET2 were initially identified in myeloid neoplasms with a deletion or uniparental disomy of this chromosomal region,1,2 and subsequently described in patients with myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPN), chronic myelomonocytic leukemia, and acute myeloid leukemia (AML).3–7 TET2 mutations are frequently acquired during progression of MPN or MDS to secondary AML8,9 and have been associated with shorter overall survival (OS) in patients with chronic myelomonocytic leukemia or AML.3,10 However, in other reports, TET2 mutations were either not significantly correlated with survival in patients with AML11 or were associated with a decreased risk of progression of MDS to AML and longer OS.12 The prognostic relevance of TET2 mutations in myeloid neoplasia thus remains controversial.
Only one recent study of TET2 mutations11 has focused specifically on primary (de novo) AML. Cytogenetically normal AML (CN-AML) represents the largest subgroup of adult primary AML,13 and within this molecularly heterogeneous group, gene mutations are increasingly used to assess prognosis and guide risk-adapted treatment.14,15 To integrate the prognostic information conveyed by cytogenetics and gene mutations and apply this information to the clinical management of patients with AML, an international expert panel on behalf of the European LeukemiaNet (ELN) recently recommended a classification scheme for adult AML based on karyotype, FLT3-internal tandem duplications (FLT3-ITD), and NPM1 and CEBPA mutations.16 According to the ELN classification, patients with CN-AML are assigned to either the favorable-risk category (CEBPA-mutated and/or mutated NPM1 without FLT3-ITD) or to the intermediate-I–risk category (all remaining patients). Mutations in TET2 and other genes were recognized as novel genetic abnormalities in the ELN report,16 and such markers may potentially be useful to refine the existing ELN categories.
We investigated the prevalence of TET2 mutations in patients with primary CN-AML, their associations with clinical and molecular characteristics, and their impact on the prognosis of the CN-AML subgroups defined by the ELN. Furthermore, to gain insights into the role of TET2 mutations in the pathobiology of AML, we derived genome-wide TET2 mutation-associated gene- and microRNA-expression signatures.
PATIENTS AND METHODS
Patients, Treatment, and Cytogenetic Studies
Pretreatment bone marrow (BM) or blood samples were obtained from 427 patients with primary CN-AML, age 18 to 83 years, who received intensive cytarabine/daunorubicin-based first-line therapy on Cancer and Leukemia Group B (CALGB) trials. For details regarding treatment protocols and sample collection, see the Data Supplement. The diagnosis of normal cytogenetics was based on the analysis of ≥ 20 metaphases in BM specimens and confirmed by central karyotype review.17 All patients provided written informed consent, and all study protocols were in accordance with the Declaration of Helsinki and approved by institutional review boards at each center.
Mutational Analyses
All coding exons of the longest known TET2 transcript were amplified from genomic DNA by polymerase chain reaction and were analyzed by direct sequencing. Synonymous sequence changes and known single nucleotide polymorphisms were not considered (Data Supplement).1,18 If available, matched buccal swab specimens or BM aspirates obtained during complete remission (CR) were analyzed to assess whether observed sequence alterations represented somatic mutations or germline variants. Patients were also characterized for FLT3-ITD19 and FLT3 tyrosine kinase domain mutations,20 MLL partial tandem duplications,21,22 and mutations in NPM1,23,24 WT1,25 CEBPA,26 and IDH1/IDH2,27 as previously reported.
Microarray Experiments
Gene-expression profiling was performed using Affymetrix oligonucleotide microarrays (Affymetrix, Santa Clara, CA), and microRNA-expression profiling was performed by using a custom microarray, as previously reported.24 Differentially expressed probe sets or probes were identified by comparing TET2-mutated and TET2 wild-type (TET2-wt) patients, using univariable significance levels of < .001 for gene-expression and < .005 for microRNA-expression profiles (Data Supplement).
Statistical Analyses
Baseline characteristics were compared between TET2-mutated and TET2-wt patients by using Fisher's exact test for categorical variables and the Wilcoxon rank sum test for continuous variables. Clinical end points were defined according to published recommendations (Data Supplement).28 For time-to-event analyses, survival estimates were calculated by using the Kaplan-Meier method, and groups in which at least 10 events had occurred were compared with the log-rank test. We constructed multivariable logistic regression models to analyze factors for the achievement of CR and multivariable Cox proportional hazards models for factors associated with survival end points. For details regarding statistical analyses, see the Data Supplement. All analyses were performed by the CALGB statistical center.
RESULTS
Prevalence and Spectrum of TET2 Mutations
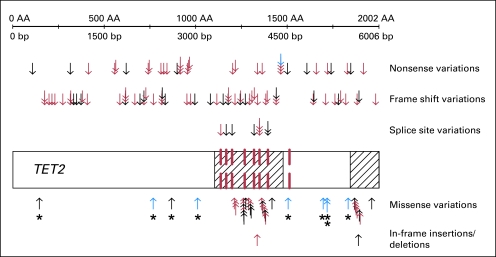
We identified variations in the TET2 coding sequence in 104 of 427 patients (Table 1). Mutations predicted to result in a truncated protein (nonsense and frame shift) occurred most frequently and were distributed throughout all coding exons. In contrast, missense changes mainly clustered in two evolutionarily conserved domains of the gene (Fig 1).1 Analyses of buccal swab or remission BM samples showed that all evaluable missense changes outside these conserved domains (n = 7) were present in the germline (Data Supplement). Therefore, as done in previous studies,1,10,12 we excluded all nine patients with missense changes outside the two conserved domains, leaving 418 patients for subsequent analyses. Thus, the prevalence of TET2 mutations in our cohort was 23% (95 of 418).
Table 1.
Overview of TET2 Sequence Variations Detected in 427 Patients With Primary Cytogenetically Normal Acute Myeloid Leukemia
| Sequence Variations | No. | % |
|---|---|---|
| Patients with wild-type TET2 | 323 | 100 |
| Patients with TET2 sequence variation | 104 | 32 |
| Single heterozygous variation | 62 | 19 |
| Two sequence variations | 35 | 10 |
| Three sequence variations | 1 | 0.3 |
| Homozygous/hemizygous variations* | 6 | 2 |
| Total No. of sequence variations | 141 | 100 |
| Missense variation | 37 | 26 |
| Within TET2 conserved domain† | 28 | 20 |
| Outside TET2 conserved domain†‡ | 9 | 6 |
| In-frame insertion/deletion | 2 | 1 |
| Nonsense variation | 34 | 24 |
| Insertion/deletion causing frame shift | 59 | 42 |
| Variation affecting splice site | 9 | 6 |
NOTE. Single-nucleotide polymorphisms occurring in healthy individuals and synonymous variations were not included in the analysis.
These six patients had sequence variations that appeared homozygous on the sequencing chromatograms, consistent with loss of heterozygosity (ie, either the remaining wild-type allele was lost or both TET2 alleles were affected by the same mutation).
The evolutionarily conserved domains within the TET2 sequence, as defined by Langemeijer et al,1 comprise codons 1104 to 1478 and 1845 to 2002.
For seven of the nine patients with missense variations outside the TET2-conserved domains, germline DNA was available for testing, and in all seven patients, the mutation was found in the germline sample. Therefore, all nine patients with such changes were excluded from analyses of clinical and molecular associations and outcomes.
Fig 1.
Localization of sequence variations in relation to the TET2 coding sequence. The cross-hatched areas mark the two evolutionarily conserved domains of TET2 (amino acids 1104 to 1478 and 1845 to 2002), and exon boundaries are shown as dashed red lines.1 Each arrow represents one of the 141 nonsynonymous sequence variations in TET2 found among 427 patients (except for known single-nucleotide polymorphisms, which were not considered). Nonsense and frame shift variations as well as variations affecting splice sites are shown in the upper part of the figure. Missense variations and in-frame insertions/deletions, which alter only one or two amino acids, are shown in the lower part. Variations that were absent in matched germline DNA and thus were proven to be somatically acquired mutations are shown by red arrows. Blue arrows represent changes that were also found in the corresponding germline sample, and black arrows represent sequence changes where corresponding germline DNA was not available. Nine of the missense changes (highlighted by asterisks) were located outside the conserved domains of TET2, and seven of them were present in the germline (two could not be tested). The nine patients harboring these changes were excluded from further analyses.
Association of TET2 Mutations With Pretreatment Demographic, Clinical, and Molecular Characteristics
TET2-mutated CN-AML patients were significantly older than TET2-wt patients (P< .001; Table 2). The prevalence of TET2 mutations gradually increased with age, from 7% in adults younger than 30 years of age to 32% in patients age 70 years or older (P for trend < .001; Data Supplement). TET2-mutated patients presented with a higher WBC (P = .04) and were more likely to be female (P = .06) than TET2-wt patients.
Table 2.
Clinical and Molecular Characteristics According to TET2Mutation Status in 418 Patients With Primary Cytogenetically Normal Acute Myeloid Leukemia
| Characteristic |
TET2Mutated(n = 95) |
TET2Wild Type(n = 323) |
P | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Age, years | < .001 | ||||
| Median | 66 | 60 | |||
| Range | 20-80 | 18-83 | |||
| Age group, years | < .001 | ||||
| < 60 | 27 | 28 | 156 | 48 | |
| ≥ 60 | 68 | 72 | 167 | 52 | |
| Female sex | 56 | 59 | 154 | 48 | .06 |
| Race/ethnicity | 1.0 | ||||
| White | 85 | 91 | 290 | 91 | |
| Nonwhite | 8 | 9 | 30 | 9 | |
| Hemoglobin, g/dL | .59 | ||||
| Median | 9.3 | 9.5 | |||
| Range | 6.5-15.0 | 4.8-13.6 | |||
| Platelet count, ×109/L | .56 | ||||
| Median | 70 | 64 | |||
| Range | 8-510 | 4-850 | |||
| WBC count, ×109/L | .04 | ||||
| Median | 33.5 | 24.5 | |||
| Range | 1.6-450.0 | 0.9-434.1 | |||
| Percentage of blood blasts | .96 | ||||
| Median | 52 | 54 | |||
| Range | 0-97 | 0-99 | |||
| Percentage of bone marrow blasts | .86 | ||||
| Median | 67 | 67 | |||
| Range | 7-97 | 7-99 | |||
| FAB category | — | ||||
| M0 | 0 | 0 | 8 | 4 | |
| M1 | 14 | 21 | 59 | 26 | |
| M2 | 22 | 32 | 64 | 29 | |
| M4 | 20 | 29 | 54 | 24 | |
| M5 | 12 | 18 | 32 | 14 | |
| M6 | 0 | 0 | 6 | 3 | |
| Extramedullary involvement | 26 | 27 | 81 | 26 | .79 |
| IDH1 | < .001 | ||||
| Mutated | 2 | 2 | 48 | 15 | |
| Wild type | 91 | 98 | 274 | 85 | |
| IDH2 | < .001 | ||||
| Mutated | 2 | 2 | 74 | 23 | |
| Codon R140 mutation | 2 | 60 | |||
| Codon R172 mutation | 0 | 14 | |||
| Wild type | 91 | 98 | 248 | 77 | |
| CEBPA | .07 | ||||
| Mutated | 20 | 21 | 42 | 13 | |
| Wild type | 74 | 79 | 278 | 87 | |
| NPM1 | .34 | ||||
| Mutated | 62 | 65 | 191 | 59 | |
| Wild type | 33 | 35 | 132 | 41 | |
| FLT3-ITD | .54 | ||||
| Present | 36 | 38 | 110 | 34 | |
| Absent | 59 | 62 | 213 | 66 | |
| ELN risk group* | .08 | ||||
| Favorable-risk | 53 | 56 | 146 | 45 | |
| Intermediate-I-risk | 42 | 44 | 177 | 55 | |
| FLT3-TKD | 1.0 | ||||
| Present | 8 | 8 | 30 | 9 | |
| Absent | 87 | 92 | 290 | 91 | |
| WT1 | .16 | ||||
| Mutated | 5 | 5 | 34 | 11 | |
| Wild type | 90 | 95 | 289 | 89 | |
| MLL-PTD | .59 | ||||
| Present | 6 | 8 | 17 | 6 | |
| Absent | 69 | 92 | 273 | 94 | |
Abbreviations: FAB, French-American-British classification; FLT3-ITD, internal tandem duplication of the FLT3 gene; ELN, European LeukemiaNet; FLT3-TKD,tyrosine kinase domain mutation in the FLT3gene; MLL-PTD, partial tandem duplication of the MLL gene.
The ELN favorable-risk group includes patients with cytogenetically normal acute myeloid leukemia with mutated CEBPA and/or mutated NPM1without FLT3-ITD. All remaining patients with cytogenetically normal acute myeloid leukemia (ie, those with wild-type CEBPA and FLT3-ITD and/or wild-type NPM1) belong to the ELN intermediate-I–risk category.
Regarding other molecular markers, IDH1 and IDH2 mutations were less frequent in TET2-mutated than in TET2-wt patients (P < .001), with mutations at IDH2 codon R172 being mutually exclusive with TET2 mutations. We also observed a trend toward a higher prevalence of CEBPA mutations among TET2-mutated patients (P = .07; Table 2).
Impact of TET2 Mutation Status on Treatment Outcomes in All Patients With CN-AML
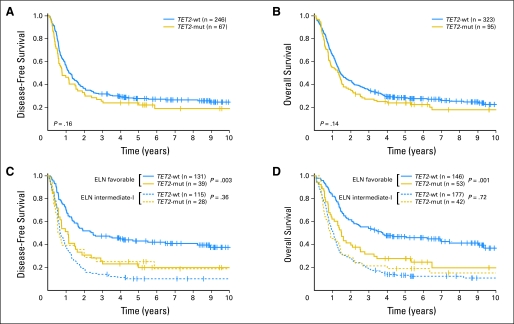
The median follow-up for patients still alive was 6.8 years (range, 2.3 to 11.7 years). Overall, TET2-mutated patients compared with TET2-wt patients showed a trend toward shorter event-free survival (EFS; P = .07; Table 3) but no significant differences in CR rate (P = .28), disease-free survival (DFS; P = .16; Fig 2A), or OS (P = .14; Fig 2B). No differences in outcomes were observed between patients with single or double TET2 mutations nor between different types of mutations (data not shown).
Table 3.
Treatment Outcomes According to TET2Mutation Status in 418 Patients With Primary Cytogenetically Normal Acute Myeloid Leukemia
| End Point |
TET2Mutated |
TET2Wild Type |
P | ||||
|---|---|---|---|---|---|---|---|
| No. | % | 95% CI | No. | % | 95% CI | ||
| All patients (N = 418) | 95 | 323 | |||||
| Event-free survival | .07 | ||||||
| Median, years | 0.6 | 0.8 | |||||
| Event-free at 3 years | 19 | 12% to 27% | 24 | 20% to 29% | |||
| Complete remission | 67 | 71 | 246 | 76 | .28 | ||
| Disease-free survival | .16 | ||||||
| Median, years | 0.8 | 1.2 | |||||
| Disease-free at 3 years | 25 | 16% to 36% | 32 | 26% to 38% | |||
| Overall survival | .14 | ||||||
| Median, years | 1.3 | 1.4 | |||||
| Alive at 3 years | 27 | 19% to 36% | 35 | 30% to 40% | |||
| ELN favorable-risk group* (n = 199) | 53 | 146 | |||||
| Event-free survival | < .001 | ||||||
| Median, years | 0.7 | 1.8 | |||||
| Event-free at 3 years | 21 | 11% to 32% | 42 | 34% to 50% | |||
| Complete remission | 39 | 74 | 131 | 90 | .007 | ||
| Disease-free survival | .003 | ||||||
| Median, years | 1.1 | 2.5 | |||||
| Disease-free at 3 years | 26 | 13% to 40% | 47 | 39% to 56% | |||
| Overall survival | .001 | ||||||
| Median, years | 1.5 | 3.8 | |||||
| Alive at 3 years | 32 | 20% to 44% | 54 | 46% to 62% | |||
| ELN intermediate-I–risk group* (n = 219) | 42 | 177 | |||||
| Event-free survival | .45 | ||||||
| Median, years | 0.5 | 0.6 | |||||
| Event-free at 3 years | 17 | 7% to 29% | 9 | 5% to 14% | |||
| Complete remission | 28 | 67 | 115 | 65 | 1.0 | ||
| Disease-free survival | .36 | ||||||
| Median, years | 0.6 | 0.7 | |||||
| Disease-free at 3 years | 25 | 11% to 42% | 14 | 8% to 21% | |||
| Overall survival | .72 | ||||||
| Median, years | 0.9 | 1.1 | |||||
| Alive at 3 years | 21 | 11% to 35% | 19 | 14% to 25% | |||
Abbreviation: ELN, European LeukemiaNet.
The ELN favorable-risk group is defined as patients with mutated CEBPA and/or mutated NPM1without internal tandem duplication of the FLT3 gene (FLT3-ITD). All remaining patients with cytogenetically normal acute myeloid leukemia (ie, those with wild-type CEBPA and FLT3-ITD and/or wild-type NPM1) belong to the ELN intermediate-I–risk category.
Fig 2.
(A) Disease-free survival and (B) overall survival of all patients with cytogenetically normal acute myeloid leukemia according to TET2 mutation status. (C) Disease-free survival and (D) overall survival of patients in the European LeukemiaNet (ELN) favorable-risk and intermediate-I–risk groups, according to TET2 mutation status. TET2-mut, mutated TET2; TET2-wt, wild-type TET2.
TET2 Mutations Are Associated With Inferior Outcomes in the ELN Favorable-Risk Group but Not in the Intermediate-I–Risk Group
The ELN guidelines introduced a standardized reporting system for genetic abnormalities, classifying patients with CN-AML into favorable-risk or intermediate-I–risk categories on the basis of specific genetic characteristics.16 Following the ELN recommendations, we investigated the prognostic impact of TET2 mutations within these categories. Among 418 patients, 199 (48%) were in the favorable-risk category (Data Supplement), and 219 (52%) were in the intermediate-I–risk category. Favorable-risk CN-AML patients had superior EFS (P < .001), a higher CR rate (P < .001), and longer DFS (P < .001) and OS (P < .001) than intermediate-I–risk patients (Data Supplement). TET2 mutations tended to be more frequent in favorable-risk than in intermediate-I–risk patients (27% v 19%; P = .08). However, types and locations of TET2 mutations were similar in both groups (data not shown).
Among favorable-risk CN-AML patients, those with TET2 mutations had shorter EFS than those with TET2-wt (P < .001; Table 3). This difference was due to a lower CR rate (P = .007) and shorter DFS (P = .003; Fig 2C) of TET2-mutated patients who also had shorter OS (P = .001; Fig 2D).
In contrast, in the intermediate-I–risk group, no significant difference in EFS (P = .45), CR rates (P = 1.0), DFS (P = .36), or OS (P = .72) was found between TET2-mutated and TET2-wt patients (Table 3). Therefore, since the adverse impact of TET2 mutations appeared to be limited to the favorable-risk subgroup, we formally tested for an interaction between TET2 mutation status and ELN risk category with regard to clinical end points. Indeed, we found statistically significant interactions between TET2 mutation status and ELN risk category for all four outcome end points (EFS, P = .001; CR rate, P = .03; DFS, P = .007; OS, P = .01), confirming a differential effect of TET2 mutations on outcome in the two ELN risk groups for CN-AML.
Multivariable Analyses
To assess whether TET2 mutations provide additional prognostic value in the context of the ELN classification and other clinical and molecular prognosticators, we constructed multivariable models including all 418 patients with CN-AML and used an interaction term to account for the differential effect of TET2 mutations in the favorable-risk and intermediate-I–risk subgroups (Table 4). In the model for EFS, TET2 mutations conferred a 71% increased risk of an event among favorable-risk patients (P = .004) after adjusting for WT1 mutation status (P = .006), WBC (P < .001), platelet count (P = .01), and age group (P < .001). TET2-mutated patients in the favorable-risk group had 38% lower odds of achieving CR (P = .03) after adjusting for WBC (P = .001), platelet count (P = .02), and age group (P = .009). In the model for DFS, favorable-risk patients with TET2 mutations had a 54% higher risk of relapse or death than TET2-wt patients (P = .05) after adjusting for WT1 mutation status (P = .03), WBC (P = .003), and age group (P < .001). In these multivariable models, among intermediate-I–risk patients, TET2 mutations were not significantly associated with EFS (P = .14), CR rate (P = .18), or DFS (P = .13).
Table 4.
Multivariable Models for the Associations Between Patient Characteristics and Treatment Outcomes
| Variable | Event-Free Survival |
Complete Remission |
Disease-Free Survival |
||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | OR | 95% CI | P | HR | 95% CI | P | |
| Interaction of TET2 status and ELN risk category* | .002 | .05 | .01 | ||||||
| Within ELN favorable-risk group: TET2 mutated v TET2wild type | 1.71 | 1.19 to 2.47 | .004 | 0.62 | 0.41 to 0.96 | .03 | 1.54 | 1.00 to 2.35 | .05 |
| Within ELN intermediate-I–risk group: TET2 mutated v TET2wild type | .14 | .18 | .13 | ||||||
| WT1(mutated v wild type) | 1.64 | 1.15 to 2.34 | .006 | N/S | 1.63 | 1.04 to 2.56 | .03 | ||
| WBC (continuous, 50-unit increase) | 1.23 | 1.13 to 1.35† | < .001 | 0.71 | 0.58 to 0.87 | .001 | 1.37 | 1.14 to 1.65† | .003 |
| Platelet count (continuous, 50-unit increase) | 1.09 | 1.02 to 1.17 | .01 | 0.83 | 0.72 to 0.97 | .02 | N/S | ||
| Age group, years (≥ 60 v < 60) | 2.00 | 1.59 to 2.51 | < .001 | 0.51 | 0.31 to 0.84 | .009 | 2.17 | 1.65 to 2.85 | < .001 |
NOTE: All multivariable models contained an interaction term to account for the interaction between European LeukemiaNet (ELN) risk group assignment and TET2 mutation status. A hazard ratio (HR) > 1 (< 1) corresponds to a higher (lower) risk for higher values of continuous variables and the first category listed of a dichotomous variable. Variables were considered for inclusion in the multivariable models if they had a univariable P value of < .2. See Data Supplement for a full list of variables evaluated in univariable analyses. Since internal tandem duplication of the FLT3 gene (FLT3-ITD), NPM1, and CEBPA mutations are integrated in the ELN risk classification, they were not additionally considered as individual variables. Several variables were considered for model inclusion. In the model for event-free survival: TET2 mutations, ELN risk group, and their interaction term; WT1 mutations, WBC, platelet count, age group (< 60 v ≥ 60 years), and race (white v nonwhite). In the model for achievement of complete remission: TET2 mutations, ELN risk group, and their interaction term; WT1 mutations, WBC, platelet count, and age group (< 60 v ≥ 60 years). In the model for disease-free survival: TET2 mutations, ELN risk group, and their interaction term; WT1 mutations, WBC, age group (< 60 v ≥ 60 years), and race (white v nonwhite).
Abbreviations: OR, odds ratio; ELN, European LeukemiaNet; N/S, not significant.
The ELN favorable-risk group is defined as patients with mutated CEBPA and/or mutated NPM1without FLT3-ITD. All remaining patients with cytogenetically normal acute myeloid leukemia (ie, those with wild-type CEBPA and FLT3-ITD and/or wild-type NPM1) belong to the ELN intermediate-I– risk category.
This variable did not meet the proportional hazards assumption. The P value corresponds to the Wald statistic of a 2-df test evaluating whether the coefficients for the variable and an artificial time-dependent covariate were equal to 0 to account for nonproportionality. The estimate of the HR is provided at 3 months.
Gene- and MicroRNA-Expression Signatures Associated With TET2 Mutations
To gain insights into the biology of TET2-mutated CN-AML, we studied mutation-associated gene-expression signatures. Within favorable-risk patients, 150 Affymetrix probe sets (representing 91 named genes) were upregulated and 63 probe sets (representing 45 named genes) were downregulated in TET2-mutated patients (n = 41) compared with TET2-wt patients (n = 93; Data Supplement). In contrast, a significant TET2 mutation-associated gene-expression signature could not be identified in intermediate-I–risk patients. Among the genes upregulated in favorable-risk TET2-mutated patients were DPPA4 (a marker of pluripotency highly expressed in hematopoietic stem and progenitor cells29), MS4A3 (a cell cycle regulator in hematopoietic cells30), NCAM1 (CD56; associated with poor outcomes in AML31), LMO4 (involved in hematopoietic stem-cell development32), APP (overexpressed in complex karyotype AML with 21q amplification33 and downregulated in NPM1-mutated CN-AML24), and the granulocytic transcription factor CEBPA and IDH1 (both frequently targeted by mutations in CN-AML15,26,27). Downregulated in favorable-risk TET2-mutated patients were the cancer-testis antigen CT45 gene family (highly expressed in CN-AML with NPM1 mutations,24 lymphoma, and epithelial cancers34), HBG1/HBG2 (hemoglobin gamma), and MLL (implicated in stem-cell self-renewal,35 frequently targeted by genomic rearrangements in AML36).
MicroRNA-expression profiles were available for a subset of older (age ≥ 60 years) patients in whom 72% of TET2 mutations were found in our study. We identified distinct microRNA-expression signatures in favorable-risk and intermediate-I–risk patients. In favorable-risk TET2-mutated patients (n = 28) compared with favorable-risk TET2-wt patients (n = 50), five microRNAs (represented by six microarray probes) were upregulated and two microRNAs were downregulated (Data Supplement). Among the upregulated microRNAs were miR-148a (targeting DNA methyltransferases, highly expressed in refractory chronic lymphocytic leukemia37) and miR-24 (stimulating myeloid cell proliferation and blocking granulocytic and erythroid differentiation38,39). One of the two downregulated microRNAs was miR-135a, which targets JAK2 and, when downregulated, is associated with shorter DFS in patients with Hodgkin's lymphoma.40 In the intermediate-I–risk group, six microRNAs were upregulated in TET2-mutated (n = 21) versus TET2-wt (n = 79) patients, and seven microRNAs (represented by nine probes) were downregulated (Data Supplement). Notably, there was no overlap between the microRNA-expression signatures identified in favorable-risk and intermediate-I–risk patients. Among the upregulated microRNAs in intermediate-I–risk TET2-mutated patients was miR-204, which targets HOXA10 and MEIS1 and shows low expression in NPM1-mutated AML.41 In contrast, miR-126, targeting HOXA9 and showing low expression in NPM1-mutated AML,24 was downregulated in intermediate-I–risk TET2-mutated patients.
DISCUSSION
Since their initial discovery, TET2 mutations have been extensively studied in MPN and MDS, and reports involving patients with AML2–4,7,12,42 have often focused on secondary AML arising from these disorders. In contrast, little is known about the prevalence and clinical relevance of TET2 mutations in primary AML. In two relatively small studies,1,11 TET2 mutations were found in 17% and 19% of patients with primary AML with various karyotypes and 22% of patients with CN-AML.11 In our larger cohort, the prevalence of TET2 mutations in primary CN-AML was 23%. Our findings that most TET2 mutations are frame shift or nonsense changes (likely resulting in a truncated protein) and that missense mutations cluster in two evolutionarily conserved domains of TET2 are in agreement with previous MDS and MPN series.1,2,5,12 We analyzed matched germline DNA samples to ascertain the somatic nature of sequence changes. In our cohort, all evaluable missense changes outside the two conserved domains were germline variants and thus likely nonpathogenic. Therefore, patients carrying these variations were excluded from our analyses.
We focused on patients with CN-AML because the prognosis of this cytogenetic subset is affected by molecular markers that are increasingly used for prognostication and risk-adapted management decisions.14–16,23–27 We demonstrated that TET2 mutations are associated with older age (as reported for MPN5) and higher pretreatment WBC and that they rarely occur together with IDH1 or IDH2 mutations. Consistent with our previous report27 that mutations of IDH2 codon R172 are mutually exclusive with other gene mutations in CN-AML, no patient concurrently harbored a TET2 mutation and an IDH2 R172 mutation.
To the best of our knowledge, our study is the largest report on the prognostic implications of TET2 mutations in primary CN-AML. Nibourel et al,11 who focused on patients achieving CR, found no differences in DFS or OS between 12 TET2-mutated and 42 TET2-wt patients with CN-AML. In another study3 of 93 patients with primary or secondary AML, TET2 mutations were associated with inferior OS, but this analysis did not consider cytogenetics or other potentially confounding variables.
With a growing number of gene mutations being identified in CN-AML, it becomes increasingly important to consider individual markers in their genetic context. The prognostic impact of one mutation may vary depending on the presence or absence of other molecular markers (eg, NPM1 mutations are associated with favorable outcomes particularly in the absence of FLT3-ITD).14,23,43,44 For clinical decision making, risk stratification algorithms integrating prognostic information conveyed by a panel of molecular markers are needed. Recently, the ELN proposed a risk stratification scheme for AML based on cytogenetics and three established molecular prognostic markers.16 The relative prognostic importance of novel molecular markers should not only be evaluated in multivariable models but also needs to be investigated in the context of accepted classification systems. This approach allows judging the potential of new markers to be readily applicable in the clinic and to be incorporated in risk-adapted management decisions for patients with AML.
The ELN classification categorizes patients with CN-AML into a favorable-risk group (CEBPA-mutated and/or mutated NPM1 without FLT3-ITD; ≈ 45% of patients with CN-AML14) and a less-favorable intermediate-I–risk group (all remaining patients with CN-AML). To examine whether TET2 mutations can be used to improve this widely accepted classification, we studied their prognostic value within the ELN risk categories of CN-AML. Among ELN favorable-risk patients, those with TET2 mutations had lower response rates and a higher risk of relapse or death than TET2-wt patients. The favorable-risk category comprises two molecularly defined subgroups of CN-AML: CEBPA-mutated patients and those with mutated NPM1 without FLT3-ITD. Notably, when separately evaluating these two genotypes, we found that TET2 mutations were associated with inferior outcomes in both subgroups (Data Supplement). In contrast, we observed no significant prognostic impact of TET2 mutations in the ELN intermediate-I–risk category or any of its molecular subsets (Data Supplement), although some of these exploratory subgroup analyses were limited by small sample sizes.
Our study included patients across a broad range of ages, potentially introducing bias in our survival analyses. However, in multivariable analyses adjusting for age group and other variables, TET2 mutations remained associated with inferior EFS, lower CR rates, and shorter DFS among favorable-risk patients. FLT3-ITD, NPM1, and CEBPA mutations do not appear in our multivariable models as individual variables, since they are already incorporated in the definition of the ELN risk categories. We did not separately consider FLT3-ITD:wt allelic ratio because the ELN favorable-risk group included only 19 patients with FLT3-ITD (all CEBPA-mutated) and because the adverse impact of a high allelic ratio has been found in younger, but not in older patients.45
Along with TET2 mutations, WT1 mutations were associated with shorter EFS and DFS after adjustment for ELN risk category and other covariables. In our model for CR rate, TET2 mutations were the only gene mutations providing additional prognostic information beyond ELN risk category. To the best of our knowledge, our study is the first to evaluate a novel molecular marker in CN-AML in the context of the current ELN classification and to suggest that TET2 (and possibly WT1) mutations are candidate markers for a refined CN-AML classification scheme.
The function of the TET2 protein is not fully understood. Its paralogue TET1 was recently shown to catalyze conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA, suggesting a role of TET proteins in epigenetic regulation.46,47 To gain insights into the biologic consequences of TET2 mutations, we studied genome-wide gene- and microRNA-expression signatures. Among favorable-risk patients, we identified a TET2 mutation-associated gene-expression signature comprising 136 differentially expressed named genes. TET2 mutations were associated with deregulation of genes involved in stem-cell self-renewal, cell cycle control, and cytokine and growth factor signaling, which may help explain their adverse prognostic impact. In contrast, no significant gene-expression signature could be identified in the intermediate-I–risk cohort.
MicroRNA-expression profiling revealed distinct TET2 mutation-associated signatures in both the favorable-risk and intermediate-I–risk groups, involving several microRNAs implicated in CN-AML and other hematologic malignancies. Interestingly, the microRNA-expression signatures in the two ELN risk groups did not overlap. Apparently, TET2 mutations affect different sets of microRNAs and genes in favorable-risk and intermediate-I–risk patients with CN-AML. These differences show that the ELN classification identifies distinct biologic subsets of CN-AML and corresponds to our finding that the prognostic impact of TET2 mutations also varies between the two ELN risk categories. Together, our results suggest that the biologic and clinical consequences of TET2 mutations in CN-AML differ between the ELN risk groups, although the mechanisms underlying this differential impact remain unclear.
In conclusion, TET2 mutations occur in > 20% of adult patients with primary CN-AML and may be useful for improving genetic risk classification schemes, such as the ELN classification, which are increasingly used in the clinic to guide personalized treatment decisions. The current ELN guidelines generally do not recommend allogeneic transplantation for favorable-risk patients in first CR,16 and none of our patients received such treatment as postremission therapy. However, our results suggest that a subset of these patients, identified by mutated TET2, do not do well with conventional postremission treatment. If our results are corroborated, some of these patients might be considered candidates for alternative therapeutic approaches.
Supplementary Material
Appendix
The following Cancer and Leukemia Group B (CALGB) institutions, principal investigators, and cytogeneticists participated in this study: Wake Forest University School of Medicine, Winston-Salem, NC: David D. Hurd, P. Nagesh Rao, Wendy L. Flejter, and Mark J. Pettenati (Grant No. CA03927); The Ohio State University Medical Center, Columbus, OH: Clara D. Bloomfield, Karl S. Theil, Diane Minka, and Nyla A. Heerema (Grant No. CA77658); North Shore-Long Island Jewish Health System, Manhasset, NY: Daniel R. Budman and Prasad R.K. Koduru (Grant No. CA35279); University of Iowa Hospitals, Iowa City, IA: Daniel A. Vaena and Shivanand R. Patil (Grant No. CA47642); Roswell Park Cancer Institute, Buffalo, NY: Ellis G. Levine and AnneMarie W. Block (Grant No. CA02599); Duke University Medical Center, Durham, NC: Jeffrey Crawford, Sandra H. Bigner, Mazin B. Qumsiyeh, John Eyre, and Barbara K. Goodman (Grant No. CA47577); Washington University School of Medicine, St. Louis, MO: Nancy L. Bartlett, Michael S. Watson, Eric C. Crawford, and Jaime Garcia-Heras (Grant No. CA77440); University of Chicago Medical Center, Chicago, IL: Hedy L. Kindler, Diane Roulston, Katrin M. Carlson, Yanming Zhang, and Michelle M. Le Beau (Grant No. CA41287); Dana-Farber Cancer Institute, Boston, MA: Harold J. Burstein, Ramana Tantravahi, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA32291); University of North Carolina, Chapel Hill, NC: Thomas C. Shea and Kathleen W. Rao (Grant No. CA47559); University of Massachusetts Medical Center, Worcester, MA: William V. Walsh, Vikram Jaswaney, Michael J. Mitchell, and Patricia Miron (Grant No. CA37135); Vermont Cancer Center, Burlington, VT: Steven M. Grunberg, Elizabeth F. Allen, and Mary Tang (Grant No. CA77406); Dartmouth Medical School, Lebanon, NH: Konstantin Dragnev, Doris H. Wurster-Hill, and Thuluvancheri K. Mohandas (Grant No. CA04326); Ft. Wayne Medical Oncology/Hematology, Ft. Wayne, IN: Sreenivasa Nattam and Patricia I. Bader; Eastern Maine Medical Center, Bangor, ME: Harvey M. Segal and Laurent J. Beauregard (Grant No. CA35406); Weill Medical College of Cornell University, New York, NY: John Leonard, Ram S. Verma, Prasad R.K. Koduru, and Susan Mathew (Grant No. CA07968); Minneapolis VA Medical Center, Minneapolis, MN: Vicki A. Morrison and Sugandhi A. Tharapel (Grant No. CA47555); Mount Sinai School of Medicine, New York, NY: Lewis R. Silverman and Vesna Najfeld (Grant No. CA04457); University of Puerto Rico School of Medicine, San Juan, PR: Eileen I. Pacheco, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton; Rhode Island Hospital, Providence, RI: William Sikov, Teresita Padre-Mendoza, Hon Fong L. Mark, Shelly L. Kerman, and Aurelia Meloni-Ehrig (Grant No. CA08025); SUNY Upstate Medical University, Syracuse, NY: Stephen L. Graziano, Larry Gordon, and Constance K. Stein (Grant No. CA21060); Christiana Care Health Services, Newark, DE: Stephen S. Grubbs, Digamber S. Borgaonkar, and Jeanne M. Meck (Grant No. CA45418); Long Island Jewish Medical Center Community Clinical Oncology Program (CCOP), Lake Success, NY: Kanti R. Rai and Prasad R.K. Koduru (Grant No. CA11028); Massachusetts General Hospital, Boston, MA: Jeffrey W. Clark, Leonard L. Atkins, Paola Dal Cin, and Cynthia C. Morton (Grant No. CA 12449); University of California at San Diego: Barbara A. Parker, Renée Bernstein, and Marie L. Dell'Aquila (Grant No. CA11789); University of Maryland Cancer Center, Baltimore, MD: Martin J. Edelman, Joseph R. Testa, Maimon M. Cohen, Judith Stamberg, and Yi Ning (Grant No. CA31983); Western Pennsylvania Hospital, Pittsburgh, PA: John Lister and Gerard R. Diggans; University of Minnesota, Minneapolis, MN: Bruce A. Peterson, Diane C. Arthur, and Betsy A. Hirsch (Grant No. CA16450); University of Missouri/Ellis Fischel Cancer Center, Columbia, MO: Michael C. Perry and Tim H. Huang (Grant No. CA12046); University of Nebraska Medical Center, Omaha, NE: Anne Kessinger and Warren G. Sanger (Grant No. CA77298); University of Illinois at Chicago: David J. Peace, Maureen M. McCorquodale, and Kathleen E. Richkind (Grant No. CA74811); Walter Reed Army Medical Center, Washington, DC: David C. Van Echo, Rawatmal B. Surana, and Digamber S. Borgaonkar (Grant No. CA26806); Georgetown University Medical Center, Washington, DC: Minnetta C. Liu, and Jeanne M. Meck (Grant No. CA77597); McGill Department of Oncology, Montreal, Quebec, Canada: J.L. Hutchison and Jacqueline Emond (Grant No. CA31809); Medical University of South Carolina, Charleston, SC: Mark R. Green, G. Shashidhar Pai, and Daynna J. Wolff (Grant No. CA03927); University of Cincinnati Medical Center, Cincinnati, OH: Orlando J. Martelo and Ashok K. Srivastava (Grant No. CA47515); Columbia-Presbyterian Medical Center, New York, NY: Rose R. Ellison and Dorothy Warburton (Grant No. CA12011); Virginia Commonwealth University Minority-Based CCOP, Richmond, VA: John D. Roberts and Colleen Jackson-Cook (Grant No. CA52784); SUNY Maimonides Medical Center, Brooklyn, NY: Sameer Rafla and Ram S. Verma (Grant No. CA25119); Southern Nevada Cancer Research Foundation CCOP, Las Vegas, NV: John Ellerton and Marie L. Dell'Aquila (Grant No. CA35421); University of California at San Francisco: Charles J. Ryan and Kathleen E. Richkind (Grant No. CA60138).
Footnotes
See accompanying article on page 1364
Supported in part by Grants No. CA101140, CA114725, CA140158, CA31946, CA33601, CA16058, CA77658, and CA129657 from the National Cancer Institute, the Coleman Leukemia Research Foundation, and the Deutsche Krebshilfe-Dr. Mildred Scheel Cancer Foundation (H.B.).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Klaus H. Metzeler, Guido Marcucci,Clara D. Bloomfield
Financial support: William Blum, Michael A. Caligiuri, Guido Marcucci, Clara D. Bloomfield
Administrative support: Richard A. Larson, Michael A. Caligiuri,Clara D. Bloomfield
Provision of study materials or patients: William Blum, Bayard L. Powell, Thomas H. Carter, Meir Wetzler, Joseph O. Moore, Jonathan E. Kolitz, Maria R. Baer, Andrew J. Carroll, Richard A. Larson, Michael A. Caligiuri, Guido Marcucci, Clara D. Bloomfield
Collection and assembly of data: Klaus H. Metzeler, Kati Maharry, Michael D. Radmacher, Krzysztof Mrózek, Dean Margeson, Heiko Becker, John Curfman, Kelsi B. Holland, Sebastian Schwind, Susan P. Whitman, Yue-Zhong Wu, William Blum, Bayard L. Powell, Thomas H. Carter, Meir Wetzler, Joseph O. Moore, Jonathan E. Kolitz, Maria R. Baer, Andrew J. Carroll, Richard A. Larson, Michael A. Caligiuri, Guido Marcucci
Data analysis and interpretation: Klaus H. Metzeler, Kati Maharry, Michael D. Radmacher, Krzysztof Mrózek, Dean Margeson, Heiko Becker, Kelsi B. Holland, Susan P. Whitman, Guido Marcucci, Clara D. Bloomfield
Manuscript writing: All authors
Final approval of manuscript: All authors
REFERENCES
- 1.Langemeijer SM, Kuiper RP, Berends M, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 2.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Wahab O, Mullally A, Hedvat C, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jankowska AM, Szpurka H, Tiu RV, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tefferi A, Pardanani A, Lim K-H, et al. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tefferi A, Levine RL, Lim K-H, et al. Frequent TET2 mutations in systemic mastocytosis: Clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23:900–904. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tefferi A, Lim K-H, Abdel-Wahab O, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdel-Wahab O, Manshouri T, Patel J, et al. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70:447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saint-Martin C, Leroy G, Delhommeau F, et al. Analysis of the ten-eleven translocation 2 (TET2) gene in familial myeloproliferative neoplasms. Blood. 2009;114:1628–1632. doi: 10.1182/blood-2009-01-197525. [DOI] [PubMed] [Google Scholar]
- 10.Kosmider O, Gelsi-Boyer V, Ciudad M, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94:1676–1681. doi: 10.3324/haematol.2009.011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nibourel O, Kosmider O, Cheok M, et al. Incidence and prognostic value of TET2 alterations in de novo acute myeloid leukemia achieving complete remission. Blood. 2010;116:1132–1135. doi: 10.1182/blood-2009-07-234484. [DOI] [PubMed] [Google Scholar]
- 12.Kosmider O, Gelsi-Boyer V, Cheok M, et al. TET2 mutation is an independent favorable prognostic factor in myelodysplastic syndromes (MDSs) Blood. 2009;114:3285–3291. doi: 10.1182/blood-2009-04-215814. [DOI] [PubMed] [Google Scholar]
- 13.Mrózek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 14.Schlenk RF, Döhner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 15.Mrózek K, Marcucci G, Paschka P, et al. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: Are we ready for a prognostically prioritized molecular classification? Blood. 2007;109:431–448. doi: 10.1182/blood-2006-06-001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 17.Mrózek K, Carroll AJ, Maharry K, et al. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: The Cancer and Leukemia Group B experience. Int J Oncol. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- 18.Sherry ST, Ward M-H, Kholodov M, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiede C, Steudel C, Mohr B, et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 20.Whitman SP, Ruppert AS, Radmacher MD, et al. FLT3 D835/I836 mutations are associated with poor disease-free survival and a distinct gene-expression signature among younger adults with de novo cytogenetically normal acute myeloid leukemia lacking FLT3 internal tandem duplications. Blood. 2008;111:1552–1559. doi: 10.1182/blood-2007-08-107946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitman SP, Ruppert AS, Marcucci G, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: A Cancer and Leukemia Group B study. Blood. 2007;109:5164–5167. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caligiuri MA, Strout MP, Schichman SA, et al. Partial tandem duplication of ALL1 as a recurrent molecular defect in acute myeloid leukemia with trisomy 11. Cancer Res. 1996;56:1418–1425. [PubMed] [Google Scholar]
- 23.Döhner K, Schlenk RF, Habdank M, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: Interaction with other gene mutations. Blood. 2005;106:3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 24.Becker H, Marcucci G, Maharry K, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paschka P, Marcucci G, Ruppert AS, et al. Wilms' tumor 1 gene mutations independently predict poor outcome in adults with cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4595–4602. doi: 10.1200/JCO.2007.15.2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcucci G, Maharry K, Radmacher MD, et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcucci G, Maharry K, Wu Y-Z, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheson BD, Cassileth PA, Head DR, et al. Report of the National Cancer Institute-sponsored workshop on definitions of diagnosis and response in acute myeloid leukemia. J Clin Oncol. 1990;8:813–819. doi: 10.1200/JCO.1990.8.5.813. [DOI] [PubMed] [Google Scholar]
- 29.Tondeur S, Pangault C, Le Carrour T, et al. Expression map of the human exome in CD34+ cells and blood cells: Increased alternative splicing in cell motility and immune response genes. PLoS One. 2010;5:e8990. doi: 10.1371/journal.pone.0008990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donato JL, Ko J, Kutok JL, et al. Human HTm4 is a hematopoietic cell cycle regulator. J Clin Invest. 2002;109:51–58. doi: 10.1172/JCI14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raspadori D, Damiani D, Lenoci M, et al. CD56 antigenic expression in acute myeloid leukemia identifies patients with poor clinical prognosis. Leukemia. 2001;15:1161–1164. doi: 10.1038/sj.leu.2402174. [DOI] [PubMed] [Google Scholar]
- 32.Meier N, Krpic S, Rodriguez P, et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133:4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 33.Baldus CD, Liyanarachchi S, Mrózek K, et al. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: Amplification discloses overexpression of APP, ETS2, and ERG genes. Proc Natl Acad Sci U S A. 2004;101:3915–3920. doi: 10.1073/pnas.0400272101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y-T, Chadburn A, Lee P, et al. Expression of cancer testis antigen CT45 in classical Hodgkin lymphoma and other B-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107:3093–3098. doi: 10.1073/pnas.0915050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMahon KA, Hiew SY, Hadjur S, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Meyer C, Kowarz E, Hofmann J, et al. New insights to the MLL recombinome of acute leukemias. Leukemia. 2009;23:1490–1499. doi: 10.1038/leu.2009.33. [DOI] [PubMed] [Google Scholar]
- 37.Ferracin M, Zagatti B, Rizzotto L, et al. MicroRNAs involvement in fludarabine refractory chronic lymphocytic leukemia. Mol Cancer. 2010;9:123. doi: 10.1186/1476-4598-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaidi SK, Dowdy CR, van Wijnen AJ, et al. Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAPK network. Cancer Res. 2009;69:8249–8255. doi: 10.1158/0008-5472.CAN-09-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Q, Huang Z, Xue H, et al. MicroRNA miR-24 inhibits erythropoiesis by targeting activin type I receptor ALK4. Blood. 2008;111:588–595. doi: 10.1182/blood-2007-05-092718. [DOI] [PubMed] [Google Scholar]
- 40.Navarro A, Diaz T, Martinez A, et al. Regulation of JAK2 by miR-135a: Prognostic impact in classic Hodgkin lymphoma. Blood. 2009;114:2945–2951. doi: 10.1182/blood-2009-02-204842. [DOI] [PubMed] [Google Scholar]
- 41.Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couronné L, Lippert E, Andrieux J, et al. Analyses of TET2 mutations in post-myeloproliferative neoplasm acute myeloid leukemias. Leukemia. 2010;24:201–203. doi: 10.1038/leu.2009.169. [DOI] [PubMed] [Google Scholar]
- 43.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 44.Thiede C, Koch S, Creutzig E, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107:4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 45.Whitman SP, Maharry K, Radmacher MD, et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. Blood. 2010;116:3622–3626. doi: 10.1182/blood-2010-05-283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szpurka H, Jankowska AM, Ng KP, et al. Involvement of TET2 in regulation of epigenetic silencing in MDS and related myeloid malignancies. Blood. 2009;114:1136. abstr 290. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.