Abstract
Campylobacter jejuni is a prevalent gastrointestinal pathogen in humans and a common commensal of poultry. When colonizing its hosts, C. jejuni comes into contact with intestinal carbohydrates, including l-fucose, released from mucin glycoproteins. Several strains of C. jejuni possess a genomic island (cj0480c–cj0490) that is up-regulated in the presence of both l-fucose and mucin and allows for the utilization of l-fucose as a substrate for growth. Strains possessing this genomic island show increased growth in the presence of l-fucose and mutation of cj0481, cj0486, and cj0487 results in the loss of the ability to grow on this substrate. Furthermore, mutants in the putative fucose permease (cj0486) are deficient in fucose uptake and demonstrate a competitive disadvantage when colonizing the piglet model of human disease, which is not paralleled in the colonization of poultry. This identifies a previously unrecorded metabolic pathway in select strains of C. jejuni associated with a virulent lifestyle.
Keywords: pathogenesis, metabolism, gut, mucus
The most common cause of gastroenteritis in the developed world, Campylobacter jejuni is a frequent cause of pediatric diarrheal episodes within developing countries that can last from a few days to several weeks (1). The symptoms vary from mild, watery diarrhea to bloody diarrhea with fever and severe abdominal pain (2). The disease usually resolves itself relatively quickly, but autoimmune reactions to the infection can lead to postinfectious sequelae, such as Guillain-Barré syndrome and reactive arthritis (2).
When colonizing its host (whether commensally in poultry and other animals, or pathogenically in humans), C. jejuni must establish itself within the mucus layer of the intestinal epithelium (3, 4). Byrne et al. (5) recently showed that the mucus layer originating from humans or chickens influences the virulence of C. jejuni and might differentially control the virulence versus commensalism equilibrium. However, it remains unknown which specific components of the mucus layer contribute to this change in phenotype.
C. jejuni lacks many of the pathways for nutrient metabolism used by other pathogens and the normal intestinal microbiota. For example, several key enzymes within the glycolytic pathway, the Entner-Doudoroff pathway, and the pentose phosphate pathway are absent (6, 7), and it has generally been assumed that C. jejuni is unable to use any form of carbohydrate as a substrate for growth. Instead, C. jejuni relies on the use of amino acids and citric acid cycle intermediates as carbon sources (8). The amino acid metabolic pathways that allow C. jejuni to use serine (9), aspartate, glutamate (10), and proline (11) have all been studied in depth. Additionally, some strains, including C. jejuni 81–176, can metabolize glutamine and asparagine (8). However, and in contrast to the generally accepted paradigm, we demonstrate in this study that certain strains of C. jejuni contain a unique pathway for the uptake and metabolism of the sugar l-fucose, which is obtained from the host during colonization of the intestine.
The importance of fucose to the intestinal microflora is well established (12–14). Fucosylated glycans are common within the digestive tract, where they are found on cell surfaces and are typically located in terminal positions on the extensively modified mucin glycoproteins (13, 15). These fucosylated glycans coating the mucins are known to act as adherence targets for many intestinal bacteria [including C. jejuni (16)], potentially contributing to the function of mucin as a barrier to intestinal pathogens (17). Fucose monomers and fucosylated glycans can serve as a chemoattractant (18) and carbon source for many species within the gut. Being so common within the intestine, fucose is an integral part of the intestinal ecology. For example, the presence of the ubiquitous commensal Bacteroides thetaiotaomicron in the gut triggers an increase in the production of fucosylated mucins by the intestinal epithelium, which is important for gut maturation (19). These bacteria then secrete fucosidases to release fucose residues from the oligosaccharides and transport the sugar into the cell for metabolism and surface presentation (12, 20). Even bacteria such as Escherichia coli, which do not express fucosidases, use this same pathway for fucose transport (21) and colonization of the bovine rectum (22).
Despite the lack of an annotated fucose metabolic pathway, fucose has long been described as influencing the behavior of C. jejuni. In 1988, Hugdahl et al. reported C. jejuni chemotaxis toward l-fucose, as well as toward intact mucins, which would have included fucosylated glycans (18). In addition, fucosylated milk oligosaccharides were used in an elegant experiment to competitively inhibit the specific attachment of C. jejuni to intestinal H-antigens (23). More recently, Korolik and colleagues have made use of glycan microarrays to demonstrate increased C. jejuni adherence to fucosylated glycans, such as those found on mucin and cell surfaces (16), structures that were suggested to be significant factors in the ability of C. jejuni to closely adhere to the cell surface and invade intestinal epithelial cells.
This study is unique in demonstrating the uptake and utilization of l-fucose by certain strains of C. jejuni via a previously undescribed pathway not found in other enteric bacteria. We further demonstrate how l-fucose transport provides a distinct competitive advantage in the pathogenesis model that is not found during commensal growth.
Results
Fucose Up-Regulates Expression of Genes Linked to Fucose Use.
Microarray experiments were used to assemble a transcriptomic profile of C. jejuni NCTC 11168 during growth in MEMα medium supplemented with 25 mM l-fucose. This process led to the identification of 74 up-regulated genes, and 52 down-regulated genes (Table S1). The most notable change from the list of up-regulated genes was an operon between cj0481 and cj0490, which exhibited six- to -ninefold up-regulation (Table S1). The putative regulator cj0480c did not show any significant change in expression. Similarly, a microarray study of C. jejuni grown in MEMα medium supplemented with 2 mg/mL porcine gastric mucin also revealed significant up-regulation (two- to fourfold) of the genes cj0481 to cj0490 (Table S2).
Identification of a Genomic Island Required for Fucose Utilization.
None of the genes cj0480c to cj0490 have any confirmed function, but cj0486 bears homology to the fucose permease gene (fucP) of other bacteria (21). This operon has been identified previously in RM1221 and NCTC 11168 as a genomic island spanning from cj0480c to cj0490 (24) and nestled in between the genes rpoC and fusA (Fig. S1) of some strains. Notably, this region is absent in strain 81–176. Although this region is not universally conserved within strains of C. jejuni, it has been linked to C. jejuni hyper-invasiveness (25, 26).
In a comparison of the amino acid sequence of Cj0486 among sequenced Campylobacter genomes, C. jejuni NCTC 11168, RM1221, CF93-6, and 84–25, and C. jejuni, subsp. doylei 269.97, all exhibited greater than 98% identity, but Campylobacter coli RM2228 and Campylobacter fetus 82–40 displayed 93% and 59% identity, respectively. All of these strains contain the whole genomic island in the same location on the chromosome, with the exception of C. fetus, which contained a similar group of genes elsewhere in the chromosome.
Cj0486 Acts as a Fucose Permease to Transport Extracellular Fucose.
To confirm the function of Cj0486, we sought to ascertain whether the C. jejuni cj0486 gene product was functional as a fucose transporter in E. coli. The C. jejuni NCTC 11168 fucP gene homolog, cj0486, was transformed into an E. coli fucP mutant (27) and successfully restored function, indicating metabolism of l-fucose (Fig. 1). This finding indicated that despite low homology between the genes (37.2%), Cj0486 is able to function as an l-fucose transporter in E. coli. Further comparison of the E. coli fucP and the C. jejuni Cj0486 protein sequence (Fig. S2) reveals conservation of Asp46, Glu135, and Asn146, all identified by Dang et al. as critical for the active transport of fucose into the cell (28). Cj0484 is annotated as a major facilitator superfamily (MFS) transporter, but does not contain these conserved residues or a significant degree of sequence similarity, suggesting it does not serve as a second l-fucose transporter.
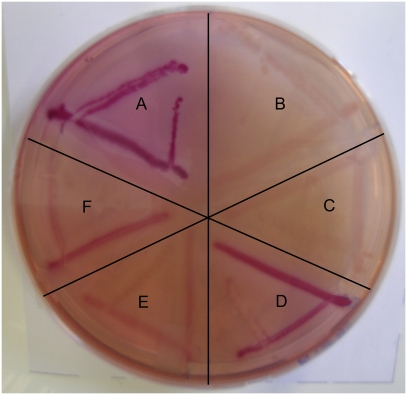
Fig. 1.
Complementation of an Escherichia coli fucP mutant. The image shows a MacConkey agar plate supplemented with 30 mM fucose. (A) Positive control, E. coli K12; (B) negative control, E. coli ΔfucP; (C) negative control, E. coli K12+pBR322; (D) positive gene-replacement control, E. coli K12+pBR322+EcfucP; (E and F) replacement with C. jejuni cj0486 in E. coli ΔfucP with pBR322 containing cj0486. Red growth indicates acidification of the medium as a result of fucose fermentation.
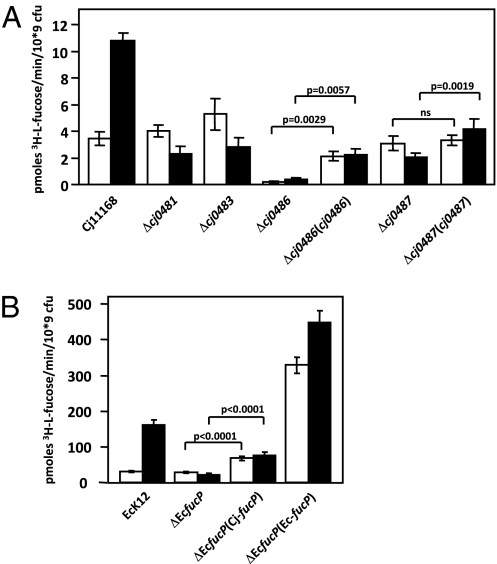
Active transport of l-fucose by C. jejuni was confirmed in 3H-l-fucose uptake experiments. In wild-type cells grown in the absence of l-fucose, the initial uptake was approximately threefold lower than in cells grown in the presence of l-fucose (Fig. 2A), consistent with E. coli (Fig. 2B), indicating that [3H]-fucose transport is inducible by its substrate. Insertion mutants were subsequently constructed into C. jejuni genes cj0481, cj0483, cj0486, and cj0487. The Δcj0486 mutant (Fig. 2) showed no significant fucose transport (< 0.2 pmols·min·109 cfu), demonstrating that Cj0486 is an essential component of the active l-fucose assimilation pathway in C. jejuni. The mutants cj0481, cj0483, and cj0487 all transport l-fucose at levels of the uniduced wild-type strain independent of the presence of fucose in the growth medium. This finding indicates that fucose transport is still occurring but not inducible in these strains (Fig. 2A). Complementation of cj0486 and cj0487 showed partial, but significant, restoration of l-fucose uptake. Quantitative RT-PCR analysis demonstrated that cj0486 and cj0487 are expressed at <11% of the wild-type levels in the complemented strains (Fig. S3), explaining the partial restoration of transport phenotypes. Similarly, complementation of the E. coli fucP mutant with cj0486 was able to significantly restore fucose uptake (Fig. 2B), consistent with the fucose fermentation assay (Fig. 1).
Fig. 2.
Fucose uptake by C. jejuni (A) and E. coli (B). The initial velocity of [3H]fucose incorporation, which was linear within the first 120 s of the assay, is shown for the indicated strains. White bars: grown in the absence of fucose (uninduced); black bars, grown in the presence of fucose (induced). SDs are displayed by error bars. P values derived from paired t tests compare the statistical differences in uptake rates between the mutants and their complements. ns, no statistically significant difference; i.e., P > 0.05.
l-Fucose Enhances C. jejuni Growth.
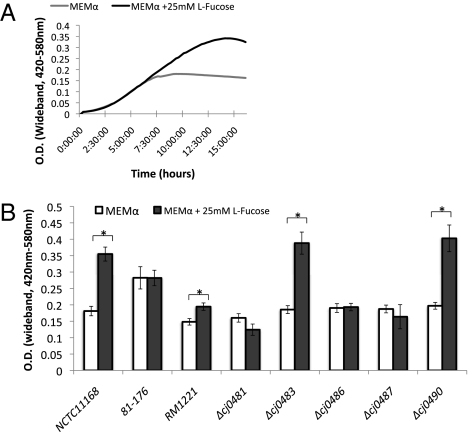
With C. jejuni demonstrating positive chemotaxis toward both l-fucose and fucosylated mucins (18) and our observation of l-fucose uptake, we sought to determine if C. jejuni has the capacity to use l-fucose for growth. Three commonly studied strains of C. jejuni, NCTC 11168, 81–176, and RM1221, were grown in MEM (MEMα) with and without 25 mM l-fucose. Two of the three strains, NCTC 11168 and RM1221, demonstrated significant increases in growth in response to the availability of l-fucose as a nutrient source (P < 0.05) (Fig. 3). The third strain, 81–176, although growing better than the other two strains [likely because of its ability to use glutamine in the growth medium (8)], did not show increased growth in the presence of l-fucose.
Fig. 3.
(A) Growth of C. jejuni NCTC 11168 in MEMα medium with and without 25 mM l-fucose over 16 h. (B) Maximum OD attained over 36 h of growth for the reference strains C. jejuni NCTC 11168, 81–176, and RM1221, and the mutant strains C. jejuni Δcj0481, Δcj0483, Δcj0486, Δcj0487, and Δcj0490. The asterisk denotes statistical significance, P < 0.05 using a paired t test. Error bars represent the SD.
Each of the previously constructed mutants (and cj0490) were grown on MEMα medium, with and without 25mM l-fucose, and growth was compared with wild-type NCTC 11168. Similar to the wild-type, mutants containing insertions in cj0483 and cj0490 (Fig. 3B) exhibited increased growth in the presence of l-fucose. However, the mutants cj0481, cj0486, and cj0487 did not show increased growth beyond normal levels in MEMα medium, indicating a loss in their ability to use l-fucose. The loss of Cj0486, the putative permease, would impair l-fucose uptake in agreement with our transport experiments. However, the roles of Cj0487, annotated as an amidohydrolase, and Cj0481, annotated as a putative dihydrodipicolinate synthase, are not currently evident. Complementation of the cj0486 and cj0487 strains did not restore growth, despite the partial restoration of l-fucose uptake (Fig. 2A). Not only were the transcript levels of cj0486 and cj0487 in the complements at very low levels (as described above), but cj0481 was not expressed in any of the mutants or complements (Fig. S3), indicating an absence of induction of the whole operon rather than polar mutations that would result in complete loss of 3H-l-fucose uptake as observed for cj0486 in Fig. 2A. These results are in agreement with the tight regulation of this operon by l-fucose induction and show that the low level of fucose assimilation into the complemented cj0486 and cj0487 strains is insufficient for induction and enhanced growth by these complemented strains.
Fucose Metabolism in C. jejuni Does Not Rely on ATP-Dependent Phosphorylation or GTP-Dependent Activation of l-Fucose.
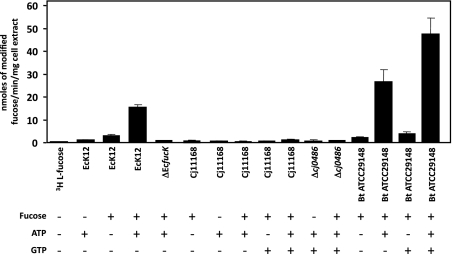
One of the key enzymes of the E. coli l-fucose metabolic pathway is the l-fuculose kinase encoded by fucK (19). This ATP-dependent kinase phosphorylates l-fuculose to l-fuculose-1-phosphate, allowing for l-fucolose-1-phosphate aldolase to convert it to l-lactaldehyde and dihydroxyacetone phosphate (Fig. S4) (29). The absence of an annotated l-fucose kinase in the C. jejuni genome would suggest that C. jejuni does not make use of this pathway. To confirm this hypothesis, we modified an approach outlined by Wang et al. (30). As predicted, sugar phosphates were not formed from 3H-l-fucose by C. jejuni, whereas they were readily formed by E. coli in an l-fuculose kinase-dependent manner (Fig. 4).
Fig. 4.
ATP-dependent 3H-l-fucose phosphorylation and GTP-dependent 3H-l-fucose activation. Whole-cell lysates (100 μg) of the indicated strains grown in the presence or absence of [3H]-l-fucose (as indicated) for 24 h and assayed for their potential to fucose under the indicated assay conditions. Ec, E. coli K12 wild-type; ΔEcfucK, E. coli K12 fucK mutant; Cj, C. jejuni NCTC11168 wild-type; Δcj0486, C. jejuni NCTC11168 Δ0486; Bt, B. thetaiotaomicron wild-type. The presence or absence of ATP and GTP are indicated by (+) and (−). Formation of modified 3H-l-fucose was found to be linear over the first 10 min of the assay. SDs are displayed by error bars. 3H-l-fucose alone did not show any significant binding to the column material.
In contrast, Bacteroides species activate l-fucose through a bifunctional l-fucokinase/GDP-fucose pyrophosphorylase (FKP) pathway, which requires ATP to generate a fucose-1-phosphate intermediate, which is then activated with GTP to generate GDP-fucose (Fig. S4) (14). Again, because C. jejuni does not possess FKP pathway homologs, 3H-modified fucose was not detected, but high levels of fucose intermediates were detected in B. thetaiotaomicron extracts incubated with ATP and augmented with added GTP (Fig. 4).
Fucose Transport Provides C. jejuni with a Competitive Advantage in Vitro and in Vivo.
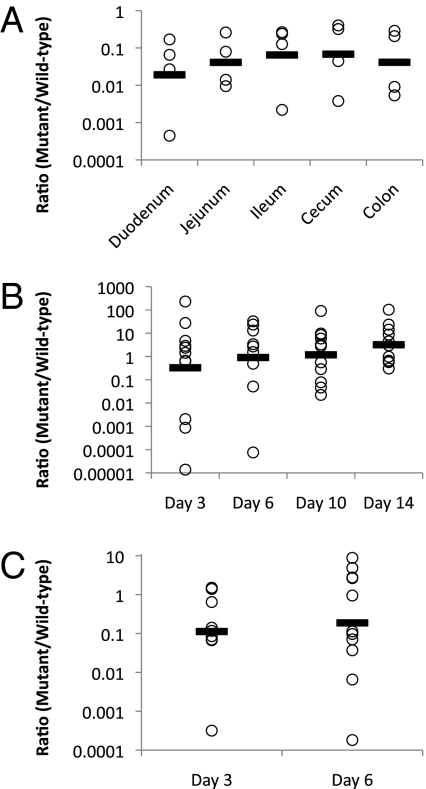
To assess the advantage of fucose transport to C. jejuni in vivo, we carried out competitive colonization assays. Wild-type NCTC 11168 and the cj0486 mutant were inoculated at a 1:1 ratio into either the pathogenic piglet or the commensal chicken model. After 3 d postinfection of neonatal piglets, samples from each section of the intestine (duodenum, jejunum, ileum, cecum, and colon) were collected and the numbers of colonizing bacteria were enumerated. The mutant to wild-type ratio recovered from the intestinal sections ranged from 0.37 (2.7-fold reduction) to 4.10−4 (2,500-fold reduction), which corresponds to an average 15- to 100-fold reduction (P < 0.002) in the mutant strain relative to the wild-type strain in each intestinal region (Fig. 5A). Thus, in the pathogenic model, the ability to transport fucose provides a strong competitive advantage for colonization.
Fig. 5.
(A) Competitive C. jejuni colonization assay in neonatal piglets. The points represent the ratio of Δcj0486 mutant/wild-type NCTC 11168 recovered from each intestinal segment indicated, 3 d postinoculation. Values are adjusted relative to the ratio of the inoculum of 0.927. A line denotes the mean for each dataset where n = 4. Statistical significance was determined by a one-way ANOVA, P < 0.002. (B) Competitive C. jejuni colonization assay of chicks over a 2-wk period postinoculation. Each point is the ratio of mutant to wild-type recovered from the cecum of a single chick. A line denotes the mean for each day, n = 9–11 per day. Statistical significance was determined by a one-way ANOVA, P = 0.82. (C) Competitive C. jejuni colonization assay in chicks receiving an l-fucose dietary supplement over a 6-d period postinoculum. Day 3, n = 9 and day 6, n = 11. Statistical significance was determined by a Student t test for each day.
Two-day-old chicks were inoculated to establish whether this phenomenon also occurred in a commensally colonized host. On days 3, 6, 10, and 14 postinoculation, ceca were extracted from the chicks and the mutant to wild-type ratio was determined. As shown in Fig. 5B, the mean of each day showed very little change above or below a competitive index of 1 (despite variability between chicks), indicating that neither the wild-type nor the mutant had a competitive advantage in this model system (P = 0.82). To determine whether l-fucose supplementation was capable of promoting a competitive advantage for C. jejuni wild-type growth in our chick model, we repeated the colonization experiment by orally feeding chicks with l-fucose three times daily at 8-h intervals. The competitive index of the cj0486 mutant was 0.11 (ninefold reduction) and 0.19 (fivefold reduction) at 3 and 7 d postinoculation, respectively (Fig. 5C). Although statistically significant for only day 3 (P < 0.05), this result indicates a notable decrease in the competitiveness of the mutant compared with wild-type, suggesting a role for fucose utilization in the early stages of colonization. The competitive index of the cj0486 mutant grown in Mueller-Hinton (MH) medium or MEM medium supplemented with 25 mM l-fucose (for 18 h) was 0.94 and 0.5 (twofold reduction), respectively. These results indicated that l-fucose confers a fitness advantage to C. jejuni both in vitro and in vivo, and the up to 50-fold difference between the in vitro and in vivo piglet competitive indices indicates an improved competitiveness of the wild-type strain in the piglet infectious model, possibly through enhanced virulence or induced pathogenic traits.
Discussion
Since early studies describing C. jejuni as an asaccharolytic organism, it was believed that C. jejuni relied completely on other molecules, such as amino acids, as substrates for growth (31). This theory was further confirmed when the first C. jejuni genome was sequenced and no known carbohydrate metabolic pathways were annotated (6). In contrast to this current dogma, we have demonstrated in this study that some strains of C. jejuni exhibit substantially increased growth in the presence of l-fucose. We have also identified a key functional protein in this pathway, Cj0486, which acts as a fucose permease to transport l-fucose into the bacterial cell. This observation is consistent with results recently obtained by Muraoka and Zhang (32), who observed the reduction of tetrazolium with a phenotypic microarray, and increased growth in minimal media by C. jejuni NCTC 11168 in the presence of l-fucose.
The use of l-fucose as a nutrient source is common to bacteria, especially among the gut microflora. Organisms such as B. thetaiotaomicron and E. coli use well-studied pathways for fucose metabolism (Fig. S4) (19). Typically, a regulator (fucR), a permease (fucP), an isomerase (fucI), a kinase (fucK), and an aldolase (fucA) are required to produce lactaldehyde and dihydroxyacetone phosphate, which is fed into the glycolytic pathway. The lactaldehyde can then be converted into lactate under aerobic conditions by lactaldehyde dehydrogenase (ald), or 1,2-propanediol under anaerobic conditions by fucO (29). However, with the exception of cj0486 (a homolog of fucP), and ald (cj0490), C. jejuni does not contain any identifiable homologs of this pathway. Instead, surrounding these two genes is a variety of other genes with largely unknown functions. These include an iclR-type regulator (Cj0480c), a putative dihydrodipicolinate synthase (Cj0481), a putative altronate hydrolase (Cj0483), an MFS transport protein (Cj0484), a short-chain dehydrogenase (Cj0485), a putative amidohydrolase (Cj0487), and a hypothetical protein (Cj0488).
We confirmed that unlike in E. coli or Bacteroides species, C. jejuni does not phosphorylate or GDP-activate l-fucose for metabolism, which would be consistent with the absence of an annotated fucK or FKP encoding gene. This finding could suggest that C. jejuni metabolizes l-fucose via an alternate pathway. The genes identified here bear some resemblance to a fucose metabolic pathway recently identified in Xanthomonas campestris (33). This pathway uses an l-fucose dehydrogenase to form l-fuconolactone, followed by an l-fuconolactonase to form l-fuconate. Further steps can catabolize l-fuconate to smaller molecules such as pyruvate and l-lactate that can be used by other pathways of the cell via the mechanism described in X. campestris (Fig. S4) (33), or the previously described l-rhamnose pathways (34).
We have shown that cj0486 complements E. coli K12 fucP, enabling the transport and subsequent metabolism of l-fucose. The observed inducible, Cj0486-dependent uptake of 3H-l-fucose by C. jejuni NCTC 11168 also provides further support that Cj0486 functions as a fucose permease. Uptake by wild-type NCTC 11168 was less than that of the E. coli fucose transporter (Fig. 3), which could reflect differences in permease activities or carbon source preferences, but was confirmed to be specific by lack of uptake in the absence of l-fucose activation. Three amino acid residues in the E. coli FucP (Asp46, Glu135, and Asn162) have previously been found to be critical for the proper functioning of FucP. These three residues are also conserved within the sequence of Cj0486, supporting the conclusion that Cj0486 serves as a fucose permease.
The fact that C. jejuni NCTC 11168 gains a growth benefit from l-fucose supplementation suggests that C. jejuni is transporting l-fucose into the cytoplasm for metabolism. Although Liu et al. recently showed that the related organism, Helicobacter pylori, can attach host-derived l-fucose to the bacterial outer surface (35); we do not currently have data demonstrating that this occurs for C. jejuni. An explanation for the down-regulation of the capsular polysaccharide putative transferases genes cj1434c and cj1438c (Table S1) in the presence of l-fucose is also not immediately clear. However, cross-talk between glycoconjugate biosynthetic pathways has been shown previously (36, 37), and it could be speculated that l-fucose provides a cue to C. jejuni to down-regulate capsular polysaccharide production.
Within the gut, the primary sources of l-fucose will be the host itself, where both host-secreted and cell surface-associated mucins (particularly in the small intestine and cecum) are heavily fucosylated. Previous studies into the fucose content of pig ileal digesta have found that as much as 74% of the fucose present is mucin-derived (38, 39). Our microarray results from C. jejuni cultures grown in the presence of porcine gastric mucin (Table S2) found a significant up-regulation of the genes cj0481 to cj0490, similar to when C. jejuni is grown with l-fucose alone, demonstrating that purified mucin also triggers this operon.
Although our mass spectrometry analyses indicate similar fucosylation levels in both chick and porcine mucins, we observed that most of the fucosylated O-glycan mucin structures from chicks are more sulfated compared with their pig counterparts (Fig. S5). Sulfation of mucin oligosaccharides may decrease the accessibility of fucose by mucin-degrading fucosidases (40) and has been demonstrated to decrease the intestinal barrier permeability to pathogens, including C. jejuni (41). When an excess of free l-fucose was added to our chick colonization experiment, we then observed a competitive advantage of wild-type C. jejuni over the fucP mutant, further suggesting that the availability of fucose in the intestine influences bacterial fitness. Thus, the availability of fucose to C. jejuni in the presence and absence of sulfation deserves further study.
Herein, we have demonstrated that a functional l-fucose permease provides a distinct competitive advantage for C. jejuni, colonizing the porcine intestine that is not found during normal colonization of the chick cecum. A 15- to 100-fold reduction in the colonization of the cj0486 mutant compared with the wild-type was observed in the piglet pathogenic model, but no significant difference was observed in the chick commensal model in the absence of added l-fucose. Differences in the microbial ecosystems between the two animals may alter the competition for these niches, making the ability to draw upon l-fucose valuable for C. jejuni colonization in the piglet, but less relevant in the chick. When the chicks were given an l-fucose supplement into their diet, the ratio of colonizing mutant and wild-type showed a distinct shift in favor of the l-fucose using wild-type strain during early stages of colonization. The results point to the conclusion that if significant amounts of l-fucose are available in the environment, the ability to acquire and use l-fucose provides a competitive advantage for C. jejuni.
Materials and Methods
Strains Used and Growth Media.
C. jejuni strains NCTC 11168, sequenced by the Sanger Centre in 2000 (6); 81–176, an isolate from an outbreak of Campylobacter in raw milk (42); and RM1221, an isolate recovered from a chicken carcass (24) were used in this study. Routine growth of C. jejuni was carried out on MH agar plates, MH biphasic flasks, or MH liquid cultures, supplemented with chloramphenicol (20 μg/mL), and kanamycin (10 μg/mL) as needed. Cultures were incubated at 37 °C in MACS microaerobic incubators with a gas concentration of 8% O2, 4% H2, 5% CO2, and 83% N2. B. thetaiotaomicron was grown anaerobically at 37 °C in trypticase yeast extract glucose broth supplemented with 10 μg/mL erythromycin. Genetic manipulations were conducted using E. coli strain K12 grown on Luria-Bertani (LB) agar plates or LB broth supplemented with ampicillin (100 μg/mL), chloramphenicol (20 μg/mL), or kanamycin (10 μg/mL) as needed, and incubated at 37 °C.
Mutant Construction and Complementation.
Mutants were constructed by the insertion of an antibiotic resistance cassette into the ORF of the gene using previously described protocols (43–46) and detailed in SI Materials and Methods. Complemented strains were constructed by the addition of the genes cj0486 to cj0490 as described by Muraoka et al. (32) and in SI Materials and Methods (Table S3).
Growth on l-Fucose.
MEMα medium (Gibco 41061) containing glutamine, but no phenol red, was supplemented with 20 μM FeSO4 and 25 mM l-fucose (Sigma-Aldrich) and filter-sterilized (0.2-μm pore size). The wild-type or mutant strains were grown overnight in MH biphasic cultures. These cultures were used to inoculate the growth medium at an OD600 of 0.01. Aliquots of this culture (300 μL) were injected into honeycomb 100-well plates designed for use with a Bioscreen C incubator/plate reader. Each growth condition was replicated in 10 wells each, as were blanks containing only MEMα medium. The plates were incubated for 1 h, at 37 °C in a MACS-VA500 workstation, under microaerobic conditions, and were sealed shut with airtight polyethylene sealing tape (Micronova Manufacturing, Inc.). The plates were then placed in a Bioscreen C plate reader and incubated at 37 °C with moderate, continuous shaking for 36 h with wideband (420–580 nm) OD readings every 15 min. The results are displayed as the maximum OD obtained during 36 h of growth, normalized against the background OD of the medium. Statistical significance was determined using Student t test with a P value ≤ 0.05.
Radioactive l-Fucose Uptake.
Bacteria were grown to exponential phase in MH or LB broth media in the presence or absence of 20 mM l-fucose and the OD was adjusted to 1.0 at 600 nm. Bacteria were washed three times in transport buffer [50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 10 mM KCl]. Uptake measurements were initiated by the addition of l-[5,6-3H]fucose (ART) together with nonradioactive fucose to a final concentration of 20 μM (40 mCi·mmol−1) to the bacterial suspension at 37 °C. Samples (1 mL) were taken at different time points (30 s to 10 min), rapidly filtered through 0.22-μm mixed cellulose (GS) filters (Millipore), and washed with 5 mL of 0.1 M LiCl to stop the reaction. Radioactivity of bacteria on the filter membrane was determined by scintillation oscillography. E. coli K12 wild-type strain and the corresponding fucP mutant were used to optimize the assay and C. jejuni NCTC 11168 and corresponding isogenic mutants were then compared in triplicate experiments.
ATP-Dependent l-Fucose Phosphorylation and GTP-Dependent l-Fucose Activation.
ATP-dependent phosphorylation and GTP-dependent activation of radiolabeled l-fucose was carried out using a modified version of the protocol by Wang et al. (30) for 20 min at 30 °C in a reaction volume of 500 μL using 20 mg of whole cell lysate in a reaction buffer composed of: 25 mM Tris/HCl (pH 7.5), 5 mM DTT, 10 mM nucleotide triphosphate (ATP, GTP, or ATP plus GTP), 5 mM of divalent cations (MgCl2 or MnCl2), and 20 μM [3H] l-fucose (40 mCi·mmol−1). Whole-cell lysates were prepared by sonication and dialyzed against 25 mM Tris/HCl (pH 7.5). DTT and MgCl2 /MnCl2 were added separately after the dialysis step. ATP, GTP, or both nucleotides were added with [3H] l-fucose to start the reaction. At different time points, 0.1-mL aliquots of the reaction mixtures were loaded onto 0.5 mL of an anion-exchange resin (Dowex anion-exchange resin 1 × 8 mesh size 200–400) prepared in a gravity-flow column (GE Healthcare; Column PD-10, empty). After three washes with 2 mL of distilled water, the column was eluted with 0.1 M HCl (3 × 1 mL). The eluate was added to 10 mL of scintillation mixture, and radioactivity was determined using a scintillation counter.
Total RNA Extraction and Microarray Hybridization and Analysis.
Total RNA was extracted using the previously described hot phenol method (43, 44, 47). Microarray hybridization and analysis was also conducted as previously described (43, 44, 47).
Competitive Assays.
Animal procedures were approved by the Animal Care and Use Committee at the University of Ottawa. The ability of the cj0486 mutant to compete against the NCTC 11168 wild-type strain was tested in both in vitro and in vivo conditions. For the porcine competitive colonization assay: male, specific pathogen-free neonatal colostrum-deprived piglets were acquired from the Canadian Food Inspection Agency. The piglets were allowed to acclimatize over a couple days to their environment and four piglets were inoculated with ∼107 cells of a 1:1 mixture of wild-type C. jejuni NCTC 11168 and the cj0486 mutant. To confirm the exact ratio of wild-type to mutant, samples of the inoculum were serially diluted, plated and enumerated. The piglets remained infected for 3 d, at which point they were killed, and samples were taken from the duodenum, jejunum, ileum, cecum, and colon. The tissue samples were homogenized, serially diluted in MH broth and plated onto Campylobacter agar base (Oxoid CM935) containing Campylobacter-selective Karmali supplements (Oxoid SR167E). The mutant strains were isolated using selective plates also containing kanamycin. The plates were incubated at 37 °C, under microaerobic conditions for 48 h. Statistical significance was determined using a one-way ANOVA.
A similar approach was used for competitive assays using the chick model. Forty-three chicks were acquired from the Canadian Food Inspection Agency's specific pathogen-free flock. These chicks were inoculated with ∼104 cells of a 1:1 mixture of NCTC 11168 and cj0486. At 3, 6, 10, and 14 d postinoculation, 10 or 11 chicks were killed by cervical dislocation and their ceca were collected and weighed. The cecal contents were resuspended into MH broth, serially diluted, and plated onto Karmali selective plates, as described above. The bacteria counts for all of the in vitro and in vivo competitive assays were determined and are represented as the log10 ratio of cj0486 cfu/wild-type cfu. Twenty additional chicks were syringe-fed 17 mg l-fucose supplements, three times daily, at 8-h intervals. They were inoculated with a 1:1 mixture of cj0486 mutant to wild-type, as described above, following their second fucose supplement. Nine and 11 chicks were killed 3 and 6 d postinoculation, respectively. Statistical significance was determined by a Student t test.
An in vitro competitive growth assay was carried out in filter-sterilized MEMα medium supplemented with 20 μM FeSO4, both with and without 25 mM l-fucose, as well as in MH medium alone. As above, the media were inoculated with a 1:1 mixture of the cj0486 mutant and wild-type and grown for 36 h with samples taken and plated at 6-h intervals.
Supplementary Material
Acknowledgments
The authors thank Tracy Raivio for access to the Escherichia coli mutant Keio collection, Anna Cunningham and Jacek Stupak for assistance with mucin purification and analyses, and Qijing Zhang for providing the pRRK plasmid construct for the creation of the cj0486 and cj0487 complements. This work was supported by the Canadian Institutes of Health Research (A.S.) and the Ontario Ministry of Agriculture and Food (C.M.S.); the Ontario Graduate Scholarship in Science and Technology (to M.S.); and a Canadian Association of Gastroenterology/AstraZeneca Research Initiative Award (to L.M.F.). C.M.S. holds an Alberta Ingenuity Scholar Award.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data have been deposited in the National Center for Biotechnology Information Geo Omnibus database.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014125108/-/DCSupplemental.
References
- 1.Coker AO, Isokpehi RD, Thomas BN, Amisu KO, Obi CL. Human campylobacteriosis in developing countries. Emerg Infect Dis. 2002;8:237–244. doi: 10.3201/eid0803.010233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galanis E. Campylobacter and bacterial gastroenteritis. CMAJ. 2007;177:570–571. doi: 10.1503/cmaj.070660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ketley JM. Pathogenesis of enteric infection by Campylobacter. Microbiology. 1997;143(1):5–21. doi: 10.1099/00221287-143-1-5. [DOI] [PubMed] [Google Scholar]
- 4.Lee A, O'Rourke JL, Barrington PJ, Trust TJ. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: A mouse cecal model. Infect Immun. 1986;51:536–546. doi: 10.1128/iai.51.2.536-546.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrne CM, Clyne M, Bourke B. Campylobacter jejuni adhere to and invade chicken intestinal epithelial cells in vitro. Microbiology. 2007;153:561–569. doi: 10.1099/mic.0.2006/000711-0. [DOI] [PubMed] [Google Scholar]
- 6.Parkhill J, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 7.Velayudhan J, Kelly DJ. Analysis of gluconeogenic and anaplerotic enzymes in Campylobacter jejuni: An essential role for phosphoenolpyruvate carboxykinase. Microbiology. 2002;148:685–694. doi: 10.1099/00221287-148-3-685. [DOI] [PubMed] [Google Scholar]
- 8.Hofreuter D, Novik V, Galán JE. Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe. 2008;4:425–433. doi: 10.1016/j.chom.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Velayudhan J, Jones MA, Barrow PA, Kelly DJ. L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect Immun. 2004;72:260–268. doi: 10.1128/IAI.72.1.260-268.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guccione E, et al. Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol Microbiol. 2008;69(1):77–93. doi: 10.1111/j.1365-2958.2008.06263.x. [DOI] [PubMed] [Google Scholar]
- 11.Leach S, Harvey P, Wali R. Changes with growth rate in the membrane lipid composition of and amino acid utilization by continuous cultures of Campylobacter jejuni. J Appl Microbiol. 1997;82:631–640. doi: 10.1111/j.1365-2672.1997.tb02873.x. [DOI] [PubMed] [Google Scholar]
- 12.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 15.Becker DJ, Lowe JB. Fucose: Biosynthesis and biological function in mammals. Glycobiology. 2003;13(7):41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- 16.Day CJ, et al. Differential carbohydrate recognition by Campylobacter jejuni strain 11168: influences of temperature and growth conditions. PLoS ONE. 2009;4:e4927. doi: 10.1371/journal.pone.0004927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cone RA. Barrier properties of mucus. Adv Drug Deliv Rev. 2009;61(2):75–85. doi: 10.1016/j.addr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Hugdahl MB, Beery JT, Doyle MP. Chemotactic behavior of Campylobacter jejuni. Infect Immun. 1988;56:1560–1566. doi: 10.1128/iai.56.6.1560-1566.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 20.Hooper LV, Gordon JI. Glycans as legislators of host-microbial interactions: Spanning the spectrum from symbiosis to pathogenicity. Glycobiology. 2001;11(2):1R–10R. doi: 10.1093/glycob/11.2.1r. [DOI] [PubMed] [Google Scholar]
- 21.Gunn FJ, Tate CG, Henderson PJ. Identification of a novel sugar-H+ symport protein, FucP, for transport of L-fucose into Escherichia coli. Mol Microbiol. 1994;12:799–809. doi: 10.1111/j.1365-2958.1994.tb01066.x. [DOI] [PubMed] [Google Scholar]
- 22.Snider TA, Fabich AJ, Conway T, Clinkenbeard KD. E. coli O157:H7 catabolism of intestinal mucin-derived carbohydrates and colonization. Vet Microbiol. 2009;136(1-2):150–154. doi: 10.1016/j.vetmic.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 24.Parker CT, Quiñones B, Miller WG, Horn ST, Mandrell RE. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J Clin Microbiol. 2006;44:4125–4135. doi: 10.1128/JCM.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fearnley C, et al. Identification of hyperinvasive Campylobacter jejuni strains isolated from poultry and human clinical sources. J Med Microbiol. 2008;57:570–580. doi: 10.1099/jmm.0.47803-0. [DOI] [PubMed] [Google Scholar]
- 26.Javed MA, et al. Transposon mutagenesis in a hyper-invasive clinical isolate of Campylobacter jejuni reveals a number of genes with potential roles in invasion. Microbiology. 2010;156:1134–1143. doi: 10.1099/mic.0.033399-0. [DOI] [PubMed] [Google Scholar]
- 27.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 29.Baldomà L, Aguilar J. Metabolism of L-fucose and L-rhamnose in Escherichia coli: Aerobic-anaerobic regulation of L-lactaldehyde dissimilation. J Bacteriol. 1988;170:416–421. doi: 10.1128/jb.170.1.416-421.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Xiao X, Saito A, Schrempf H. Streptomyces olivaceoviridis possesses a phosphotransferase system that mediates specific, phosphoenolpyruvate-dependent uptake of N-acetylglucosamine. Mol Genet Genomics. 2002;268:344–351. doi: 10.1007/s00438-002-0749-3. [DOI] [PubMed] [Google Scholar]
- 31.Karmali MA, Roscoe M, Fleming PC. Modified ammonia electrode method to investigate D-asparagine breakdown by Campylobacter strains. J Clin Microbiol. 1986;23:743–747. doi: 10.1128/jcm.23.4.743-747.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muraoka WT, Zhang Q. Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J Bacteriol. 2011;193:1065–1075. doi: 10.1128/JB.01252-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yew WS, et al. Evolution of enzymatic activities in the enolase superfamily: L-fuconate dehydratase from Xanthomonas campestris. Biochemistry. 2006;45:14582–14597. doi: 10.1021/bi061687o. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe S, Saimura M, Makino K. Eukaryotic and bacterial gene clusters related to an alternative pathway of nonphosphorylated L-rhamnose metabolism. J Biol Chem. 2008;283:20372–20382. doi: 10.1074/jbc.M801065200. [DOI] [PubMed] [Google Scholar]
- 35.Liu TW, et al. Role for alpha-L-fucosidase in the control of Helicobacter pylori-infected gastric cancer cells. Proc Natl Acad Sci USA. 2009;106:14581–14586. doi: 10.1073/pnas.0903286106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerry P, Szymanski CM. Campylobacter sugars sticking out. Trends Microbiol. 2008;16:428–435. doi: 10.1016/j.tim.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Bernatchez S, et al. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J Biol Chem. 2005;280:4792–4802. doi: 10.1074/jbc.M407767200. [DOI] [PubMed] [Google Scholar]
- 38.Lien KA, Sauer WC, Fenton M. Mucin output in ileal digesta of pigs fed a protein-free diet. Z Ernahrungswiss. 1997;36(2):182–190. doi: 10.1007/BF01611398. [DOI] [PubMed] [Google Scholar]
- 39.Monsma DJ, Vollendorf NW, Marlett JA. Determination of fermentable carbohydrate from the upper gastrointestinal tract by using colectomized rats. Appl Environ Microbiol. 1992;58:3330–3336. doi: 10.1128/aem.58.10.3330-3336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson AM, Wright DP. Bacterial glycosulphatases and sulphomucin degradation. Can J Gastroenterol. 1997;11:361–366. doi: 10.1155/1997/642360. [DOI] [PubMed] [Google Scholar]
- 41.Dawson PA, et al. Reduced mucin sulfonation and impaired intestinal barrier function in the hyposulfataemic NaS1 null mouse. Gut. 2009;58:910–919. doi: 10.1136/gut.2007.147595. [DOI] [PubMed] [Google Scholar]
- 42.Hofreuter D, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palyada K, et al. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics. 2009;10:481. doi: 10.1186/1471-2164-10-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl Environ Microbiol. 2008;74:1583–1597. doi: 10.1128/AEM.01507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guerry P, Yao R, Alm RA, Burr DH, Trust TJ. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 1994;235:474–481. doi: 10.1016/0076-6879(94)35163-5. [DOI] [PubMed] [Google Scholar]
- 46.Karlyshev AV, Wren BW. Development and application of an insertional system for gene delivery and expression in Campylobacter jejuni. Appl Environ Microbiol. 2005;71:4004–4013. doi: 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palyada K, Threadgill D, Stintzi A. Iron acquisition and regulation in Campylobacter jejuni. J Bacteriol. 2004;186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.