Abstract
Pax transactivation domain-interacting protein (PTIP) is a ubiquitously expressed, nuclear protein that is part of a histone H3K4 methyltransferase complex and is essential for embryonic development. Methylation of H3K is an epigenetic mark found at many critical developmental regulatory genes in embryonic stem cells (ES) and, together with H3K27 methylation, constitutes a bivalent epigenetic signature. To address the function of PTIP in ES cells, we generated ES cell lines from a floxed ptip allele and deleted PTIP function with Cre recombinase. The ptip−/− ES cell lines exhibited a high degree of spontaneous differentiation to trophectoderm and a loss of pluripotency. Reduced levels of Oct4 expression and H3K4 methylation were observed. Upon differentiation, ptip−/− embryoid bodies showed reduced levels of marker gene expression for all three primary germ layers. These results suggest that the maintenance of H3K4 methylation is essential and requires PTIP function during the in vitro propagation of pluripotent ES cells.
Keywords: PTIP, Oct-4, epigenetics, spontaneous differentiation
Introduction
Embryonic Stem (ES) cells can remain pluripotent in culture and contribute to all tissues including the germ line when reintroduced into a developing embryo. Pluripotency requires the continued expression of Oct4 and nanog, two transcriptional factors that regulate a network of genes in ES cells 1. Differentiation into specialized cell types was thought to be stable and irreversible. However, the reintroduction of a limited number of genes, including oct4, sox2, c-myc, and klf4, and subsequent selection for nanog expression can dedifferentiate somatic cells into an ES cell like state 2, 3. Differentiation and loss of pluripotency may be driven, at least in part, by epigenetic modifications of chromatin, including histone methylation at specific lysine residues. Consistent with the idea that ES cell chromatin is epigenetically plastic, the patterns of histone methylation at key regulatory loci in ES cells show low levels of both active and inactive epigenetic marks, that are resolved upon differentiation into fully active or fully repressed marks depending upon cell lineage 4, 5.
The mammalian homologues of the Drosophila Trithorax and Polycomb group genes encode the histone modification machinery that specifies active or inactive regions of the genome. Genetic studies in mice reveal an essential role for histone methyltransferases and their associated factors in early embryonic development, as cells assume a more differentiated fate and loose pluripotency 6–11. However, the roles of histone methyltransferase complexes in maintaining growth and pluripotency of cultured ES cells have not been well studied.
Methylation of histone H3 at lysine 4 (H3K4) is a key modification that correlates with gene expression and is thought to promote assembly of nucleosome remodeling complexes required for transcription, elongation, and splicing 12, 13]. In ES cells, H3K4 trimethylation is present at the transcription initiation sites of many genes and is coupled to RNA polymerase occupancy, even if in most cases transcriptional elongation and mRNA expression does not progress 14. The mammalian homologues of Trithorax are the MLL family of H3K4 histone methyltransferases, which are co-purify with the accessory proteins WDR5, RBBP5, and ASH2L similar to the yeast Set1 COMPAS methyltransferase complex 15, 16. The BRCT-domain containing protein PTIP is a novel component of the MLL2 methyltransferase complex 17, 18 and is essential for embryonic development post gastrulation 19. PTIP interacts with the developmental regulatory transcription factor Pax2 and promotes assembly of the MLL2 histone H3K4 methyltransferase complex at a Pax2 DNA binding site 20. However, PTIP is likely to interact with other DNA binding proteins to impact patterns of histone modification at many loci, as both germline null and conditional mutants show reduced levels of H3K4 di- and trimethylation in affected tissues. These data suggest that PTIP is critical for linking the MLL2 complex to specific DNA binding transcription factors during differentiation, such that H3K4 methylation is regulated in a locus and tissue specific manner.
The developmental defects observed in ptip−/− homozygous embryos are evident at the time of gastrulation and result in a disorganized mass of poorly differentiated cells. Many ptip−/− nuclei exhibit free DNA ends, are stuck in the cell cycle, and, exhibit reduced levels of global H3K4 methylation 19, 20. Even at earlier stages, blastocyst explants from E3.5 showed clear inhibition of inner cell mass proliferation in ptip−/− embryos. However, these experiments did not assess the role of PTIP in maintaining ES cell pluripotency. If H3K4 methylation was important for maintaining potency then the loss of PTIP may affect global levels and lead to a reduction in differentiation potential of stem cells. In order to test this hypothesis, we derived ES cell lines from mice carrying one conditional floxed ptip alleles and one null allele (ptipFl/−) and generated a PTIP loss of function mutation in ES cell lines with Cre recombinase under conditions that should retain pluripotency. The ES cells lacking PTIP underwent spontaneous differentiation despite the optimal media with feeder cells and recombinant leukemia inhibitory factor (LIF). The loss of ES cell pluripotency was assayed by real time QRT-PCR, Western blotting, alkaline phosphatase staining, TROMA1 immunostaining, and chromatin immunoprecipitation. The data suggest a crucial role for PTIP and H3K4 methylation in maintaining the undifferentiated state and pluripotency in ES cells in culture.
Materials and methods
Generation of PTIP-deficient mouse embryonic stem cells
To address the functions of PTIP in ES cells, ptip floxed mice were generated as described previously21. Mice carrying one floxed allele were intercrossed to generate homozygous ptipfl/fl mice that are viable and fertile. Three to five week old females of ptipfl/fl were superovulated by injecting intraperitoneally with Pregnant mare serum gonadotrophin and Human chorionic gonadotrophin two days before and on the day of mating with ptipfl/− mice, respectively. Three days later, blastocysts were flushed out of the uterine horns and cultured in ES cell growth medium described in our previous work (Kim, 2005). The ptipfl/− or ptipfl/fl ES cells were selected for infection with Cre-expressing adenovirus. The ES cells were split onto MEF feeders and infected with Cre-expressing adenovirus (Ad-Cre) or adenovirus alone (Ad) at 1000:1 MOI (Multiplicity of Infection) for 4 hrs and fresh media was added after removal of the media with the virus. After 2 days, colonies were picked and transferred to individual wells of a 96-well plate with MEFs and rLIF. After 2 additional days, ES cells were split into 24 well plates. Genotyping was done at passage 4–6 to see if ptip is deleted by the Cre-expressing adenovirus. At that passage, two clones were identified as PTIP-deficient ES cells and were expanded in 6 well plates for an additional 2–4 passages before being used for making embryoid body and other assays.
The ptipfl/− ES cells were also used for generating proteins, RNAs, and chromatin from without clonal selection. After trypsinization, 1×106 cells were plated onto gelatin coated 60 mm plates with rLIF and infected with adenoviral vectors at an MOI of 500. Cells were harvested after 4 days and deletion of PTIP monitored by western blotting.
Immunocytochemistry and Alkaline phosphatase staining
ES cells were seeded onto glass coverslips and allowed to grow for up to 3 days. Growth medium was removed and the cells were fixed in 4% paraformaldehyde for 10 min. The primary antibodies used were anti-rabbit Oct4 (Santa Cruz Biotechnology; Santa Cruz, CA) or mouse anti-TROMA-1, diluted 1:100 each in 0.1% Tween 20 in PBS (PBST) plus 2% goat serum and applied for 1 hr at room temperature. Secondary antibody (anti-rabbit IgG-FITC conjugated or anti-mouse IgG-TRITC, diluted 1:200 diluted in PBST plus 2% goat serum was applied for 1 hour at room temperature. Cells were further counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories, Burlingame, CA) and imaged on a Nikon ES800 fluorescent scope with a SPOT digital camera.
Alkaline phosphatase substrate solution was prepared using Sigma Fast™ fast red tablet sets (Sigma, F-4523) as manufacturer’s instruction. ES cells on glass coverslips were incubated in the solution at 37°C incubator for 1 hr and monitored for the development of a red reaction product indicating alkaline phosphatase activity.
Western blotting
Cells were lysed in PK-lysis buffer (Cai et al., 2002) and protein levels were quantified by the Bio-Rad colorometric assay (Bio-Rad, Hercules, CA). Histone proteins were acid extracted as described (AbCam). SDS/PAGE sample buffer was added and samples were boiled for 5 min. Samples were run on a 10% polyacrylamide gel, transferred to PVDF membrane (Perkin Elmer, Boston, MA), and blocked with 5% non-fat milk in Tris-buffered saline. Membranes were immunoblotted with anti-rabbit Oct-3/4, anti-mouse β-actin, anti-rabbit trimethyl H3K4 (AbCam), and mono-,di-,trimethyl H3K4 (AbCam) antibodies. For quantitation, secondary antibodies were coupled to fluorescent conjugated secondary antibodies and a Li-Cor Odessey infrared imager were used.
Real-time reverse transcription polymerase chain reaction
Total RNA extraction and reverse transcription-PCR analysis were performed as described previously (Kim, 2005). The real time PCR primer pairs were as follows: Oct-4 AGCTGCTGAAGCAGAAGAGG, GGTTCTCATTGTTGTCGGCT; Hand-1 GGATGCACAAGCAGGTGAC, CACTGGTTTAGCTCCAGCG; Vimentin AACACCCGCACCAAC, TGTCCCGCTCCACCTCGAC; Enolase TGCTAAGGCCCTTTTCTGTT, GACTAGGCACCCCTATTCCA; α-feto protein CACTGCTGCAACTCTTCGTA, CTTTGGACCCTCTTCTGTGA; Hnf-4 ACACGTCCCCATCTGAAG, CTTCCTTCTTCATGCCAG; Nkx2.5 AGCAACTTCGTGAACTTTG, GATCCGGTCCTAGTGTGGA; GATA-4 GGTTCCCAGGCCTCTTGCAATGCGG, AGTGGCATTGCTGGAGTTACCGCTG; PTIP CCGAAGTTCCAGAGGAGCTA, GTCCCCATCCTCGAAATGA; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) TCCGCCCCTTCTGCCGATG, CACGGAAGGCCATGCCAGTGA.
Chromatin Immunoprecipitation
ChIP was performed in triplicate according to published protocols from Upstate Biotech with minor modifications. Cells were fixed for 10 min at 25°C with 1% formaldehyde in culture medium. Cross-linking was stopped by the addition of glycine to 0.125 M. Cells were washed twice with ice-cold PBS, scraped, and harvested by centrifugation. The cell pellet was washed in PBS, resuspended in cell lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5%NP40, and protease inhibitors), incubated at 4°C for 5 minutes and centrifuged for 5 minutes at 3000 X g. The nuclei were resuspended in nuclei lysis buffer (50 mM Tris-HCl, pH 8.1, 10 mM EDTA, 1% SDS, protease inhibitors) and were sonicated on ice with three 20 s pulses using a microtip probe sonicator (Branson Sonifier 250) with output control set to 1. Sonicated lysates were clarified by centrifugation at 4°C fro 15 minutes. 20 μg of chromatin was diluted in IP dilution buffer (0.01% SDS, 1.1% Triton-X100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1 and 167 mM NaCl) and preclarified with 80 ul protein A-agarose or protein G-agarose (Upstate). The precleared chromatin preparation was then precipitated with 2–5 μg antibodies against tri-methyl H3K4 (Abcam) or mono-, di-, trimethyl H3K4 (Upstate). After overnight incubation at 4°C, 60 μl protein A-agarose or protein G-agarose were added and the incubation was continued for 1 h. The beads were sequentially washed two times in IP dilution buffer, two times in TSE-500 wash buffer (0.1% SDS, 1% Trition X-100, 2nM EDTA, 20 mM Tris-HCl, pH 8.1,500 mM NaCl), two times in LiCl buffer (100 mM Tris-HCl, pH 8.1, 500 mM LiCl, 1% NP-40, 1% Na deoxycholate), and finally two times in Tris-EDTA. Bound complexes were eluted by vortexing beads twice for 15 min at 25°C in 250 μl of elution buffer (50 mM Na bicarbonate and 1% SDS). 5M NaCl was added to a final concentration of 0.2M to the pooled eluates and cross-links reversed by incubating samples at 65°C overnight. The samples were digested with proteinase K for 1 hr at 56°C and phenol-chloroform extracted. The precipitated DNA was reconstituted in sterile water and Real-time PCR quantitation of precipitated genomic DNA relative to inputs was performed in triplicate using IQ SYBR GREEN mastermix (Bio-rad) in an iCycler (Bio-rad). Oct4, Sox2, and Pax3 genomic sequences were analyzed by real-time PCR (Bio-Rad). The data are represented as Mean +/− one standard error of the mean of two independent experiments.
Results
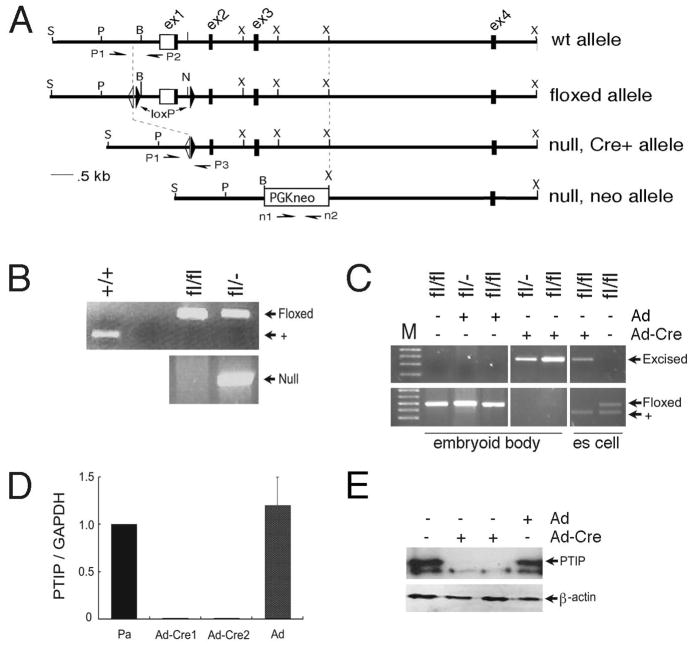
The function of PTIP in epigenetic maintenance of embryonic stem cell cultures was addressed by deleting PTIP in ES cell lines derived from ptipfl/− or ptipfl/fl mouse blastocysts using Cre recombinase mediated excision. We were able to derive stable, parental ES cell lines from a cross of ptipfl/fl and ptipfl/− animals that were homozygous fl/fl or heterozygous fl/−. These cells were grown on MEFs with rLIF and maintained a pluripotent ES cell phenotype for many generations. For generating clonal lines, the ptipfl/− ES parental cell lines (Pa) were infected with either Cre-expressing Adenovirus or Adenovirus alone to delete exon 1 and the 5′ regulatory region of the ptip gene (Fig. 1). Two early passage ptip−/− cell lines were established that deleted the ptip floxed allele (Ad-Cre1 and Ad-Cre2). At passage 8–10, neither cell line had detectable amounts of PTIP mRNA or protein, as measured by RT-PCR and western blotting respectively (Fig. 1C, D).
Fig 1. Generation of ptip−/− ES cells.
PCR genotyping of ES cells with allele specific primer pairs. A) Schematic of the different alleles used in this study. The floxed allele contains loxP sites flanking exon 1 (ex1); a single flip recombinase site is also present (open triangle). The germline null allele deletes exons 1–3 and still contains PGK-neo, as described previously. Primers P1 and P2 were used to detect the wild-type and floxed alleles, primers P1 and P3 detect the Cre excised allele, and primers n1 and n2 detect the neo-containing null allele. B) Wild-type and ES cell lines derived from a ptipfl/fl x ptipfl/− mating were genotyped with allele specific primer pairs before Cre mediated excision. Top panel indicates the wild-type (+) and floxed alleles. The bottom panel shows the germline null allele. C) Genotype of ES cells after infection with Cre expressing adenovirus (Ad-Cre). As indicated, cells were cultured as embryoid bodies, without feeders, or as ES cells on feeders and LIF. Note, wild-type allele is due to feeder cells. D) Relative mRNA expression determined by real time qPCR. Parental ptipfl/− ES cells (Pa), ptip−/− derivatives (Ad-Cre1, Ad-Cre2), and parental cells infected with Ad only are shown. E) PTIP protein expression as determined by Western blotting from ptipfl/− ES cells infected with the control (Ad) or Cre (Ad-Cre) expressing adenovirus as indicated.
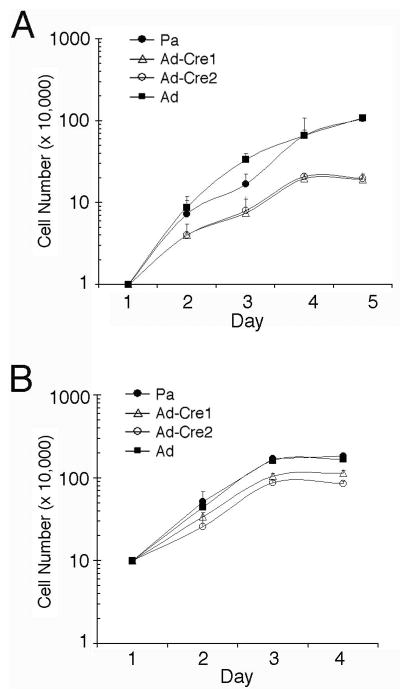
Our previous work with complete ptip−/− embryos showed a defect in proliferation in blastocyst outgrowth culture 19. However, these outgrowths did not contain feeder cells or rLIF to maintain pluripotency. To determine if ptip−/− ES cells can grow under conditions that maintain pluripotency, cell proliferation was measured after seeding either at low (1×104/ml, Fig 2A) or high (1×105/ml, Fig 2B) density in rLIF and mouse embryo fibroblasts (MEFs) feeder cells (Fig. 2). At low density, wild type ES cells grew up to 1×106/ml by 5 days whereas ptip−/− ES cells grew to no more than 3×105/ml. At high density, differences in growth rates were less pronounced as ptip−/− ES cells grew nearly to 1×106/ml when seeded at high density.
Fig 2. Growth curves of ES cells cultured on MEF.
(A) The ES cells were seeded at 104 cells/ml and counted daily thereafter. (B) The ES cells were seeded at 105 cells/ml and counted daily, reaching saturation within 3 days. Parental ES ptipfl/− cell lines (Pa) or ptipfl/− ES cells infected with control adenovirus (Ad) were compared to ptipfl/− ES cells infected with Cre recombinase expressing virus (Ad-Cre1, Ad-Cre2).
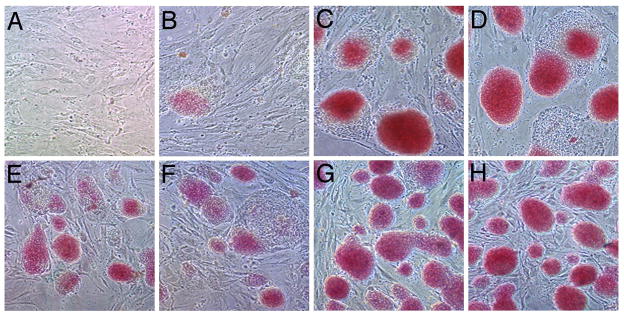
Given that ES cells are self-renewing, ptip−/− cells might loose their self-renewal capacity when seeded at low density, but maintain self-renewal, at least in part, at higher densities. Self-renewal maintains the pluripotent state and suppresses spontaneous differentiation. Thus, cells were subjected to alkaline phosphatase staining, which stains only the undifferentiated ES cell colonies (Fig. 3). Colonies from ptip−/− ES cells seeded at low density stained poorly for alkaline phosphase and were generally small with a more flattened phenotype (Fig 3B–D). At higher densities, ptip−/− ES cells stained positive for alkaline phosphatase but intensities were weaker and colony size was reduced compared to wild-type ES cells (Fig 3E–H). These data suggest that ptip−/− ES cells undergo spontaneous differentiation even in the presence of MEFs and rLIF.
Fig 3. Alkaline phosphatase staining of ptip deficient ES cells.
Panels A–D are 5 days after seeding at 104 cells/ml and panels E-H are 3 days after seeding at 105 cells/ml. All cells were cultured with MEFs and LIF. A) MEFs only. B) ptipfl/− Ad-Cre1 line. C) Parental line ptipfl/−. D) Parental ptipfl/− line infected with Ad only. E) ptipfl/− Ad-Cre1 line. (F) ptipfl/− Ad-Cre2 line. G) Parental ptipfl/− line. H) Parental ptipfl/− line infected with Ad only.
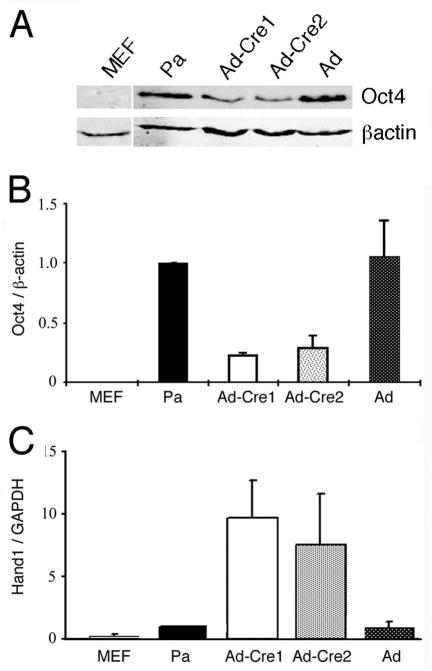
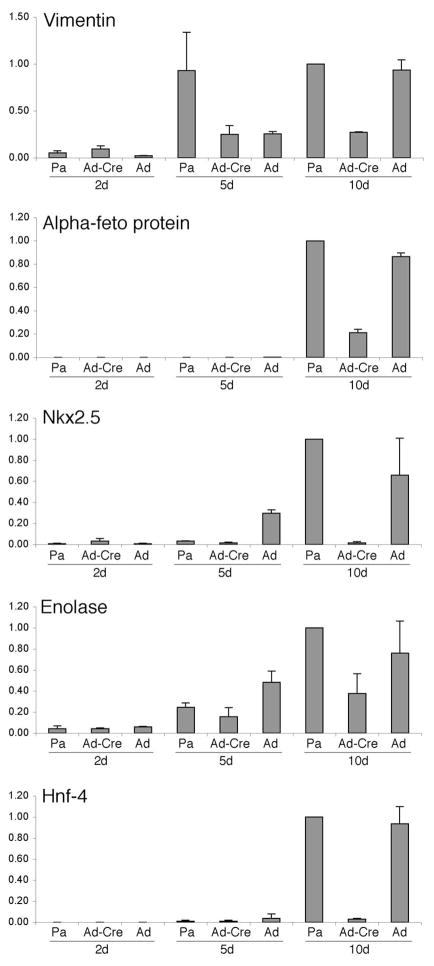
The maintenance of pluripotency in ES cells depends on the continued expression of the transcription factor Oct4. In ptip−/− ES cell lines, Oct4 mRNA and protein levels are reduced, exhibiting 3–4 fold less protein (Fig. 4A, B). However, the ptip−/− ES cells show a marked 7–9 fold increase in expression of the trophectodermal marker Hand1, as measured by quantitative real time RT-PCR (Fig. 4C). These data are consistent with the hypothesis that ptip−/− ES cells spontaneously differentiate into the trophectodermal lineage, perhaps in part by downregulating Oct4. This spontaneous loss of Oct4 may not affect the ability of PTIP deficient cells to differentiate after removal of LIF, since Oct4 is generally downregulated upon formation of the three primary germ layers. To examine this directly, we cultured cells under conditions that favored embryoid body (EBs) formation and examined the cultures for gene expression over time. The EBs obtained from PTIP- cells were much smaller than the PTIP+ EBs and expressed reduced levels of differentiation markers (Fig. 5). Markers for all three germ layers were used, including ectoderm (Vimentin and Enolase), endoderm (α-fetoprotein and Hnf-4), and mesoderm (Nkx2.5). Given the reduced levels of expression of all markers coupled with the increase in Hand1, it is likely that PTIP-deficient ES cells are unable to differentiate into the normal germ layers, again implying that they have lost much of their pluripotency and prefer the trophectodermal lineage.
Fig 4. Oct4 and Hand1 expression in PTIP-deficient ES cells.
A) Western blotting of Oct4 and β-actin from parental ES cells and PTIP deleted cells. B) Quantitative protein measurements of Oct4 normalized to β-actin, using flourescent conjugated secondary antibodies and a LYCOR reader. C) Real time RT-PCR of Hand1 normalized with GAPDH. Parental ptipfl/− ES line (Pa), control adenovirus infected ptipfl/− ES cells (Ad), and ptipfl/− Cre recombinase expressing cells (Ad-Cre1, Ad-Cre2) were compared. All data were produced from three independent experiments. Error bars are one standard deviation from the mean.
Fig. 5. Expression of Lineage markers in Embryoid Bodies.
Real time RT-PCR analysis of genes expressed in differentiating embryoid bodies over time. Markers for all three germ layers were used, including ectoderm (Vimentin and Enolase), endoderm (β-feto protein and Hnf4), and mesoderm (Nkx2.5). Parental ES ptipfl/− cells (Pa), Ad-Cre 1 infected cells (Ad-Cre), and adenoviral vector infected cells (Ad) were used to generate embryoid bodies in hanging drops then cultured for 2, 5, or 10 days in Petrie dishes. Total RNA was isolated and RT-PCR were normalized to GAPDH. Averages for three readings are shown; error bars represent one standard deviation from the mean.
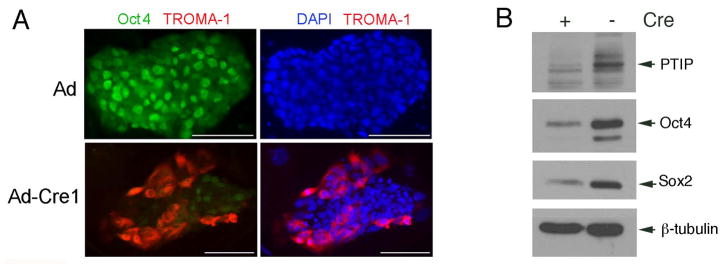
Finally, we assessed this effect at the single cell level by staining parental and ptip−/− ES cell colonies with antibodies against Oct4 and the trophectodermal marker TROMA1 (Fig 6). Wild type ES cells exhibited strong nuclear staining with anti-Oct4 and very little staining with anti-TROMA1. Consistent with the western blotting and RT-PCR data, ptip−/− ES cell colonies were heterogenous with some cells Oct4 positive and TROMA1 negative, yet many other cells positive for TROMA1 and negative for Oct4. Over 90% ptip−/− ES cell colonies cultured at high density contained TROMA-1 positive cells, whereas no wild type ES cell colonies exhibited detectable staining with the same antibody (data not shown). Therefore, we conclude that a fraction of ptip−/− ES cells differentiated spontaneously into trophectodermal cells over time. This spontaneous differentiation affected the stability of the ES cell clones after multiple passages. By passage 12–15, the self-renewing capacity of the ES cell clones was limited, making long term culture of the PTIP null cells difficult.
Fig 6. Immunocytochemistry of PTIP-deficient ES cells.
ES cell colonies were stained with anti-Oct4 (green) and anti-TROMA1 (red) as indicated. Right panels were counterstained with DAPI.
Given the potential for clonal effects after Ad-Cre infection and clonal selection, we also examined the expression of ES cell markers in populations of ptipfl/fl ES cells that had been infected with Cre and control virus. Cells grown on feeders with rLIF were trypsinized and plated on gelatin coated plates with rLIF only, infected with Cre or control virus, and harvested 96 hours post infection. PTIP excision was monitored by western blotting of lysates (Fig. 7); expression of pluripotency markers were also assessed (Fig. 7). The data indicate the loss of Oct4 and Sox2 expression is coincident with PTIP excision in Cre infected cells. These data are consistent with the changes in gene expression observed in clonal isolates after Cre mediated excision, at low passage numbers.
Fig. 7. Protein Expression Analysis in Mixed ES Cell Populations.
Parental ptipfl/fl ES cells were infected with Ad-Cre or Adenovirus alone and cultured for 96 hours in rLIF on gelatinized plates. Total protein lysates were assayed by Western blotting for the proteins indicated.
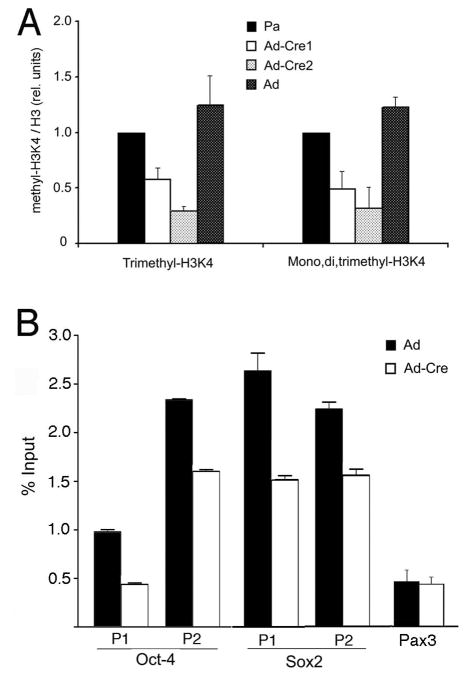
The PTIP protein is part of a histone H3, lysine 4 (H3K4) methyltransferase complex that includes the proteins MLL2, MLL3, ASH2L, WDR5, and RBBP5 17, 18, 20. In a cell culture models, PTIP is necessary for the assembly of an H3K4 methyltransferase complex in response to binding of the developmental regulatory protein Pax2 to its DNA recognition site. Consistent with this model, ptip−/− embryonic tissues show a reduction in global levels of di- and trimethylation at H3K4, but not at lysine 20 of histone H4. To test whether global levels of H3K4 methylation were affected we did quantitative western blotting of parental and ptip−/− ES cell histones after acid extraction from clonal cells at lower passages (Fig. 8A). Antibodies against trimethyl-H3K4 or mono-di-trimethyl-H3K4 showed a 2–3 fold reduction in total methylation compared to wild type ES cell, implying that the mutant ES cells have altered histone H3K4 methylation patterns at many loci.
To examine whether the reduction in H3K4 methylation could occur at specific loci, chromatin immunoprecipitation experiments were performed using antibodies against methyl H3K4 and PCR primer pairs corresponding to putative promoter elements (Fig 8B). Populations of Ad-Cre infected ES cells were prepared without clonal selection, as assayed in Fig. 7. Since expression was reduced in ptip−/− ES cells, the oct4 and sox2 promoters were examined, as well as the Pax3 genes which encodes a developmental regulator. In ptip−/− ES cells, the oct4 and sox2 promoter sequences showed a reduction in H3K4 trimethylation compared to wild-type ES cells. The Pax3 promoter did not show any measurable change in H3K4 methylation, consistent with its low expression levels in both PTIP+ and PTIP- cells (data not shown). Given the heterogenous nature of the ptip−/− ES cells, this reduction in oct4 H3K4 methylation is likely to reflect the proportion of cells that have turned down oct4 completely.
Discussion
Pluripotent ES cells can maintain the undifferentiated state in culture indefinitely, whereas in vivo pluripotency is a transient state reserved for the inner cell mass of the blastocyst, the epiblast cells of the post-implantation embryo, and the germ cells (for review see 22). The maintenance of ES cell pluripotency requires a defined transcriptional regulatory circuit, consisting of Oct4, Sox2 and Nanog 23, and epigenetic factors, such as Polycomb repressive complexes 24 and histone demethylases 25. The chromatin signatures of critical regulatory genes in ES cells exhibit both H3K27 and H3K4 methylation 4, 5, 26, epigenetic marks that were thought to correlate with gene silencing and activation exclusively. Furthermore, acetylation at H3K9 and promoter occupancy by RNA polymerase II indicate that many non-expressed genes in ES cells are in an accessible state, suggesting that polymerase elongation is the critical regulatory event 14. Despite its prevalence at many loci, the necessity of H3K4 methylation complexes in ES cells has not been demonstrated. In this report, the function of PTIP is examined in cultured ES cells. Given that PTIP is an integral component of the MLL2 H3K4 methyltransferase complex 17, 18, 20, our data suggest that maintenance of H3K4 methylation is necessary for continued proliferation of pluripotent ES cells in culture.
During mouse development, deletion of the H3K4 methyltransferases ALL/MLL19, 27 or MLL2 (which is homologous to human MLL4)11 results in post gastrulation lethality, characterized by the misregulation of Hox gene expression. Despite multiple cell autonomous defects, both mll1−/− and mll2−/− embryos progressed well past gastrulation with axial patterning and germ layer differentiation easily recognizable at E8.5. In contrast, ptip−/− embryos showed significant developmental arrest by E7.5, resulting in a small and disorganized tissue mass by E8.519, 20. Mouse embryonic stem cells lacking either MLL128 or MLL211 have also been reported. For the mouse MLL2 mutant ES cells, differentiation was delayed, but endodermal lineages and self-renewal seemed unaffected29. The severity of the ptip−/− phenotype suggests that the protein is interacting with more than just one H3K4 methyltransferase. The presence of MLL2 and/or MLL3 in the PTIP complex and the global reduction of H3K4 methylation observed in ptip−/− embryos would support this hypothesis17, 18, 20. Thus, the available evidence suggests an earlier or more integral function for PTIP in early growth and differentiation than for any single H3K4 methyltransferase described to date.
The culture of ES cells is an artificial system that is able to suppress the transient nature of pluripotency. Through the use of feeder cells and LIF, factor such as Oct4, Nanog, and Sox2 are maintained indefinitely. Direct loss of Oct4 by gene targeting 30 or RNA interference 31 promotes trophoblast differentiation at the expense of inner cell mass cells. Thus, the reduction of Oct4 protein over time could account for much of what is observed in the ptip−/− ES cells. This reduction of Oct4 and loss of pluripotency may be less critical in vivo because of maternal contributions and the limited number of cell divisions required of the pluripotent cells. Thus, the status of H3K4 methylation in vivo, once established in the ICM, may not require much maintenance or modification until gastrulation. However, the loss of PTIP in ES cells cultured in vitro may result in a gradual decrease of H3K4 methylation marks due to the increased number of cell divisions incurred.
While the necessity of Oct4 expression for maintaining pluripotency in ES cells or establishing pluriotency in iPS cells is clear, the regulation of oct4 is less well understood. Evidence for regulation by the spalt homologue Sall4 includes direct binding to a distal oct4 enhancer and transactivation 32. More recently, the orphan nuclear receptor, Esrrb, was also shown to bind to oct4 promoter sequences to maintain expression in ES cell lines 33 and can function to reprogram fibroblasts into iPS cells when expressed with Oct4 and Sox2 34. However, it is not clear whether loss of these factors is ultimately responsible for oct4 down regulation upon differentiation.
Our previous studies with PTIP indicate that it is necessary for linking the DNA binding protein Pax2 to the MLL2/3 histoneH3K4 methyltransferase complex 20. In Xenopus animal caps, the PTIP homologue Swift was shown to bind to P-Smad to direct mesoderm development 35. It is unlikely that PTIP regulates oct4 through a Pax protein, as Pax genes are not expressed at high levels in ES cells. Indeed, we were unable to immunoprecipitate PTIP directly to the oct4 promoter sequences that show reduced levels of H3K4 methylation. This may reflect a lack of sensitivity of our anti-PTIP antibodies. Alternatively, the reduction of H3K4 methylation could reflect the decreased transcription levels from the oct4 locus due to the loss of an activator protein that is directly under the control of PTIP. However given the global reduction of H3K4 methylation observed in the PTIP– ES cells and in the germline and neural stem cell specific PTIP mutants described previously 20, the data strongly argue for a direct affect on the epigenetic marks of many genes by the PTIP containing complexes.
In summary, loss of PTIP in cultured ES cells results in reduced levels of H3K4 methylation, including at the oct4 and sox2 promoter, reduced Oct4 protein and mRNA expression and spontaneous differentiation towards the trophectoderm lineage. These results suggest that the continued activities of H3K4 methylation complexes are needed to maintain ES cell pluripotency in vitro.
Acknowledgments
We thank Elizabeth Hughes and Thom Saunders of the University of Michigan Transgenic Animal Core Facility for their help in generating the conditional allele and the ES cell lines. This work was supported by NIH grants DK054740 and DK073722 to G. R. D. and DK02803 and DK039255 to S. R. P. D.K. was the recipient of an American Foundation for Urological Research Fellowship.
References
- 1.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 5.O’Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 6.O’Carroll D, Erhardt S, Pagani M, et al. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and cellular biology. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faust C, Lawson KA, Schork NJ, et al. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development (Cambridge, England) 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- 8.Voncken JW, Roelen BA, Roefs M, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu BD, Hess JL, Horning SE, et al. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- 10.Terranova R, Agherbi H, Boned A, et al. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaser S, Schaft J, Lubitz S, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development (Cambridge, England) 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 12.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 13.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Current opinion in cell biology. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guenther MG, Levine SS, Boyer LA, et al. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura T, Mori T, Tada S, et al. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Molecular cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 16.Miller T, Krogan NJ, Dover J, et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Issaeva I, Zonis Y, Rozovskaia T, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Molecular and cellular biology. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho YW, Hong T, Hong S, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. The Journal of biological chemistry. 2007 doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho EA, Prindle MJ, Dressler GR. BRCT domain-containing protein PTIP is essential for progression through mitosis. Molecular and cellular biology. 2003;23:1666–1673. doi: 10.1128/MCB.23.5.1666-1673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel SR, Kim D, Levitan I, et al. The BRCT-Domain Containing Protein PTIP Links PAX2 to a Histone H3, Lysine 4 Methyltransferase Complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, Wang M, Cai Q, et al. Pax transactivation-domain interacting protein is required for urine concentration and osmotolerance in collecting duct epithelia. J Am Soc Nephrol. 2007;18:1458–1465. doi: 10.1681/ASN.2006060625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 23.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 25.Loh YH, Zhang W, Chen X, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes & development. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nature cell biology. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 27.Milne TA, Briggs SD, Brock HW, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 28.Ernst P, Fisher JK, Avery W, et al. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6:437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 29.Lubitz S, Glaser S, Schaft J, et al. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Molecular biology of the cell. 2007;18:2356–2366. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 31.Velkey JM, O’Shea KS. Oct4 RNA interference induces trophectoderm differentiation in mouse embryonic stem cells. Genesis. 2003;37:18–24. doi: 10.1002/gene.10218. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Tam WL, Tong GQ, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nature cell biology. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Zhang J, Wang T, et al. Esrrb activates Oct4 transcription and sustains self-renewal and pluripotency in embryonic stem cells. The Journal of biological chemistry. 2008;283:35825–35833. doi: 10.1074/jbc.M803481200. [DOI] [PubMed] [Google Scholar]
- 34.Feng B, Jiang J, Kraus P, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nature cell biology. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu K, Bourillot PY, Nielsen SJ, et al. Swift is a novel BRCT domain coactivator of Smad2 in transforming growth factor beta signaling. Molecular and cellular biology. 2001;21:3901–3912. doi: 10.1128/MCB.21.12.3901-3912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]