Abstract
Aminoacyl-tRNA synthetases perform a critical step in translation by aminoacylating tRNAs with their cognate amino acids. Although high fidelity of aminoacyl-tRNA synthetases is often thought to be essential for cell biology, recent studies indicate that cells tolerate and may even benefit from tRNA misacylation under certain conditions. For example, mammalian cells selectively induce mismethionylation of nonmethionyl tRNAs, and this type of misacylation contributes to a cell’s response to oxidative stress. However, the enzyme responsible for tRNA mismethionylation and the mechanism by which specific tRNAs are mismethionylated have not been elucidated. Here we show by tRNA microarrays and filter retention that the methionyl-tRNA synthetase enzyme from Escherichia coli (EcMRS) is sufficient to mismethionylate two tRNA species, 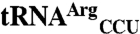 and
and 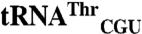 , indicating that tRNA mismethionylation is also present in the bacterial domain of life. We demonstrate that the anticodon nucleotides of these misacylated tRNAs play a critical role in conferring mismethionylation identity. We also show that a certain low level of mismethionylation is maintained for these tRNAs, suggesting that mismethionylation levels may have evolved to confer benefits to the cell while still preserving sufficient translational fidelity to ensure cell viability. EcMRS mutants show distinct effects on mismethionylation, indicating that many regions in this synthetase enzyme influence mismethionylation. Our results show that tRNA mismethionylation can be carried out by a single protein enzyme, mismethionylation also requires identity elements in the tRNA, and EcMRS has a defined structure-function relationship for tRNA mismethionylation.
, indicating that tRNA mismethionylation is also present in the bacterial domain of life. We demonstrate that the anticodon nucleotides of these misacylated tRNAs play a critical role in conferring mismethionylation identity. We also show that a certain low level of mismethionylation is maintained for these tRNAs, suggesting that mismethionylation levels may have evolved to confer benefits to the cell while still preserving sufficient translational fidelity to ensure cell viability. EcMRS mutants show distinct effects on mismethionylation, indicating that many regions in this synthetase enzyme influence mismethionylation. Our results show that tRNA mismethionylation can be carried out by a single protein enzyme, mismethionylation also requires identity elements in the tRNA, and EcMRS has a defined structure-function relationship for tRNA mismethionylation.
Keywords: methionine, aminoacylation, mistranslation, arginine, threonine
Aminoacyl-tRNA synthetases (aaRSs) play a critical role in translation by catalyzing the aminoacylation of tRNAs with their cognate amino acids (1). The aminoacylated tRNAs are used by the ribosome to decode mRNA codons. Misacylation of a tRNA with a noncognate amino acid could result in mistranslation of corresponding codons, thereby introducing mutated residues into proteins (2–4). Extensive studies have shown how the catalytic sites and editing domains of various aaRS ensure extremely high fidelity of aminoacylation on the order of 10-4 and 10-5 (5–8). Extensive work using purified enzymes has also determined a vast library of tRNA identity elements that aaRSs use to discriminate their cognate from noncognate tRNAs (9).
However, an increasing body of recent work has revealed that misacylation and mistranslation may be more prevalent in cells than would be expected (10, 11). Furthermore, there is increasing evidence that a certain level of misacylation may even be beneficial to the cell (12, 13).
tRNA identity elements have been largely studied using biochemical techniques, such as filter-based aminoacylation assays; although powerful, these assays have certain practical limitations when comparing large numbers of different tRNA species (14). Typically, for each aminoacylation reaction a single tRNA species and a single enzyme species are assayed at a time. When a pool of tRNA is used in such an assay, the resulting signal is believed to arise solely from the cognate tRNA in the sample. This technique has been remarkably successful in determining critical identity elements for each aaRS and has allowed the study of aaRS evolution across the three domains of life (9, 15). However, the specific nature of the assay reduces its efficiency for identifying misacylation. The in vitro transcription or purification of all noncognate tRNAs that could be substrates for a particular aaRS enzyme becomes prohibitively time-consuming and thus the actual scale of misacylation has not been sufficiently explored. Additionally, tRNA transcripts lack posttranslational modifications that may influence misacylation (16).
Emerging techniques such as tRNA microarrays can complement traditional aminoacylation assays and resolve the aminoacylation state of individual tRNA species in complex samples. tRNA microarrays are glass slides with oligonucleotide probes attached to their surface. Each probe sequence is complementary to a particular tRNA, and hybridization conditions are sufficiently stringent to allow discrimination between two tRNAs that differ by eight or more nucleotides/modifications (17, 18). An array can contain spots for all different tRNAs for a given organism. In practice, each array contains six or more spots for each tRNA as well as spots corresponding to tRNAs from other organisms as negative controls. The products of aminoacylation reactions using total tRNA and a radiolabeled amino acid are hybridized to the array, and subsequent phosphorimaging reveals which tRNA species have been aminoacylated. This technique screens for all tRNA species that are aminoacylated with the labeled amino acid in parallel allowing detection of both aminoacylated cognate tRNA and misacylated tRNA from the same sample. An obvious benefit of this “shotgun” approach is that it does not require selecting candidate tRNAs prior to testing for misacylation, thus reducing any observational bias from studying only those noncognate tRNA species that appear similar to the cognate species. Additionally, there is no need to transcribe, purify, and assay each tRNA individually, because a pool of total tRNA can be assayed in a single microarray. Once misacylated tRNAs are identified by microarrays, filter-based aminoacylation can be used to explore the effect of single nucleotide differences between wild-type and mutant tRNA transcripts to determine potential identity elements for misacylation.
Microarray assays have revealed extensive mismethionylation in mammalian cells (13). In unstressed cells, approximately 1% of the methionylated tRNAs were noncognate and mismethionylation was further increased by up to 10-fold when cells were exposed to various forms of oxidative stress. Some methionine residues were hypothesized to serve a protective function for cellular proteins against inactivation by reactive oxygen species (19, 20). Mismethionylation and subsequent utilization of mismethionylated tRNAs in translation could enable cells to generate protein libraries with protective methionine residues introduced at different positions. However, the previous study did not identify enzyme(s) responsible for mismethionylating tRNAs or how particular tRNAs from the cellular pool of noncognate tRNAs were selectively mismethionylated. Methionyl-tRNA synthetase (MRS) seemed a likely candidate for mismethionylation of noncognate tRNAs.
Because aaRSs are among the most ancient proteins and are universally conserved in living organisms, we decided in this work to explore whether mismethionylation might also be present in the bacterial domain of life. We applied microarray techniques to detect mismethionylation by Escherichia coli methionyl-tRNA synthetase (EcMRS) and identified specific, noncognate tRNAs for mismethionylation. We then used traditional filter-based aminoacylation assays with wild-type and mutant tRNA transcripts to confirm the role of EcMRS in mismethionylation and to elucidate a key identity element used by EcMRS to selectively mismethionylate these tRNAs. We also examined the involvement of EcMRS functional domains in controlling mismethionylation.
Results
Full-Length EcMRS and N547MRS Mismethionylate 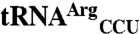 and
and 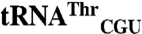 .
.
EcMRS was overexpressed in E. coli with an amino-terminal His6 tag and purified using affinity chromatography to homogeneity. The pure enzyme was used to aminoacylate total tRNA isolated from E. coli with [35S]-methionine. The reaction was assayed with a microarray containing probes for all E.coli tRNAs and numerous yeast tRNA probes as controls. As expected, strong [35S] signals are present for spots corresponding to E. coli elongator (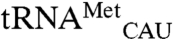 ) and initiator methionine tRNA (
) and initiator methionine tRNA (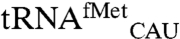 ). In addition to these cognate tRNAs, signal was also seen for E. coli
). In addition to these cognate tRNAs, signal was also seen for E. coli 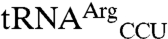 and
and 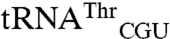 spots (Fig. 1A). Setting the sum of all 35S signals as 100%, the signal for tRNAMet (elongator plus initiator),
spots (Fig. 1A). Setting the sum of all 35S signals as 100%, the signal for tRNAMet (elongator plus initiator), 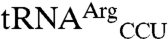 , and
, and 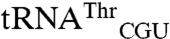 corresponds to 93.6%, 1.3%, and 5.1%, respectively.
corresponds to 93.6%, 1.3%, and 5.1%, respectively.
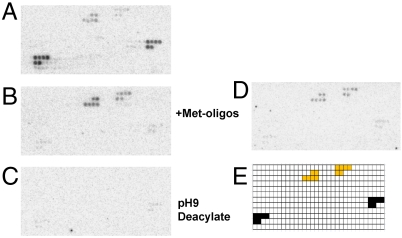
Fig. 1.
Microarray analysis of tRNA mismethionylation with E. coli MRS and total E. coli tRNA. (A–C) Full-length MRS. The aminoacylation reaction mixture was hybridized without (A) or with (B) the presence of excess, free oligonucleotide probes for the initiator and elongator tRNAMet. The same sample was deacylated first before hybridization (C). (D) Truncated MRS. The array includes the presence of excess Met-oligo probes. (E) The array key shows the location of tRNAMet probes in black and misacylated-tRNA probes in orange.
We performed two controls to ensure that the observed signal for the noncognate tRNAs is derived from bona fide mismethionylated tRNAs. First, to rule out cross-hybridization of cognate methionine tRNAs, an excess of free oligonucleotide probes for 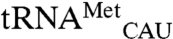 and
and 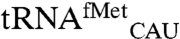 were added to the reaction mixture prior to array hybridization. As expected, this significantly reduced the signals for both tRNAMet spots as the excess oligos sequestered these tRNAs and prevented their hybridization to microarrays. However, signals for
were added to the reaction mixture prior to array hybridization. As expected, this significantly reduced the signals for both tRNAMet spots as the excess oligos sequestered these tRNAs and prevented their hybridization to microarrays. However, signals for 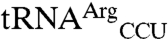 and
and 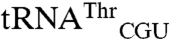 spots were not affected, indicating that these signals were not due to cross-hybridization (Fig. 1B). Second, to confirm that the signals from noncognate tRNAs are derived from aminoacyl tRNAs, the reaction mixture was incubated at pH 9 to hydrolyze the amino acid-tRNA ester linkage prior to array hybridization. As expected, signals for cognate and noncognate tRNAs were completely lost (Fig. 1C).
spots were not affected, indicating that these signals were not due to cross-hybridization (Fig. 1B). Second, to confirm that the signals from noncognate tRNAs are derived from aminoacyl tRNAs, the reaction mixture was incubated at pH 9 to hydrolyze the amino acid-tRNA ester linkage prior to array hybridization. As expected, signals for cognate and noncognate tRNAs were completely lost (Fig. 1C).
Both the dimeric full-length EcMRS (Fig. 1 A–C) and a truncated monomeric form of EcMRS (Fig. 1D) consisting of the N-terminal 547 amino acids mismethionylate the same noncognate tRNAs. This truncated form (N547MRS) is catalytically active and has been used extensively in enzyme kinetic studies (21). The mismethionylation levels for N547MRS are reduced by approximately 2-fold compared to the full-length EcMRS. These results indicate that monomeric EcMRS is sufficient for mismethionylation of 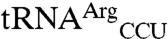 and
and 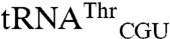 and the carboxy-terminal residues 548–677 are not required for the mismethionylation activity.
and the carboxy-terminal residues 548–677 are not required for the mismethionylation activity.
Anticodon Nucleotides Constitute an Identity Element for Mismethionylation.
To elucidate how EcMRS specifically recognizes its mismethionylated tRNAs, we compared the sequences of 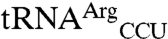 and
and 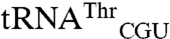 both to tRNAArg and tRNAThr isoacceptors that were not mismethionylated and to elongator tRNAMet (Fig. 2). The non-mis-methionylated
both to tRNAArg and tRNAThr isoacceptors that were not mismethionylated and to elongator tRNAMet (Fig. 2). The non-mis-methionylated 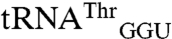 differs from
differs from 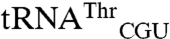 by only 20 bases. The entire D stem and loop are shared between the two isoacceptors and their acceptor stems differ only by a single base pair. However, there is greater heterogeneity in the T stem and anticodon stem (Fig. 2B).
by only 20 bases. The entire D stem and loop are shared between the two isoacceptors and their acceptor stems differ only by a single base pair. However, there is greater heterogeneity in the T stem and anticodon stem (Fig. 2B). 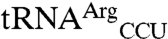 and the non-mis-methionylated isoacceptor
and the non-mis-methionylated isoacceptor 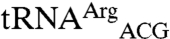 also have different anticodon stems but their T stem and loop are more similar than their acceptor stem or D stem and loop (Fig. 2C). We also compared the mismethionylated tRNAs to cognate
also have different anticodon stems but their T stem and loop are more similar than their acceptor stem or D stem and loop (Fig. 2C). We also compared the mismethionylated tRNAs to cognate 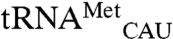 (Fig. 2A) to search for shared identity elements not found on the non-mis-methionylated isoacceptors. The most obvious distinction is the presence of C34N35U36 anticodon sequences among the cognate and mismethionylated tRNAs (A35 for tRNAMet, G35 for tRNAThr, and C35 for tRNAArg; E. coli does not possess a tRNA with C34U35U36). The CAU anticodon was previously shown to be the critical identity element for EcMRS-catalyzed methionylation of cognate tRNAMet (14).
(Fig. 2A) to search for shared identity elements not found on the non-mis-methionylated isoacceptors. The most obvious distinction is the presence of C34N35U36 anticodon sequences among the cognate and mismethionylated tRNAs (A35 for tRNAMet, G35 for tRNAThr, and C35 for tRNAArg; E. coli does not possess a tRNA with C34U35U36). The CAU anticodon was previously shown to be the critical identity element for EcMRS-catalyzed methionylation of cognate tRNAMet (14).
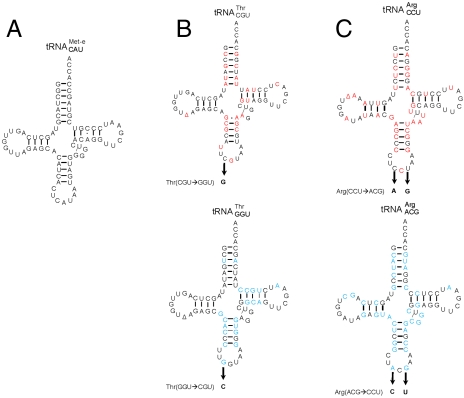
Fig. 2.
Comparison of mismethionylating and non-mis-methionylating tRNA sequences. (A) Elongator tRNAMet, (B) 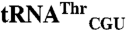 and
and 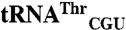 . (C)
. (C) 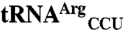 and
and 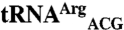 . Differences between tRNAMet and mismethionylating
. Differences between tRNAMet and mismethionylating 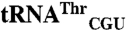 and
and 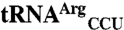 are in red. Differences between mismethionylating and non-mis-methionylating tRNA isoacceptors are in blue. The anticodon mutants are indicated by arrows followed by corresponding sequence changes.
are in red. Differences between mismethionylating and non-mis-methionylating tRNA isoacceptors are in blue. The anticodon mutants are indicated by arrows followed by corresponding sequence changes.
To test the importance of anticodon nucleotides in mismethionylation, we transcribed a series of tRNAs and tested their methionylation activity using the full-length EcMRS. Transcripts were made of the wild-type sequences of 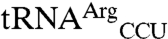
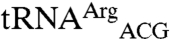 ,
, 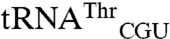 ,
, 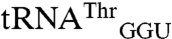 , and
, and 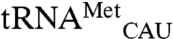 . Four variant transcripts contain anticodon swaps between tRNAThr and tRNAArg isoacceptors:
. Four variant transcripts contain anticodon swaps between tRNAThr and tRNAArg isoacceptors: 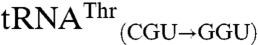 and
and 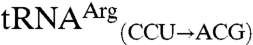 possess the anticodon of the non-mis-methionylated isoacceptor, whereas
possess the anticodon of the non-mis-methionylated isoacceptor, whereas 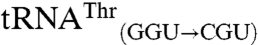 and
and 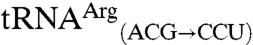 possess the anticodon of the mismethionylated isoacceptor. Filter-based aminoacylation assays, in which aminoacyl-tRNA is precipitated with trichloroacetic acid onto filter paper for scintillation counting, were used to confirm the microarray results and to test tRNA variants for mismethionylation. The in vitro transcribed
possess the anticodon of the mismethionylated isoacceptor. Filter-based aminoacylation assays, in which aminoacyl-tRNA is precipitated with trichloroacetic acid onto filter paper for scintillation counting, were used to confirm the microarray results and to test tRNA variants for mismethionylation. The in vitro transcribed 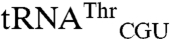 and
and 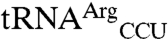 revealed clear mismethionylation above background samples that contained EcMRS but no tRNA (Fig. 3). The mismethionylation level was about 10% of that for cognate
revealed clear mismethionylation above background samples that contained EcMRS but no tRNA (Fig. 3). The mismethionylation level was about 10% of that for cognate 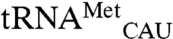 transcript at saturation. These results also show that tRNA modifications are not required for mismethionylation as the tRNA transcripts lack all modifications present in the E. coli tRNA used in the microarray assay.
transcript at saturation. These results also show that tRNA modifications are not required for mismethionylation as the tRNA transcripts lack all modifications present in the E. coli tRNA used in the microarray assay.
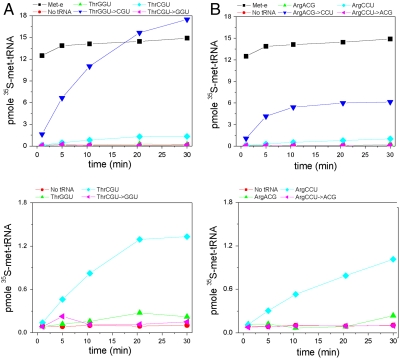
Fig. 3.
Mismethionylation of tRNA transcripts using full-length EcMRS. (A) tRNAThr. (B) tRNAArg. Charging levels of all transcripts are shown in the top graph, whereas the y axis of the bottom graph is 1/10th of the y axis of the top graph.
The substitution of non-CNU anticodons into these mismethionylated isoacceptors completely abolished mismethionylation (Fig. 3). Both 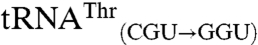 and
and 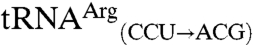 variants showed levels of mismethionylation indistinguishable from background. The activity of all tRNAThr and tRNAArg transcripts was confirmed in aminoacylation assays using an E. coli aminoacyl-tRNA synthetase mixture and [3H]threonine or [3H]arginine, respectively (Fig. S1). These variant tRNAs had therefore lost methionine-acceptance activity, but were still capable of efficiently accepting their cognate amino acid.
variants showed levels of mismethionylation indistinguishable from background. The activity of all tRNAThr and tRNAArg transcripts was confirmed in aminoacylation assays using an E. coli aminoacyl-tRNA synthetase mixture and [3H]threonine or [3H]arginine, respectively (Fig. S1). These variant tRNAs had therefore lost methionine-acceptance activity, but were still capable of efficiently accepting their cognate amino acid.
Transcripts of both 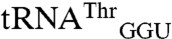 and
and 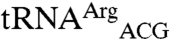 were not mismethionylated by EcMRS above background levels; but when CNU anticodons were introduced into these tRNAs they showed robust methionine acceptance (Fig. 3). Both
were not mismethionylated by EcMRS above background levels; but when CNU anticodons were introduced into these tRNAs they showed robust methionine acceptance (Fig. 3). Both 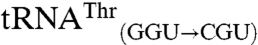 and
and 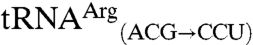 are readily methionylated, reinforcing the view that the anticodon is an important identity element for mismethionylation. Unexpectedly, mismethionylation levels at saturation for both
are readily methionylated, reinforcing the view that the anticodon is an important identity element for mismethionylation. Unexpectedly, mismethionylation levels at saturation for both 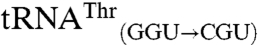 and
and 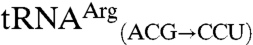 mutants are 13-fold and 6-fold higher than those for their respective wild-type transcripts,
mutants are 13-fold and 6-fold higher than those for their respective wild-type transcripts, 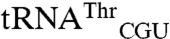 and
and 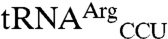 (Fig. 3). This result indicates that although the anticodon serves as a key identity element, sequences outside of this region can significantly influence the degree of mismethionylation. As all tRNAThr and tRNAArg transcripts are efficiently threonylated and arginylated, it appears that
(Fig. 3). This result indicates that although the anticodon serves as a key identity element, sequences outside of this region can significantly influence the degree of mismethionylation. As all tRNAThr and tRNAArg transcripts are efficiently threonylated and arginylated, it appears that 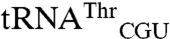 and
and 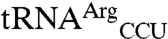 include overlapping identity elements, allowing these tRNAs to be efficient substrates of more than one aminoacyl-tRNA synthetase.
include overlapping identity elements, allowing these tRNAs to be efficient substrates of more than one aminoacyl-tRNA synthetase.
We also tested all transcripts with the monomeric N547MRS construct, and similar results to the full-length EcMRS were obtained in all cases (Fig. S2).
In summary, our studies with tRNA transcripts show that the identity of the anticodon nucleotides is a crucial recognition element for mismethionylation by EcMRS. However, the body sequence of the mismethionylatable tRNAs imposes constraints that limit the mismethionylation levels at saturation. Non-mis-methionylated 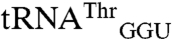 and
and 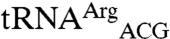 , which lack CNU anticodons, appear to have less evolutionary pressure to tune their body sequences to constrain their methionylation by EcMRS.
, which lack CNU anticodons, appear to have less evolutionary pressure to tune their body sequences to constrain their methionylation by EcMRS.
EcMRS Variants Exhibit Altered Mismethionylation Levels.
Having identified EcMRS-mediated mismethionylation, we wanted to identify residues in the enzyme that play a role in mismethionylation. We purified a total of 21 monomeric EcMRS variants to compare against the N547MRS protein that mismethionylated 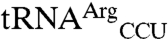 and
and 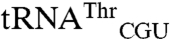 in our assays. These included single amino acid substitutions, a double substitution that generates a disulfide bond, and a six-residue peptide swap. Substitutions are distributed across the four structural domains of MRS: Rossmann fold active site, connective polypeptide, stem contact fold, and anticodon-binding domain (Fig. 4A). Substitutions included those at highly conserved residues previously shown to have severe impacts on methionyl-adenylate formation (D369) or anticodon recognition (W461) and subsequent reduced levels of cognate
in our assays. These included single amino acid substitutions, a double substitution that generates a disulfide bond, and a six-residue peptide swap. Substitutions are distributed across the four structural domains of MRS: Rossmann fold active site, connective polypeptide, stem contact fold, and anticodon-binding domain (Fig. 4A). Substitutions included those at highly conserved residues previously shown to have severe impacts on methionyl-adenylate formation (D369) or anticodon recognition (W461) and subsequent reduced levels of cognate 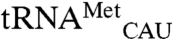 methionylation (22, 23). Other substitutions were made at residues not directly involved in either anticodon binding or catalysis, but hypothesized to participate in functional coupling of these domains (24, 25). For example, F277 on the solvent-exposed side of the Rossmann fold was identified in a molecular dynamics study as being correlated with motion of W461 (25), and Q211 and Q213 in the connective polypeptide were found mutated in a genetic screen for EcMRS variants able to methionylate a suppressor tRNA (26).
methionylation (22, 23). Other substitutions were made at residues not directly involved in either anticodon binding or catalysis, but hypothesized to participate in functional coupling of these domains (24, 25). For example, F277 on the solvent-exposed side of the Rossmann fold was identified in a molecular dynamics study as being correlated with motion of W461 (25), and Q211 and Q213 in the connective polypeptide were found mutated in a genetic screen for EcMRS variants able to methionylate a suppressor tRNA (26).
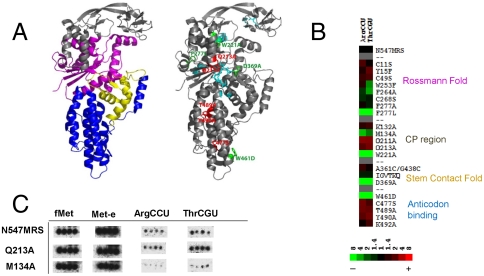
Fig. 4.
Comparison of mismethionylation activity of EcMRS variants. (A) Location of protein mutation superimposed in the crystal structure [Protein Data Bank ID code 1QQT (28)]. In the left structure, the key regions of EcMRS including Rossmann fold (magenta), connective polypeptide (CP region, gray), stem contact fold (yellow), and anticodon-binding domain (blue) are depicted (28). In the right structure, residues mutated for charging analysis are color-coded according to the change from N547MRS levels of percent mismethionylated tRNA. Residues corresponding to > 2-fold increases are in red, residues corresponding to > 6-fold reduction are in green, and residues showing milder effects are in cyan. (B) Heat map of mutant effects on the 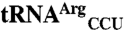 and
and 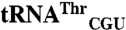 mismethionylation. Scale reflects fold change from N547MRS levels of percent misacylation (100% = total 35S signal of both cognate and mismethionylated tRNA). (C) Representative spots for N547MRS, a mutant showing increased mismethionylation, Q213A, and a mutant showing decreased mismethionylation, M134A.
mismethionylation. Scale reflects fold change from N547MRS levels of percent misacylation (100% = total 35S signal of both cognate and mismethionylated tRNA). (C) Representative spots for N547MRS, a mutant showing increased mismethionylation, Q213A, and a mutant showing decreased mismethionylation, M134A.
Differences between wild-type N547MRS and variants for mismethionylating tRNA were determined by microarray assay for each variant (Fig. 4 B and C). Under the condition tested, total [35S] signal corresponding to cognate plus mismethionylated tRNA appeared reduced for many of the variant arrays and was confirmed by trichloroacetic acid precipitation and phosphoimaging, which showed approximately 1.5-fold reduction for W221A and D369A and 4-fold reduction of the W461D variant. Twelve substitutions affected aminoacylation of cognate and mismethionylated tRNA similarly; they did not exhibit striking changes in percent of mismethionylated tRNA. Four variants (W221A, F277L, D369A, and W461D) displayed at least a 6-fold reduction in mismethionylation percentage, whereas five variants (Q211A, Q213A, C477S, T489A, and Y490A) showed at least a 2-fold increase in mismethionylation (Fig. 4 A and B). Variant-derived changes in mismethionylation of 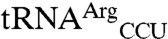 and
and 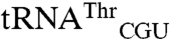 appear correlated in the direction of the change and to a lesser extent its magnitude (Fig. 4B).
appear correlated in the direction of the change and to a lesser extent its magnitude (Fig. 4B).
Four variants that significantly interfere with mismethionylation are in the anticodon-binding domain. The cocrystal structure of Aquifex aeolicus methionyl-tRNA synthetase with 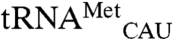 is useful for exploring EcMRS-tRNA anticodon interactions (27). The highly conserved residue corresponding to EcMRS W461 directly stacks with the
is useful for exploring EcMRS-tRNA anticodon interactions (27). The highly conserved residue corresponding to EcMRS W461 directly stacks with the 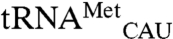 anticodon base C34, which in turn stacks with A35 and A38. A35 also hydrogen bonds with bases C32, U33 and residue Asn391. Substitution of A35 in mismethionylated tRNA may disturb these stabilizing interactions and make noncognate tRNA more sensitive than cognate tRNA to the W461D variant that further destabilize anticodon binding. In contrast to the W461D variant that decreased mismethionylation, substituting three residues in the α12 helix of EcMRS’s anticodon-binding domain increased mismethionylation. Cys-477 is near the residues that directly interact with the anticodon, whereas T489 and Y490 are more distal but also located on the same helix (28). Thus, the ∼2-fold increase in mismethionylation seen for these three mutants might be mediated through positioning of the α12 helix.
anticodon base C34, which in turn stacks with A35 and A38. A35 also hydrogen bonds with bases C32, U33 and residue Asn391. Substitution of A35 in mismethionylated tRNA may disturb these stabilizing interactions and make noncognate tRNA more sensitive than cognate tRNA to the W461D variant that further destabilize anticodon binding. In contrast to the W461D variant that decreased mismethionylation, substituting three residues in the α12 helix of EcMRS’s anticodon-binding domain increased mismethionylation. Cys-477 is near the residues that directly interact with the anticodon, whereas T489 and Y490 are more distal but also located on the same helix (28). Thus, the ∼2-fold increase in mismethionylation seen for these three mutants might be mediated through positioning of the α12 helix.
The five other MRS variants that strongly affect mismethionylation are located far from the anticodon-binding domain. Residues Q211, Q213, and W221 are located on the α4 helix of the connective polypeptide; D369 is in the stem contact fold, and F277 is in the Rossmann fold (28). Substitutions at W221, F277, and D369 result in a decrease of mismethionylation. Asp-369 is highly conserved among MRSs; the carboxylate side chain is in close contact to Lys-295 in the enzyme active site, and the D369A variant is defective in methionyl-adenylate formation (23). Similarly, F277A exhibits decreased amino acid activation. For these variants, it is not surprising that cognate tRNA aminoacylation would be decreased, but the reduction in percent mismethionylation suggests that catalytic deficiencies in methionyl-adenylate formation may not evenly impact cognate and noncognate tRNA methionylation. In contrast, W221A decreases mismethionylation but does not affect methionyl-adenylate formation, suggesting a role in coupling of anticodon binding and catalysis.
Substitution of some residues at the interface between anticodon binding and catalytic domains has minimal effects on mismethionylation. For example, the doubly substituted A361C/G438C variant introduces a disulfide bond designed to limit domain–domain flexibility (29) but showed a limited effect on mismethionylation. Similarly, replacement of EcMRS residues 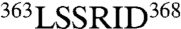 , which form the loop of the stem contact fold motif, with the structurally equivalent
, which form the loop of the stem contact fold motif, with the structurally equivalent 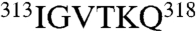 residues of E. coli glutaminyl-tRNA synthetase showed little effect on mismethionylation. This replacement also had minimal effect on cognate tRNA aminoacylation efficiency, consistent with the hypothesis that the class Ia/Ib stem contact fold orients the tRNA core on the enzyme without making base-specific contacts (23).
residues of E. coli glutaminyl-tRNA synthetase showed little effect on mismethionylation. This replacement also had minimal effect on cognate tRNA aminoacylation efficiency, consistent with the hypothesis that the class Ia/Ib stem contact fold orients the tRNA core on the enzyme without making base-specific contacts (23).
Discussion
Role of EcMRS in Mismethionylation.
This work shows that the bacterial enzyme EcMRS is sufficient to mismethionylate two tRNA species. To confer substrate selection, EcMRS-mediated mismethionylation likely requires that the enzyme’s anticodon-binding domain accommodates other tRNA species. The strong reduction in mismethionylation seen for the key anticodon-binding residue mutant, W461D, indicates at least partial sharing of the enzyme’s anticodon recognition elements for cognate and noncognate tRNA. Although the catalytic core would also need the 3′ end of the noncognate tRNA to be appropriately positioned within the active site, the amino acid activation step and the transfer of methionine to the 3′ end of tRNA is likely analogous to the reaction performed on cognate tRNAMet. The accommodation of noncognate tRNA species might be intrinsic to the native enzyme. Alternatively, as these enzymes have been overexpressed and purified from E. coli, a low level of posttranslational modification(s) at a yet-to-be mapped site could generate a low-fidelity form of EcMRS that specifically aminoacylates noncognate tRNAs.
EcMRS variant-derived changes in mismethionylation levels found in this work indicate that there is potential for decoupling methionylation levels of cognate versus noncognate tRNA. Regulation of mismethionylation independent from cognate methionylation may provide benefits to the cell. In the case of mammalian mismethionylation, oxidative stress increases mismethionylation by up to 10-fold and is hypothesized to provide a protective effect from reactive oxygen species (13). In E. coli, the C477S substitution increased tRNA mismethionylation in vitro, indicating that the alteration of this residue in vivo, possibly by oxidation, might influence the accommodation of noncognate tRNA. When EcMRS and N547MRS were treated with hydrogen peroxide in the absence of any reducing agents, no clear change in mismethionylation patterns was observed in vitro. This result suggests that a potential low-fidelity form of EcMRS, if derived from oxidation of Cys or Met residues, cannot be easily generated in vitro. This contrasts with E. coli threonyl-tRNA synthetase whose fidelity has been shown to be reduced by oxidation in vitro (30). However, this does not preclude the generation of a small fraction of oxidized EcMRS in vivo where additional pathways or oxidative enzymes could mediate oxidation.
Given the spatially disperse nature of the variants tested, some of these substitutions may be acting more generally on mismethionylation by disrupting communication networks within MRS (24, 31). In particular, residues Q211 and Q213 may be part of a network that mediate anticodon binding to amino acid transfer in the enzyme active site. In a genetic screen designed to identify EcMRS residues that promote methionylation of a suppressor tRNA (corresponding to the elongator tRNAMet with a CUA amber anticodon), Q211 and Q213 were found mutated to Arg and Pro, respectively (26). This earlier in vivo result suggests that these residues in the connective polypeptide may serve to discriminate against tRNAs lacking the cognate CAU anticodon through long-range coupling to MRS’s anticodon-binding domain.
Mismethionylation Identity Elements.
The use of the anticodon as an identity element for mismethionylation is consistent with the importance of the anticodon nucleotides for cognate tRNA discrimination by MRS and many other aaRSs. Both mismethionylated E. coli tRNAs share two of three anticodon nucleotides with tRNAMet. Two unexpected results for mismethionylating tRNAs suggest that misacylation may have evolved to be maintained at a particular level that benefits the cell without catastrophically compromising protein synthesis fidelity. First, both 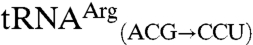 and
and 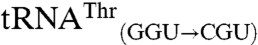 variants are far better substrates for EcMRS than wild-type
variants are far better substrates for EcMRS than wild-type 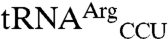 and
and 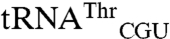 . Second, both
. Second, both 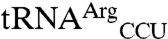 and
and 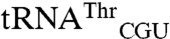 have a low level of mismethionylation at saturation, consistent with these substrates being tuned to allow a substoichiometric amount of mismethionylation. In vitro aminoacylation assays reveal that the wild-type and variant tRNAs have not lost arginine or threonine acceptance. These results suggest a lack of evolutionary pressure to maximize methionine-acceptance activity of the mismethionylated tRNA and may point to a necessary balance between gain of oxidative protection and loss of native proteome function.
have a low level of mismethionylation at saturation, consistent with these substrates being tuned to allow a substoichiometric amount of mismethionylation. In vitro aminoacylation assays reveal that the wild-type and variant tRNAs have not lost arginine or threonine acceptance. These results suggest a lack of evolutionary pressure to maximize methionine-acceptance activity of the mismethionylated tRNA and may point to a necessary balance between gain of oxidative protection and loss of native proteome function.
In terms of mismethionylation identity, our results indicate that the anticodon functions as a critical identity element for allowing mismethionylation while additional nucleotides outside of the anticodon have the potential to alter this ability. Permissive body sequences alone are insufficient to confer methionine acceptance, however. 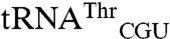 and
and 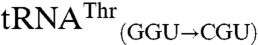 differ by only 19 nucleotides, and it is not obvious which of these nucleotides alters methionylation efficiency. The acceptor stem would seem a likely place to look for candidate nucleotides that could influence the positioning of the 3′ end of the tRNA in the active site. However, the acceptor stems of these two tRNAs differ by a single 3–70 base pair and the minimally methionylated
differ by only 19 nucleotides, and it is not obvious which of these nucleotides alters methionylation efficiency. The acceptor stem would seem a likely place to look for candidate nucleotides that could influence the positioning of the 3′ end of the tRNA in the active site. However, the acceptor stems of these two tRNAs differ by a single 3–70 base pair and the minimally methionylated 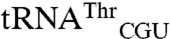 shares a C3-G70 base pair with cognate
shares a C3-G70 base pair with cognate 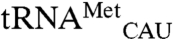 (Fig. 2). Alternatively, significant variation between the anticodon stems may influence the positioning of the anticodon on the enzyme or alter tRNA structural deformations that have been shown to occur upon EcMRS anticodon binding (27).
(Fig. 2). Alternatively, significant variation between the anticodon stems may influence the positioning of the anticodon on the enzyme or alter tRNA structural deformations that have been shown to occur upon EcMRS anticodon binding (27).
Nature of the Amino Acid Substitution.
The use of C34N35U36 identity element in other bacteria would constrain MRS-mediated mismethionylation to threonine, arginine, and possibly lysine in species with 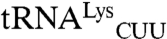 . Unlike methionine, these three amino acids are polar and in the case of arginine and lysine, positively charged. The substitution of methionine for similarly sized aliphatic amino acids such as isoleucine or leucine would appear to be a more conservative replacement that would likely minimize the disturbance of protein function. Instead, the substitution of arginine and threonine (and possibly lysine) with methionine in proteins may be driven by the cooption of the anticodon-binding domain of the MRS for recognizing mismethionylated tRNA. Cells may tolerate a low proportion of catalytically inactive or misfolded proteins in its tRNA mismethionylation-derived protein pool, as other proteins would contain extra methionine in positions with minimal disturbance to catalytic activity or structure. These extra Met residues could serve a protective effect against oxidative stress (19).
. Unlike methionine, these three amino acids are polar and in the case of arginine and lysine, positively charged. The substitution of methionine for similarly sized aliphatic amino acids such as isoleucine or leucine would appear to be a more conservative replacement that would likely minimize the disturbance of protein function. Instead, the substitution of arginine and threonine (and possibly lysine) with methionine in proteins may be driven by the cooption of the anticodon-binding domain of the MRS for recognizing mismethionylated tRNA. Cells may tolerate a low proportion of catalytically inactive or misfolded proteins in its tRNA mismethionylation-derived protein pool, as other proteins would contain extra methionine in positions with minimal disturbance to catalytic activity or structure. These extra Met residues could serve a protective effect against oxidative stress (19).
The nature of the mismethionylated tRNA species in E. coli provides a way for the organism to mitigate the potential for amino acid substitutions through codon usage in each gene. Mismethionylated 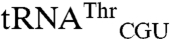 reads only the ACG codon and
reads only the ACG codon and 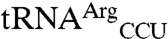 reads only the AGG codon. Threonine and arginine residues can thus be specified by codons corresponding to either mismethionylated or non-mis-methionylated tRNA isoacceptors. By using codons corresponding to non-mis-methionylated tRNAs for key catalytic or phosphorylation sites, cells would be better able to tolerate even high levels of mismethionylation. Bioinformatic approaches that look for correlations between catalytically important residues and codons corresponding to non-mis-methionylated versus mismethionylated tRNA isoacceptors might suggest whether such a codon-selection strategy is in use.
reads only the AGG codon. Threonine and arginine residues can thus be specified by codons corresponding to either mismethionylated or non-mis-methionylated tRNA isoacceptors. By using codons corresponding to non-mis-methionylated tRNAs for key catalytic or phosphorylation sites, cells would be better able to tolerate even high levels of mismethionylation. Bioinformatic approaches that look for correlations between catalytically important residues and codons corresponding to non-mis-methionylated versus mismethionylated tRNA isoacceptors might suggest whether such a codon-selection strategy is in use.
Relation to Mammalian Mismethionylation.
This work shows mismethionylation in bacteria, but its relation to the mismethionylation observed in mammalian cells is unclear. Mismethionylation patterns of E. coli and mammals in vitro do not correspond to each other. For example, the dominant in vitro mismethionylated tRNA species for the mammalian system is 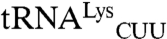 , but mismethionylation of
, but mismethionylation of 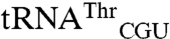 and
and 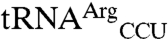 was not observed (13). Bacterial EF-Tu does not deliver some misacylated tRNA to the ribosome due to an imbalance of EF-Tu binding affinities for tRNA and amino acid, whereas mammalian elongation factors appear more permissive for misacylated tRNA (32–34). To our knowledge, tRNA misacylated with methionine has not been studied for EF-Tu binding. Therefore it remains to be determined whether, as in mammalian cells, the mismethionylated tRNAs found in this study are accepted by the E. coli ribosome.
was not observed (13). Bacterial EF-Tu does not deliver some misacylated tRNA to the ribosome due to an imbalance of EF-Tu binding affinities for tRNA and amino acid, whereas mammalian elongation factors appear more permissive for misacylated tRNA (32–34). To our knowledge, tRNA misacylated with methionine has not been studied for EF-Tu binding. Therefore it remains to be determined whether, as in mammalian cells, the mismethionylated tRNAs found in this study are accepted by the E. coli ribosome.
The EcMRS used in this study may not exhibit its full mismethionylation potential, which could be influenced by posttranslational modifications or interactions with other proteins or allosteric regulators. Alternatively, bacterial mismethionylation may exist in a simple form relying on subtle accommodations by MRS for the few tRNACNU species. Mammalian mismethionylation may represent a more complex phenomenon than bacterial mismethionylation. The MRS in mammalian cells is usually associated with a large multisynthetase complex involving nine different aaRSs as well as three other proteins that interact with components of multiple cellular signaling pathways (35). Various controls of these associations offer pathways for influencing mismethionylation that may not be available to bacterial cells.
In human cells, even basal mismethionylation involves more tRNA species (eight tRNAs) than mismethionylation observed in vitro (six tRNAs), and the number of mismethionylated tRNA species is further expanded to 26 upon oxidative stress. Further studies of mammalian MRS-mediated mismethionylation are needed to determine if mammalian mismethionylation is an evolutionary extension of bacterial mismethionylation or represents the convergent evolution of a distinct mismethionylation mechanism.
Materials and Methods
Protein Expression and Purification.
Full-length EcMRS, N547MRS, and all MRS mutants were overexpressed in E. coli with N-terminal His6 tags and purified as previously described (29).
Microarray Analysis.
The basic features of tRNA microarray analysis using radioactive detection have been described previously for the determination of mismethionylation of tRNA in mammalian cells (13, 18). Bacterial mismethionylation was assessed with arrays containing 41 probes for E. coli tRNA and 23 probes from yeast to serve as controls. The E. coli probes uniquely hybridize with each tRNA isotype of the K12 E. coli strain although some probes hybridize with multiple isodecoders, tRNA possessing the same anticodon but having different sequences. Each array contained six repeats for each probe.
Aminoacylation of tRNA for microarray analysis was performed at 37 °C for 15 min in 20 mM K-Hepes (pH 7.6), 10 mM MgCl2, 5 mM DTT, 4 mM ATP, 150 mM NH4Cl, 0.1 mM Na2EDTA, 1 μCi/μL L-[35S]methionine, 1 mg/mL total E. coli tRNA (Sigma-Aldrich), and 0.25–1 μM purified MRS enzyme.
Array hybridization on a Genomic Solutions Hyb4 station with 10-μg total RNA, deacylation, and blocking with excess methionine probe for the microarray analysis controls were performed as previously described (13).
In Vitro Transcription.
E. coli 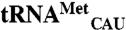 ,
, 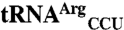 ,
, 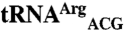 ,
, 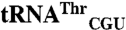 , and
, and 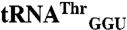 sequences were obtained from the genomic tRNA database (36).
sequences were obtained from the genomic tRNA database (36). 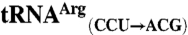 ,
, 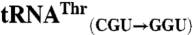 ,
, 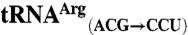 , and
, and 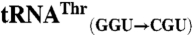 sequences were generated by swapping the anticodon triplet bases between the wild-type isoacceptor sequences. Transcripts were generated by in vitro transcription of overlapping oligonucleotides and purified as previously described (23, 37). The concentration of active tRNA molecules was determined by aminoacylation plateaus (Fig. S1) from reactions that included the tRNA transcript, radiolabeled cognate amino acid, and 1 mg/mL total E. coli aminoacyl-tRNA synthetase (Sigma-Aldrich) under conditions that have been previously described (38, 39).
sequences were generated by swapping the anticodon triplet bases between the wild-type isoacceptor sequences. Transcripts were generated by in vitro transcription of overlapping oligonucleotides and purified as previously described (23, 37). The concentration of active tRNA molecules was determined by aminoacylation plateaus (Fig. S1) from reactions that included the tRNA transcript, radiolabeled cognate amino acid, and 1 mg/mL total E. coli aminoacyl-tRNA synthetase (Sigma-Aldrich) under conditions that have been previously described (38, 39).
Filter-Based Aminoacylation Reactions.
Filter-based aminoacylation reactions were performed at 37 °C in 20 mM K-Hepes (pH 7.6), 100 μM methionine, 10 mM MgCl2, 5 mM DTT, 4 mM ATP, 150 mM NH4Cl, 0.1 mM Na2EDTA, 0.5 μCi/mL L-[35S] methionine, 0 or 2 μM tRNA transcripts, and 0.3 μM EcMRS or N547MRS enzyme as reported (14).
Supplementary Material
Acknowledgments.
This work was supported in part by National Institutes of Health (T.P.) and by National Science Foundation (Contract MCB-0448243 to R.W.A). T.E.J. is a recipient of the Yen Fellowship at the Institute of Biophysical Dynamics at the University of Chicago.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019033108/-/DCSupplemental.
References
- 1.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Ling J, Yadavalli SS, Ibba M. Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA. 2007;13:1881–1886. doi: 10.1261/rna.684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Min B, et al. Protein synthesis in Escherichia coli with mischarged tRNA. J Bacteriol. 2003;185:3524–3526. doi: 10.1128/JB.185.12.3524-3526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nangle LA, De Crecy Lagard V, Doring V, Schimmel P. Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J Biol Chem. 2002;277:45729–45733. doi: 10.1074/jbc.M208093200. [DOI] [PubMed] [Google Scholar]
- 5.Cochella L, Green R. Fidelity in protein synthesis. Curr Biol. 2005;15:R536–540. doi: 10.1016/j.cub.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Cusack S. Aminoacyl-tRNA synthetases. Curr Opin Struct Biol. 1997;7:881–889. doi: 10.1016/s0959-440x(97)80161-3. [DOI] [PubMed] [Google Scholar]
- 7.Martinis SA, Boniecki MT. The balance between pre- and post-transfer editing in tRNA synthetases. FEBS Lett. 2010;584:455–459. doi: 10.1016/j.febslet.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schimmel P, Ribas de Pouplana L. Formation of two classes of tRNA synthetases in relation to editing functions and genetic code. Cold Spring Harb Sym. 2001;66:161–166. doi: 10.1101/sqb.2001.66.161. [DOI] [PubMed] [Google Scholar]
- 9.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 11.Ruan B, et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA. 2008;105:16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes AC, et al. A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 2007;8:R206. doi: 10.1186/gb-2007-8-10-r206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulman LH, Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988;242:765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- 15.Ribas de Pouplana L, Schimmel P. Aminoacyl-tRNA synthetases: potential markers of genetic code development. Trends Biochem Sci. 2001;26:591–596. doi: 10.1016/s0968-0004(01)01932-6. [DOI] [PubMed] [Google Scholar]
- 16.Muramatsu T, et al. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem. 1988;263:9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- 17.Dittmar KA, Mobley EM, Radek AJ, Pan T. Exploring the regulation of tRNA distribution on the genomic scale. J Mol Biol. 2004;337:31–47. doi: 10.1016/j.jmb.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 18.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci USA. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oien DB, Moskovitz J. Substrates of the methionine sulfoxide reductase system and their physiological relevance. Curr Top Dev Biol. 2008;80:93–133. doi: 10.1016/S0070-2153(07)80003-2. [DOI] [PubMed] [Google Scholar]
- 21.Mellot P, Mechulam Y, Le Corre D, Blanquet S, Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989;208:429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh G, Pelka H, Schulman LH. Identification of the tRNA anticodon recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1990;29:2220–2225. doi: 10.1021/bi00461a003. [DOI] [PubMed] [Google Scholar]
- 23.Casina VC, Lobashevsky AA, McKinney W, Brown CL, Alexander RW. Role for a conserved structural motif in assembly of a class I aminoacyl-tRNA synthetase active site domain. Biochemistry. 2011;50:763–769. doi: 10.1021/bi101375d. [DOI] [PubMed] [Google Scholar]
- 24.Alexander RW, Schimmel P. Domain-domain communication in aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 2001;69:317–349. doi: 10.1016/s0079-6603(01)69050-0. [DOI] [PubMed] [Google Scholar]
- 25.Budiman ME, Knaggs MH, Fetrow JS, Alexander RW. Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase. Proteins. 2007;68:670–689. doi: 10.1002/prot.21426. [DOI] [PubMed] [Google Scholar]
- 26.Meinnel T, et al. Selection of suppressor methionyl-tRNA synthetases: Mapping the tRNA anticodon binding site. Proc Natl Acad Sci USA. 1991;88:291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol. 2005;12:931–932. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 28.Mechulam Y, et al. Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J Mol Biol. 1999;294:1287–1297. doi: 10.1006/jmbi.1999.3339. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee P, Warf MB, Alexander R. Effect of a domain-spanning disulfide on aminoacyl-tRNA synthetase activity. Biochemistry. 2009;48:10113–10119. doi: 10.1021/bi9012275. [DOI] [PubMed] [Google Scholar]
- 30.Ling J, Soll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107:4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander RW, Schimmel P. Evidence for breaking domain-domain functional communication in a synthetase-tRNA complex. Biochemistry. 1999;38:16359–16365. doi: 10.1021/bi991948c. [DOI] [PubMed] [Google Scholar]
- 32.Asahara H, Uhlenbeck OC. The tRNA specificity of Thermus thermophilus EF-Tu. Proc Natl Acad Sci USA. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asahara H, Uhlenbeck OC. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu.GTP. Biochemistry. 2005;44:11254–11261. doi: 10.1021/bi050204y. [DOI] [PubMed] [Google Scholar]
- 34.Sanderson LE, Uhlenbeck OC. The 51–63 base pair of tRNA confers specificity for binding by EF-Tu. RNA. 2007;13:835–840. doi: 10.1261/rna.485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SG, Ewalt KL, Kim S. Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci. 2005;30:569–574. doi: 10.1016/j.tibs.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherlin LD, et al. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA. 2001;7:1671–1678. [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan B, et al. A unique hydrophobic cluster near the active site contributes to differences in borrelidin inhibition among threonyl-tRNA synthetases. J Biol Chem. 2005;280:571–577. doi: 10.1074/jbc.M411039200. [DOI] [PubMed] [Google Scholar]
- 39.Tamura K, Himeno H, Asahara H, Hasegawa T, Shimizu M. In vitro study of E. coli tRNA(Arg) and tRNA(Lys) identity elements. Nucleic Acids Res. 1992;20:2335–2339. doi: 10.1093/nar/20.9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.