Abstract
DNA topoisomerases control DNA topology by breaking and rejoining DNA strands via covalent complexes with cleaved DNA substrate as catalytic intermediates. Here we report the structure of Escherichia coli topoisomerase I catalytic domain (residues 2–695) in covalent complex with a cleaved single-stranded oligonucleotide substrate, refined to 2.3-Å resolution. The enzyme-substrate intermediate formed after strand cleavage was captured due to the presence of the D111N mutation. This structure of the covalent topoisomerase-DNA intermediate, previously elusive for type IA topoisomerases, shows distinct conformational changes from the structure of the enzyme without bound DNA and provides detailed understanding of the covalent catalysis required for strand cleavage to take place. The portion of cleaved DNA 5′ to the site of cleavage is anchored tightly with extensive noncovalent protein–DNA interactions as predicted by the “enzyme-bridged” model. Distortion of the scissile strand at the -4 position 5′ to the cleavage site allows specific selectivity of a cytosine base in the binding pocket. Many antibacterial and anticancer drugs initiate cell killing by trapping the covalent complexes formed by topoisomerases. We have demonstrated in previous mutagenesis studies that accumulation of the covalent complex of bacterial topoisomerase I is bactericidal. This structure of the covalent intermediate provides the basis for the design of novel antibiotics that can trap the enzyme after formation of the covalent complex.
Keywords: protein–DNA complex, topA, DNA relaxation, cytosine recognition
A hallmark in reactions of all DNA topoisomerases is the formation of a covalent intermediate, in which an enzyme tyrosyl residue is covalently linked to a DNA phosphoryl group (1). These enzymes play a vital role in modulating DNA supercoiling and in untangling intracellular DNA during various cellular transactions (2). Structural information on the covalent intermediates between members of the topoisomerase family and their DNA substrates is important not only in understanding how these enzymes manage the passage of DNA strands or double helices through one another, but also in the design of antimicrobial and antitumor agents that target the DNA topoisomerases. Compounds that result in the accumulation of the covalent complexes formed by type IIA and type IB topoisomerases, referred collectively as topoisomerase poisons, have been utilized extensively in antibacterial and anticancer therapy (3–5). All bacteria contain at least one type IA topoisomerase, usually topoisomerase I, along with at least one type IIA topoisomerase, usually DNA gyrase (6). We have shown in previous genetic studies that trapping of bacterial topoisomerase I covalent complex intermediate due to mutations inhibiting DNA rejoining can result in rapid bacterial cell death (7, 8) in a mechanism involving reactive oxygen species (9). Bacterial topoisomerase I is thus a validated target that could be utilized for discovery of leads for antibacterial drugs that stabilize its covalent complex (10). The emergence of bacterial pathogens resistant to current antibacterial therapies represents a global public health challenge. Lack of drugs that can be used to treat multidrug resistant gram negative pathogens (11–14) results in alarming incidences of infections resistant to virtually all currently available antibiotics (15, 16). Antibacterial lead compounds against previously unutilized targets are urgently needed for the development of the next generation of antibiotics.
Structure of the covalent complex of a type IA topoisomerase that shows the crucial interactions with the 5′ and 3′ ends of cleaved DNA has not been available previously (17). This represents a significant barrier for the elucidation of the catalytic mechanism and antibiotics discovery against this target. Here we report the structure of Escherichia coli topoisomerase I N-terminal catalytic domain (residues 2–695) in covalent complex with a substrate oligonucleotide stabilized by a mutation in the topoisomerase–primase (TOPRIM) motif that inhibits DNA rejoining (8). The structure, refined to nominal resolution of 2.3 Å, captured the enzyme during catalysis after initial strand cleavage and before stand rejoining. The structure of the covalent intermediate displays a conformation distinct from the previously determined structures of E. coli topoisomerase I catalytic domain without bound DNA (17). A structure of type IB human topoisomerase I in covalent complex with cleaved duplex DNA is available (18). However, type IA and type IB topoisomerases differ mechanistically not only in the covalent linkage to 5′ and 3′ phosphoryl ends of cleaved DNA, respectively, but also in the interaction with the free DNA end formed after DNA cleavage that is not covalently linked to the enzyme in the strand passage mechanism. This previously undescribed structure of a type IA topoisomerase covalent complex provided not only detailed understanding of the catalytic mechanism but also the structural basis for the design of antibiotics that can stabilize this important covalent intermediate.
Results
Structure of Covalent Complex.
Previous studies in our laboratory have demonstrated that a mutation of the strictly conserved acidic residue D111 (D111N) in E. coli topoisomerase I TOPRIM motif slowed down the rejoining step of the catalysis and consequently led to an extremely lethal phenotype (8). A full-length enzyme with this D111N mutation could not be expressed, but the N-terminal 67-kDa transesterification domain with this mutation was purified and shown to form a stabilized covalent complex (8). We incubated this purified 67-kDa enzyme with a 13mer oligo (5′-AATGCGCT↓TTGGG-3′) with a strong topoisomerase cleavage site indicated by the arrow. The reaction was quenched with excess EDTA to chelate the magnesium ions required for religation. The catalytic intermediate formed by the mutant enzyme was separated from the free enzyme and crystallized. The structure of the complex (Fig. 1) was solved by molecular replacement method with the wild-type apo structure (19) [Protein Data Bank (PDB) ID code 1ECL] and subsequently refined to 2.3-Å resolution with a final R factor of 23% (Rwork) and 27% (Rfree) (Table S1). Crystals of the D111N mutant form of the protein alone were also obtained for structural determination, and the structure was refined to 1.9-Å resolution with a final R factor of 22% (Rwork) and 25% (Rfree) (Table S1). The conservative Asp to Asn substitution at position 111 did not result in noticeable change in the overall structure when the mutant apo structure was compared to that of the wild-type form (rmsd of 0.5 Å for all atoms).
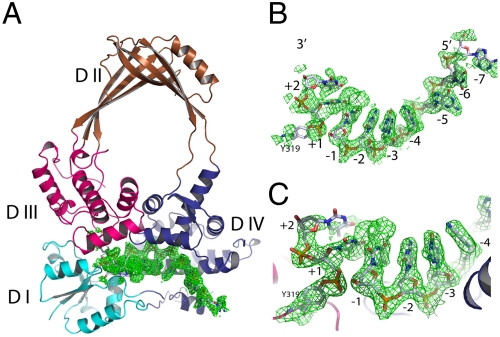
Fig. 1.
Structure of the topoisomerase I and DNA covalent complex. (A) Ribbon representation of the overall structure with four domains shown in different colors. The bound DNA fragments are shown with electron density map (green, 2fo-fc map at 1.5σ cutoff). (B) Annealed difference map (fo-fc, 1.5σ cutoff) around the DNA fragments. The difference map was calculated in the absence of bound DNA. (C) Closer view of the active site of the covalent intermediate. The continuous density (final 2fo-fc map) from Y319 to the 3′ dinucleotide (TT) is shown at 1.0σ cutoff. A clear density break of scissile bond is shown.
The overall structure of the protein in the covalent complex maintains the toroidal shape characteristic of type IA topoisomerases (Fig. 1A). The density of the ssDNA is well-defined after initial annealing refinement without a built-in DNA model (Fig. 1B) with a clear break at the expected position. The covalent bond between the catalytic residue Tyr319 and one DNA fragment is unmistakable as demonstrated by the continuous density from the tyrosine side chain to the phosphate group of the nucleotide (Fig. 1C). However, the spacing between the 5′ and 3′ ends of the cleaved DNA would not accommodate passage of another DNA single strand or duplex through the break without additional conformational change. Of the 13 bases from the original oligo, 7 of them are clearly defined by the electron density in the initial analysis, with reasonable density for one additional base at each end of the substrate. Therefore, a total of nine bases were modeled in the final structure. There are additional densities toward both 5′ and 3′ ends. However, they are not sufficiently well-defined to warrant further base models. The cleavage site of this oligo was established previously in our laboratory and the electron densities are in good agreement with two T nucleotides seen for the fragment 3′ to the cleavage site covalently linked to Tyr319, whereas seven bases can be seen for the fragment 5′ to the cleavage site bound within domain IV.
Conformational Changes from the apo Structure.
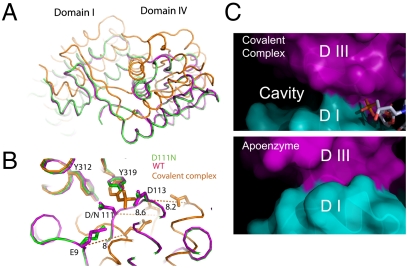
The enzyme–DNA complex structure corresponds to the covalent intermediate formed during catalysis. According to the “enzyme-bridged” strand passage model (19–21), domain II and domain III are about to move to create the space for the passage of the other strand. In comparison with the apo structure, the relative orientations of domains are similarly positioned when looking into the ssDNA binding groove (Fig. S1). The interface between domains III and IV does not change significantly either when the structures of the covalent complex and apo enzyme are compared. When individually aligned, domains II and III are virtually identical to these domains in the wild-type apo structure with rmsd of 0.55 Å and 0.87 Å, respectively, for Cα atoms. The relative orientation of these two domains is also retained in comparison with that of the apo structure (rmsd of 0.86 Å). However, from the side view of the structures, the domain orientations of the covalent complex undergo significant rearrangement compared with the apo structure (Fig. 2A and Fig. S2a). When domains II and III are aligned, domain I in the complex moved down about 2 Å and back about 6 to 10 Å (Fig. S2b and Movies S1 and S2). The relative domain movements result in rearrangement of the three key acidic residues in the TOPRIM domain (E9, D111, and D113) (Fig. 2B). When domains III are aligned, the nucleophilic tyrosine residue (Y319) and its neighboring Y312 are positioned similarly, whereas the three acidic residues shifted more than 8 Å in the covalent complex in comparison with their positions in the apo structure. Furthermore, in the covalent complex structure, domain I and domain III have limited contact due to domain I movement, which creates a significant cavity (15 × 10 × 12 Å3, Fig. 2C) between them. Domain IV undergoes significant rearrangement as well with rmsd of 1.5 Å (Cα) when aligned with domain IV of the apo enzyme. The top portion of domain IV rotates backward about 5°, whereas the bottom two helices moved backward about 6 Å (Fig. S2c). The domain rearrangements created a deep groove for ssDNA binding.
Fig. 2.
Structural alignment of the wild-type enzyme (cyan), D111N mutant (green), and the enzyme–DNA covalent complex (orange). (A) Domain I and domain IV shift significantly upon the formation of the enzyme–DNA covalent complex when domain IIIs are aligned, whereas the mutation of D111N exhibits no disruption of the overall structure. (B) Active site residues undergo a dramatic reconfiguration upon the formation of the covalent complex. When the domains III are aligned, the three critical acidic residues (E9, D111, and D113) shift about 8 Å away. (C) The domain rearrangement upon the formation of the complex (Top) creates a clear cavity between domains I and III in comparison with the apo enzyme (Bottom). The DNA fragments bound in the covalent complex are drawn as sticks for reference. The structural alignment is optimized for the overlapping of domains II and III. The shift of domain I upon the formation of the covalent complex reduces the interaction between domain I and domain III. The buried surface area between domain I and domain III decreases from 1,057 Å2 for the apo enzyme to 411 Å2 for the covalent complex.
Binding of DNA Substrate.
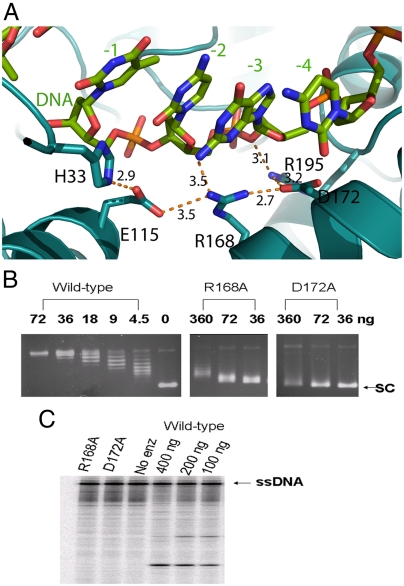
The portion of the oligonucleotide 5′ to the cleavage site binds in a deep groove formed at the interface of domains I and III as well as within domain IV, in a similar orientation as the oligonucleotide bound noncovalently to topoisomerase III (22) (Fig. S3). The phosphate groups bind deep into the groove through hydrogen bonds and salt bridges with the protein. Four of the phosphate groups (positions +1, -1, -2, and -4) are held in position by four arginine side chains (R321, R493, R195, and R507; Fig. S4). R195 and R321 have been shown in previous mutagenesis studies to be required for DNA cleavage by E. coli topoisomerase I (23–25). In addition, there is a network of ionic and hydrogen bond interactions involving the deoxyribose oxygens, E115, R168, and D172 (Fig. 3A) to hold the DNA substrate in place. Alanine substitution of E115 has been shown to reduce DNA cleavage by E. coli topoisomerase I (23, 24). The structure shown here supports a role of E115 independent of potential interaction with divalent ions. We expressed and purified R168A and D172A mutants of E. coli topoisomerase I. Results from biochemical assays showed that these mutant enzymes have > 80-fold loss of relaxation activity when compared to wild type (Fig. 3B). Cleavage assay using 5′-end labeled single-stranded DNA showed no detectable DNA cleavage activity (Fig. 3C). These biochemical results provide further evidence that these residues are critical for positioning the substrate for DNA cleavage to take place.
Fig. 3.
Interaction with ribose rings of DNA substrate at −2 and −3 position. (A) Network of hydrogen bonds involving E115, R168, and D172 and the bound oligonucleotide. The interactions among H33, E115, R168, D172, and R195 stabilize the side-chain conformations of these residues, which in turn orient the DNA substrate in proper conformation for cleavage or rejoining. (B) Comparison of relaxation activity of wild-type E. coli topoisomerase I and R168A, D172A mutant enzymes by agarose gel electrophoresis. The amount of enzyme (ng) added to each reaction is shown. SC: supercoiled plasmid DNA. (C) Assay of cleavage of 5′-32P-labeled ssDNA by R168A and D172A mutant topoisomerase I (400 ng each) and wild-type enzyme (amounts shown). After electrophoresis in sequencing gel, the labeled substrate and cleavage products were visualized with PhosphorImager.
Molecular Basis for Selectivity of a Cytosine Base at the -4 Position.
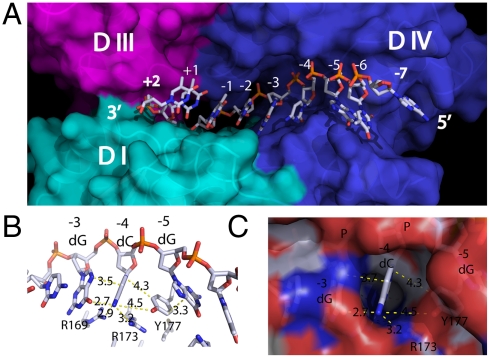
Bacterial topoisomerase I and topoisomerase III enzymes have distinct cleavage sequence selectivity (26). Bacterial topoisomerase I enzymes have been shown to cleave DNA with a cytosine nucleotide 4 bases 5′ to the cleavage site. The basis of this sequence selectively was previously unknown but is now revealed in the structure of the covalent complex. Seven bases (from -6 to +1) form hydrogen bonds with the topoisomerase residues. These bases are relatively buried into the binding groove, precluding them from base pairing in the formation of double-stand DNA. This segment of the DNA substrate mainly interacts with domain IV plus a few hydrogen bonds with domain I (Fig. 4A). Even though the oligonucleotides in the previously determined structures of topoisomerase III and topoisomerase I complexes follow the conformation of B-DNA, a difference in the oligonucleotide substrate conformation can be observed between bases -4 and -5. In this covalent complex structure of topoisomerase I, there is a distinctive kink between -4 and -5 position in the phosphate–deoxyribose chain. This kink disrupted the stacking of the bases. Instead, the side chain of Y177 forms a wedge between cytosine (-4) and guanine (-5) (Fig. 4B). The aromatic phenol ring (Y177) stacks parallel to the guanine (-5) and almost perpendicular to the cytosine (-4). As a matter of fact, this unique kink as well as residue 177 forms a sterically restrictive cavity at the -4 position that can accommodate only a cytosine (Fig. 4C). R169 and R173 also interact with the cytosine base via hydrogen bonds. These interactions explain the substrate selectivity of E. coli topoisomerase I recognizing a cytosine at the -4 position for cleavage (27, 28). Alignment of residues in this region showed that R169, R173, and Y177 are conserved in bacterial as well as archael topoisomerase I and reverse gyrase sequences with either a tyrosine or phenylalanine found at the position corresponding to Y177 (shown in blue in Fig. S5). The cleavage sites of these type IA topoisomerases all have a cytosine at the -4 position. Topoisomerase III enzymes have different amino acid residues present at positions corresponding to R169, R173, and Y177 in the alignment (Fig. S5) and cleave DNA with different sequence selectivity. In contrast, R168 and D172, which are involved in interaction with the deoxyribose, are conserved in all type IA topoisomerases (shown in red in Fig. S5).
Fig. 4.
Structural determinants for substrate binding and specificity. (A) Surface representation of the DNA binding groove. The cleavage site sits on the border between domain III and domain IV. (B) The phenol side chain of Y177 wedges between bases -4 and -5 and forms π-interactions among them. (C) The cavity formed at the -4 position can accommodate only a cytosine.
Catalytic Mechanism of DNA Cleavage/Religation.
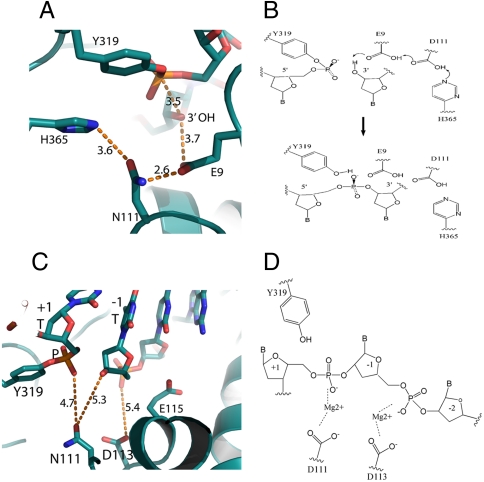
Our structure captured the protein–DNA interaction before the rejoining step and provides a clear indication of the importance of the strictly conserved glutamate residue E9 in DNA cleavage–religation by type IA topoisomerases. Previously, Perry and Mondragon (29) have proposed a mechanism in which E9 acts as a general base in DNA cleavage, assisted by charge relay through D111 and H365. In the present structure, the positions of E9, D111, and H365 are consistent with such a scheme (Fig. 5 A and B). However, replacing E9 by a glutamine did not abolish the DNA cleavage activity of the enzyme (24), and thus further studies are needed to clarify the precise role of E9.
Fig. 5.
Mechanism of catalysis. (A) The distances among the proposed catalytic residues are optimal for potential shuttling of the protons. (B) Proposed mechanism of religation involving residues E9, D111, and H365. (C) The side chains of D111 and D113 are about 5 Å away from the scissile phosphate and +1 position phosphate, respectively. The distances can accommodate two magnesium ions. (D) Proposed function of magnesium ions. The two magnesium ions coordinate with phosphates as well as the 3′-hydroxy group (after cleavage) to hold the DNA chains in position for catalysis.
The acidic triad DxDxE residues in the TOPRIM domain of E. coli topoisomerase I are known to be involved in divalent ion coordination and DNA cleavage–religation (8, 30). The side chain of D113 is about 5 Å away from the -1 phosphate, whereas the side chain of D111 (seen as N111 in this structure) is about 5 Å away from the +1 phosphate and 5 Å away from the 3′ nucleophilic hydroxyl group after cleavage (Fig. 5C). The presence of magnesium ions would help to hold the scissile bond in position for cleavage (Fig. 5D). Crystal structures of Saccharomyces cerevisiae topoisomerase II with divalent ions bound to side chains of aspartate or glutamate (31, 32) have led to a unified model of the role of two divalent ions in DNA cleavage–religation by type IIA and type IA topoisomerases (32). After cleavage by topoisomerase I, the ribose at the +1 position possesses no interactions with the enzyme, and therefore the presence of Mg2+ would be critical to hold the 3′ hydroxyl group in position for the rejoining.
Discussion
An enzyme-bridged model has long been proposed for DNA strand passage catalyzed by both type IA and type II topoisomerases (20). In this model, the free DNA end on the cleaved strand not covalently linked via the phosphoryl–tyrosine linkage is held tightly by noncovalent interactions with the enzyme for type IA topoisomerases, instead of being allowed to rotate around the noncleaved strand in the “controlled rotation” strand passage mechanism of type IB topoisomerases (33). This accounts for the change in linking number in steps of one for type IA topoisomerase (34, 35) but in multiples of one in each catalytic cycle for type IB topoisomerases (36, 37). The controlled rotation of human topoisomerase I is proposed to be brought about by ionic interactions between the DNA phosphates and a positively charged DNA-proximal surface on the enzyme (33). In this structure of the type IA covalent complex, the cleaved oligonucleotide 5′ to the cleavage site is buried in a deep groove, and the ribose and phosphates are held with a network of hydrogen bonds and salt bridges. In addition, a binding pocket specific for a cytosine base at the -4 position relative to the site of cleavage was created by stacking interaction with an aromatic residue (Y177) and a kink in the phosphodiester backbone. The extensive single-stranded region in DNA from negative supercoiling may facilitate the distortion of the scissile strand. The significance of the base specific interactions contributed by R169, R173, and Y177 in catalysis of removal of negative supercoiling will be further explored in future site-directed mutagenesis studies.
It should be noted that the phosphodiester bond closest to the active site Tyr328 in the “open structure” of wild-type topo III or Phe328 of mutant topo III noncovalently bound to ssDNA is not the cleavage site expected from the sequence selectivity of topo III (22, 38). It is not clear if sequence selectivity for positioning the expected scissile phosphate for cleavage by topo III would involve further changes in the topo III–DNA complex.
The structure of the covalent complex of DNA topoisomerase I also demonstrated the precise domain rearrangements to accommodate the covalent bond formation and catalysis. For the structures of E. coli topoisomerase III bound noncovalently to ssDNA (22, 38), the transition of the “closed structure” (apo enzyme) to the open structure is accompanied by significantly increased potential for domain II mobility as indicated by the increase in its temperature factors, which would suggest that domain II is ready to open the cavity for strand passage. In our topoisomerase I covalent complex structure, the temperature factors of domains II, III, and IV (48.9 Å2, 40.8 Å2, and 40.5 Å2, respectively) are all below the overall temperature factor of the whole protein (50.2 Å2), whereas the temperature factor of domain I increased drastically (80.5 Å2). This is also supported by the fact that the electron density of domain I is less well-defined than the rest of the protein. The temperature factor distribution of the mutant structure without DNA is similar to that of the wild-type structure. Granted, temperature factor variations could be a result of crystal packing differences. However, in our structure we suspect that the conformational change in domain I may play an important role in catalysis. This could suggest that there might be differences in the strand passage mechanism between topoisomerase I and topoisomerase III despite the sequence and structural similarities between these two enzymes. The physiological function of E. coli topoisomerase I is to remove excess negative supercoiling from DNA, and it has a much weaker decatenation activity when compared to E. coli topoisomerase III (26). It is possible that the strand passage mechanisms of these two enzymes have evolved to be optimal for these two separate catalytic activities.
The structure of the topoisomerase I covalent complex provides the potential target sites for the design of pharmaceuticals to stabilize this reaction intermediate. For inhibitors of the type IB human topoisomerase I, binding at the interface of the topoisomerase–DNA covalent complex is often observed (39). Such interfacial interactions may also inhibit DNA rejoining by type IA topoisomerases. Alternatively, to avoid potential toxic effects of DNA binders, small molecules that bind to the interface cavity between domains I and III unique to the covalent complex may also stabilize the covalent complex, thus trapping the reaction intermediate. This covalent complex structure should greatly facilitate future in silico screening and modeling studies for discovery of antibacterial drugs targeting topoisomerase I.
Materials and Methods
Protein Purification and Crystallization.
Recombinant protein of E. coli topoisomerase I 67 kDa N-transesterification domain mutant D111N was expressed (8) and purified as previously reported (40). An additional step of gel filtration chromatography (Sephacryl S-200) was included at the end of purification. To obtain the covalent complex, mutant protein was incubated with a synthetic oligonucleotide (supplied by Sigma-Aldrich) of sequence 5′AATGCGCT↓TTGGG3′ (topoisomerase I preferred cleavage site indicated by arrow) at a molar ration of 1∶3 (protein∶oligi) in buffer A (20 mM potassium phosphate, pH 7.4, 1 mM DTT, 1 mM EDTA, 10% glycerol) with 5 mM MgCl2 at 16 °C for 5 h. The reaction was terminated by the addition of 5 mM EDTA. The resulting reaction mixture was loaded onto a DE52 column (from Whatman) equilibrated with buffer A. After extensive washing to remove the free protein, the bound covalent complex was eluted with buffer A containing 250 mM NaCl and concentrated to 10 mg/mL with the Bio-Scale Mini Macro-Prep DEAE cartridge (from Bio-Rad) for crystallization. Initial crystallization conditions were identified with Crystal Screen kits (Hampton Research). The covalent complex crystals used for data collection were obtained with polyethylene glycol 4000 as precipitant (100 mM acetate, pH 5.0, 0.2 M ammonium sulfate, 25% PEG 4000), whereas the apo-enzyme crystals were obtained with 2 M ammonium sulfate as precipitant.
Data Collection, Structure Solution, and Refinement.
The covalent complex crystals were frozen in liquid propane in the presence of 10% glycerol as cryoprotectant, whereas the apo-enzyme crystals had 2 M lithium sulfate as cryoprotectant. Diffraction data were collected on the X-ray Operations and Research beamline 19ID at the Advanced Photon Source, Argonne National Laboratory, and processed with the HKL-3000 system (41). Both crystals belong to the space group P212121. The covalent complex crystals were incubated with various metal ions, and none of them induced any structural or cell dimensional changes, likely because the pH of 5.0 was too low for the acidic residues in the active site to bind the divalent ions. The crystals soaked with 0.2 mM mercury chloride gave rise to the highest resolution and therefore were used for the final refinement of the structures. Initial molecular replacement using the wild-type apo structure (PDB ID code 1ECL) with the CCP4 package (42) produced a clear solution and the densities of domains II and III were well-defined. However, electron density for domain I was poor. The apo structure was then divided into two parts with one part containing domains II and III as well as part of domain IV (residues 185 to 560) and the other part containing domain I and the last two helices of domain IV (N-terminal and C-terminal combined). Further molecular replacement with these two separate structures using crystallography and NMR system (CNS) (43) produced clear solutions with excellent densities. After simulated annealing with CNS, the density of the oligonucleotide was clearly interpretable. The structures were refined with CNS. The D111N apo structure was solved with direct molecular replacement using the wild-type apo structure (PDB ID code 1ECL), and refinements were carried out with CNS. The coordinates and structure factors were deposited at the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank under PDB ID codes 3PX7 (D111N covalent complex) and 3PWT (D111N apo-enzyme).
Analysis of Alanine Substitution at R168 and D172.
Full-length E. coli topoisomerase I enzymes with R168A or D172A mutation introduced by oligonucleotide-directed mutagenesis were expressed and purified to > 95% homogeneity as described previously (44). Relaxation activity was assayed in a reaction volume of 20 μL with 10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.1 mg/mL gelatin, 6 mM MgCl2, and 0.5 μg of CsCl gradient-purified supercoiled plasmid DNA at 37 °C for 30 min. Single-stranded DNA substrate generated by PCR with 5′-32P label was used for DNA cleavage assay as previously described for other topoisomerase I mutants (45).
Supplementary Material
Acknowledgments.
We thank Dr. Jingyang Feng for purification and relaxation assay of mutant topoisomerases, and Dr. James C. Wang and Dr. James M. Berger for helpful suggestions. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357. This work was supported by grants from the US Public Health Service [National Institutes of Health Grants AI-069313 and GM-054226 (to Y.T.)].
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank, www.pdb.org/pdb/home/home.do [RCSB ID codes 3PX7 (D111N covalent complex) and 3PWT (D111N apo-enzyme)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1100300108/-/DCSupplemental.
References
- 1.Schoeffler AJ, Berger JM. DNA topoisomerases: Harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradbury BJ, Pucci MJ. Recent advances in bacterial topoisomerase inhibitors. Curr Opin Pharmacol. 2008;8:574–581. doi: 10.1016/j.coph.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng B, Shukla S, Vasunilashorn S, Mukhopadhyay S, Tse-Dinh YC. Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J Biol Chem. 2005;280:38489–38495. doi: 10.1074/jbc.M509722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng B, et al. Asp-to-Asn substitution at the first position of the DxD TOPRIM motif of recombinant bacterial topoisomerase I is extremely lethal to E. coli. J Mol Biol. 2009;385:558–567. doi: 10.1016/j.jmb.2008.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu IF, Annamalai T, Sutherland JH, Tse-Dinh YC. Hydroxyl radicals are involved in cell killing by the bacterial topoisomerase I cleavage complex. J Bacteriol. 2009;191:5315–5319. doi: 10.1128/JB.00559-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tse-Dinh YC. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res. 2009;37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engel LS. The dilemma of multidrug-resistant gram-negative bacteria. Am J Med Sci. 2010;340:232–237. doi: 10.1097/MAJ.0b013e3181e939c3. [DOI] [PubMed] [Google Scholar]
- 12.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adams MD, Chan ER, Molyneaux ND, Bonomo RA. Genome-wide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:3569–3577. doi: 10.1128/AAC.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres C. Up against the wall. Nat Med. 2010;16:628–631. doi: 10.1038/nm0610-628. [DOI] [PubMed] [Google Scholar]
- 15.Rolain JM, Parola P, Cornaglia G. New delhi metallo-beta-lactamase (NDM-1): Towards a new pandemia? Clin Microbiol Infec. 2010;16:1699–1701. doi: 10.1111/j.1469-0691.2010.03385.x. [DOI] [PubMed] [Google Scholar]
- 16.Bush K. Alarming beta-lactamase-mediated resistance in multidrug-resistant enterobacteriaceae. Curr Opin Microbiol. 2010;13:558–564. doi: 10.1016/j.mib.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Baker NM, Rajan R, Mondragon A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009;37:693–701. doi: 10.1093/nar/gkn1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 19.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 20.Brown PO, Cozzarelli NR. Catenation and knotting of duplex DNA by type 1 topoisomerases: A mechanistic parallel with type 2 topoisomerases. Proc Natl Acad Sci USA. 1981;78:843–847. doi: 10.1073/pnas.78.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger JM, Fass D, Wang JC, Harrison SC. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc Natl Acad Sci USA. 1998;95:7876–7881. doi: 10.1073/pnas.95.14.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 23.Zhu CX, Roche CJ, Papanicolaou N, DiPietrantonio A, Tse-Dinh YC. Site-directed mutagenesis of conserved aspartates, glutamates and arginines in the active site region of Escherichia coli DNA topoisomerase I. J Biol Chem. 1998;273:8783–8789. doi: 10.1074/jbc.273.15.8783. [DOI] [PubMed] [Google Scholar]
- 24.Chen SJ, Wang JC. Identification of active site residues in Escherichia coli DNA topoisomerase I. J Biol Chem. 1998;273:6050–6056. doi: 10.1074/jbc.273.11.6050. [DOI] [PubMed] [Google Scholar]
- 25.Cheng B, Feng J, Mulay V, Gadgil S, Tse-Dinh YC. Site-directed mutagenesis of residues involved in G strand DNA binding by Escherichia coli DNA topoisomerase I. J Biol Chem. 2004;279:39207–39213. doi: 10.1074/jbc.M405891200. [DOI] [PubMed] [Google Scholar]
- 26.Viard T, de la Tour CB. Type IA topoisomerases: A simple puzzle? Biochimie. 2007;89:456–467. doi: 10.1016/j.biochi.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Tse YC, Kirkegaard K, Wang JC. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J Biol Chem. 1980;255:5560–5565. [PubMed] [Google Scholar]
- 28.Dean F, et al. Escherichia coli type-1 topoisomerases: Identification, mechanism, and role in recombination. Cold Spring Harb Sym Quant Biol. 1983;47(Pt 2):769–777. doi: 10.1101/sqb.1983.047.01.088. [DOI] [PubMed] [Google Scholar]
- 29.Perry K, Mondragon A. Biochemical characterization of an invariant histidine involved in Escherichia coli DNA topoisomerase I catalysis. J Biol Chem. 2002;277:13237–13245. doi: 10.1074/jbc.M112019200. [DOI] [PubMed] [Google Scholar]
- 30.Zhu CX, Tse-Dinh YC. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J Biol Chem. 2000;275:5318–5322. doi: 10.1074/jbc.275.8.5318. [DOI] [PubMed] [Google Scholar]
- 31.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 34.Dekker NH, et al. The mechanism of type IA topoisomerases. Proc Natl Acad Sci USA. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dekker NH, et al. Thermophilic topoisomerase I on a single DNA molecule. J Mol Biol. 2003;329:271–282. doi: 10.1016/s0022-2836(03)00320-6. [DOI] [PubMed] [Google Scholar]
- 36.Stivers JT, Harris TK, Mildvan AS. Vaccinia DNA topoisomerase I: Evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 37.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 38.Changela A, DiGate RJ, Mondragon A. Structural studies of E. coli topoisomerase III-DNA complexes reveal a novel type IA topoisomerase-DNA conformational intermediate. J Mol Biol. 2007;368:105–118. doi: 10.1016/j.jmb.2007.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pommier Y. DNA topoisomerase I inhibitors: Chemistry, biology, and interfacial inhibition. Chem Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorokin EP, et al. Inhibition of Mg2+ binding and DNA religation by bacterial topoisomerase I via introduction of an additional positive charge into the active site region. Nucleic Acids Res. 2008;36:4788–4796. doi: 10.1093/nar/gkn460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 42.Collaborative Computational Project N4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 43.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Zhu CX, Tse-Dinh YC. Overexpression and purification of bacterial DNA topoisomerase I. Methods Mol Biol. 1999;94:145–151. doi: 10.1385/1-59259-259-7:145. [DOI] [PubMed] [Google Scholar]
- 45.Cheng B, Feng J, Gadgil S, Tse-Dinh YC. Flexibility at Gly-194 is required for DNA cleavage and relaxation activity of Escherichia coli DNA topoisomerase I. J Biol Chem. 2004;279:8648–8654. doi: 10.1074/jbc.M312095200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.