Abstract
Harnessing the new bioremediation and biotechnology applications offered by the dissimilatory metal-reducing bacteria, Shewanella oneidensis MR-1, requires a clear understanding of its transcription machinery, a pivotal component in maintaining vitality and in responding to various conditions, including starvation and environmental stress. Here, we have reconstituted the S. oneidensis RNA polymerase (RNAP) core in vivo by generating a co-overexpression construct that produces a long polycistronic mRNA encoding all of the core subunits (α, β, β′, and ω) and verified that this reconstituted core is capable of forming fully functional holoenzymes with the S. oneidensis σ factors σ70, σ38, σ32, and σ24. Further, to demonstrate the applications for this reconstituted core, we report the application of single-molecule fluorescence resonance energy transfer (smFRET) assays to monitor the mechanisms of transcription by the S. oneidensis σ70-RNAP holoenyzme. These results show that the reconstituted transcription machinery from S. oneidensis, like its Escherichia coli counterpart, “scrunches” the DNA into its active center during initial transcription, and that as the holoenzyme transitions into elongation, the release of σ70 is non-obligatory.
Keywords: Shewanella oneidensis, RNA polymerase, σ Factor, Co-overexpression, Single-molecule spectroscopy, Alternating-laser excitation
Shewanella oneidensis MR-1 is a Gram-negative, facultative anaerobic, metal-reducing bacterium, whose versatile metabolic and respiratory capabilities have made it a model organism for bioremediation studies, due to its ability to reduce toxic metal and organic pollutants, such as fumarate, nitrate, dimethyl sulfoxide (DMSO), trimethylamine N-oxide (TMAO), Fe(III), Cr(VI), and U(VI) [1–3], to soluble complexes or solid phase associated minerals, like U(IV) nanoparticles [4]. In order to utilize this vast array of electron acceptors, S. oneidensis has developed an expanded suite of regulatory genes that allow for the organism’s rapid response to its diverse and fluctuating environment [5]. An extensive amount of studies have focused on unraveling the S. oneidensis genome [5,6] and its gene expression profile under various physiological conditions, such as pH [7], exposure to terminal electron acceptors [8], and heat shock [9]. However, since transcription initiation is the first step of gene expression and is an important check point of gene regulation, it is imperative to characterize the S. oneidensis RNA polymerase (RNAP)4 complex and its associated factors.
Like other prokaryotic systems, the S. oneidensis transcription machinery is composed of an RNAP core, consisting of two αs, a β, a β′, and an ω subunits, and a suite of transcription initiation factors (σ factors), of which six putative σ factor have been identified, σ70, σ54, σ38, σ32, σ27, and σ24. As in Escherichia coli, the RNAP core interacts with one of its σ factors to form an RNAP holoenzyme that is directed to a specific DNA promoter region by the sequence specific determinants of the σ factor [10], forming a transcription initiation complex. This initiation complex formation is a critical component of gene expression regulation and allows S. oneidensis to adaptively respond to external stimuli.
Recent reports have demonstrated that S. oneidensis RNAP core can be purified by immunoaffinity [11] and by FlAsH-ethylenediamine affinity [12]; however, yields for these preparations were unsatisfactory because RNAP was not overexpressed in S. oneidensis for the immunoaffinity method, and in the case of FlAsH affinity purification, the tagged-α subunit was overexpressed, but the other subunits (β, β′, and ω) were endogenously expressed at low levels. In addition, endogenous expression does not allow easy genetic manipulation of the RNAP subunits. To establish a rapid and reliable method for significant quantities of S. oneidensis RNAP core, we have created a polycistronic expression plasmid that co-overexpresses all the RNAP subunits, using E. coli as a host. This multi-subunit co-overexpression scheme has been successfully applied to the purification of RNAP of other organisms [13–15], and this scheme allows large-scale production, facile genetic manipulations, and produces in vivo assembled, fully functional, S. oneidensis RNAP core, without the misfolding and loss of activity that can be associated with in vitro reconstitution [16,17]. Additionally, we expressed and purified four of the six putative S. oneidensis σ factors (σ70, σ38, σ32, and σ24), formed their respective holoenzyme transcription complexes, and evaluated the activity and specificity of these complexes on predicted promoter sequences. To demonstrate the myriad of applications for this reconstituted core, we extended existing single-molecule fluorescence assays to primary holoenzyme of S. oneidensis, σ70-RNAP, and examined the mechanism of transcription initiation and subunit composition of this machinery during transcription initiation and elongation.
Recent single-molecule biophysical studies of E. coli σ70-RNAP have demonstrated that the mechanism of transcription initiation proceeds through the “scrunching” of downstream DNA into the active center of the RNAP [18,19]. During each cycle of abortive initiation, RNAP pulls downstream DNA into itself, pulling in 1 bp per phosphodiester bond formed and accommodating the accumulated DNA as single stranded bulges within the unwound region. When the abortive RNA is released, the RNAP releases the “scrunched” DNA, and the initial transcription state is recovered. While scrunching is thought to be a general mechanism for all polymerases, it has only been observed for one other polymerase, T7 bacteriophage RNAP [20]. Additionally, single-molecule fluorescence assays have also been utilized to determine the fate of σ70 during the transition from transcription initiation to elongation. These studies of E. coli σ70-RNAP have demonstrated its non-obligatory release of σ70 during the transition from transcription initiation to elongation [21–23], providing support for a stochastic σ release model, where σ is released stochastically during elongation rather than at a specific RNA nascent chain length [24,25].
Examining both the mechanism of transcription initiation and the extent of σ70 retention by S. oneidensis highlights the applications for reconstituted transcription machinery and provides evidence that the transcription mechanisms and σ release models found for these organisms can be generalized to other bacterial species.
Material and methods
Design of the polycistronic co-overexpression plasmid
The co-overexpression plasmid pYK201 (Fig. 1A) consists of a polycistronic artificial operon rpoA-rpoB-rpoC-rpoZ, under the control of a single IPTG-inducible T7 promoter and T7 transcription terminator derived from pET-22b(+) (Novagen); a separate ribosome-binding site (rbs) was inserted to proceed each open reading frame (ORF). First, expression construct of each subunit were generated from clones of each ORF (gifts from Dr. M. Uljana Mayer [PNNL]). The intermediate plasmids involved in the construction of pYK201 are listed in Table 1. The plasmids were maintained in NovaBlue cells (Novagen). The primers for PCR amplifications are listed in Supplementary Table 1; all PCR fragments were generated by Pfu polymerase (Stratagene). The steps leading to the final 15 kb polycistronic plasmid are described below.
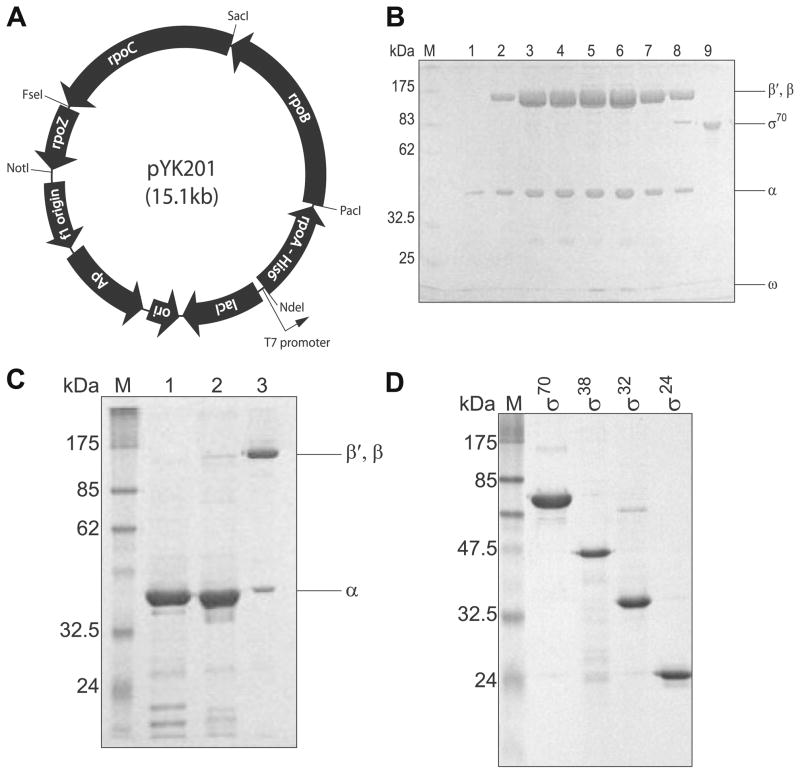
Fig. 1.
Cloning and purification of S. oneidensis RNAP. (A) Schematic diagram of the S. oneidensis RNAP co-overexpression construct. (B) SDS–PAGE gel of the purified S. oneidensis RNAP core. (Lane M) Protein molecular weight marker (New England Biolabs); (lanes 1–8) S. oneidensis RNAP core fractions from anion exchange chromatography; (lane 9) purified S. oneidensis σ70. (C) SDS–PAGE gel of IMAC-based pull-down assay from cell extract containing overexpressed α subunits. (Lane M) Protein molecular weight marker; (lane 1) S. oneidensis α overexpression; (lane 2) E. coli α overexpression; (lane 3) S. oneidensis RNAP core. (D) SDS–PAGE gel of purified σ factors. (Lane M) Protein molecular weight marker; (lane 1) σ70; (lane 2) σ38; (lane 3) σ32; (lane 4) σ24. Gels were Coomassie stained.
Table 1.
Plasmids for the polycistronic overexpression plasmid pYK201, and the σ subunits.
| Plasmid | Description | His6 tag position |
|---|---|---|
| pSH101 | α + C-terminal His6 tag + PacI inserted between NdeI and NcoI of pET-22b(+) | C |
| pSH102 | β + SacI inserted between NcoI and NdeI of pET-16b | |
| pSH103 | β′ + NotI inserted between NcoI and NdeI of pET-16b, changed start codon from GTG to ATG | |
| pSH104 | ω + NotI inserted between NcoI and NdeI of pET-16b | |
| pSH202 | rbs site + β from pSH102 inserted between PacI and SacI of pSH101 | |
| pSH203 | rbs site + β′ + FseI from pSH103 inserted between SacI and NotI of pSH202 | |
| pYK201 | rbs site + ω of pSH104 inserted between FseI and NotI of pSH203 | C (of α) |
| pSH001 | σ70 inserted between NheI and XhoI of pTrcHis A | N |
| pNG045 | σ70 inserted between NheI and EcoRI of pET-28b | N |
| pNG201 | σ70ΔCys inserted between EcoRI and NotI of pET-DUET | N |
| pNG203 | σ70, N399C inserted between EcoRI and NotI of pET-DUET | N |
| pNG079 | σ38 inserted between NcoI and EcoRI of pETM-11 | N |
| pNG080 | σ38 inserted between NheI and EcoRI of pTrcHis A | C |
| pNG076 | σ32 inserted between NdeI and NotI of pET-22b(+) | C |
| pNG077 | σ32 inserted between NheI and EcoRI of pTrcHis A | C |
| pNG083 | σ24 inserted between NcoI and NotI of pETM-11 | N |
| pNG126 | σ24 in pTrcHis-TOPO | N |
The rpoA gene (SO0256) was modified to contain a 5′ NdeI site and a 3′ His6 tag sequence, followed by a stop codon, PacI and NcoI sites sequentially, by add-on PCR using primers 0256F1 and 0256R1. The resulting PCR product was cloned as a NdeI/NcoI fragment between the NdeI and NcoI sites of pET-22b(+), to generate pSH101. PCR product of the rpoB gene (SO0224) flanked with a 5′ NcoI site and 3′ SacI/NdeI sites was produced by add-on PCR using primers 0224F1 and 0224R1; this PCR product was inserted between the NcoI and NdeI sites of pET-16b (Novagen) to create pSH102. Add-on PCR product of the rpoC gene (SO0225) flanked with a 5′ NcoI site and 3′ NotI/NdeI sites was produced with primers 0225F1 and 0225R1, then inserted between the NcoI and NdeI sites of pET-16b to create pSH103; in concert, ATG was added in front of the endogenous GTG start codon for higher translation yield, the ATG sequence was included in the add-on PCR primer 0225F1. Then, add-on PCR product of the rpoZ gene (SO0360) flanked with a 5′ NcoI site and 3′ NotI/NdeI sites was inserted between the NcoI and NdeI sites of pET-16b to create pSH104; primers for the PCR were 0360F1 and 0360R1.
For construction of the co-overexpression construct, each coding sequence (rpoB in pSH102, rpoC in pSH103 and rpoZ in pSH104) including an upstream rbs site was amplified by add-on PCR. The PCR products were flanked with unique restriction sites: 5′ PacI site and 3′ SacI site for rpoB (primers 0224F2 and 0225R2); 5′ SacI site and 3′ FseI/NotI sites for rpoC (primers 0225F2 and 0225R2); and 5′ FseI site and 3′ NotI site for rpoZ (primers 0360F2 and 0360R1). The PCR product of the rpoB gene was inserted between the PacI and SacI sites of pSH101 to yield pSH202. Then, the PCR product of the rpoC gene was inserted between the SacI and NotI sites of pSH202 to produce pSH203. Finally, the co-overexpression plasmid pYK201 was created after the PCR product of rpoZ gene was inserted between the FseI and NotI sites of pSH203.
Construction of expression plasmids of the σ factors and single-cysteine σ70 mutant
For the construction of the σ factors’ expression plasmids, each σ subunit gene fragment was PCR-amplified from the S. oneidensis genome with the primers listed in Supplementary Table 1. The digested fragments were then inserted into the corresponding sites of pTrcHis A (Invitrogen), pET-22b(+), pET-M11 (EMBL) [26], or pTrcHis-TOPO (Invitrogen). The resulting expression vectors for the rpoD (SO1284), rpoS (SO3432), rpoH (SO4583), and rpoE (SO1342) genes are listed in Table 1.
The single-cysteine mutant of σ70 (N399C) was prepared with the Quikchange site-directed mutagenesis kit (Stratagene). A pET-DUET (Novagen) expression vector containing the σ70 gene (rpoD, SO1284) under control of the T7 promoter was used as a backbone for all subcloning experiments. Prior to creating the single-cysteine derivative, the two native Cys residues C294 and C343 of wild-type S. oneidensis σ70 were replaced with structurally similar serine residues, creating a Cys(−) σ70, referred to as σ70ΔCys (pNG201), and no negative effect on transcription activity was observed with the serine replacements (data not shown), consistent with E. coli σ70ΔCys [27,28]. Then, a cysteine residues at residue 399 in σ70 was introduced using the pNG201 as the template plasmid for site-directed mutagenesis, creating σ70,N399C (pNG203). The DNA sequences of the single-cysteine σ70 mutant constructs and that of σ70ΔCys were confirmed by DNA sequencing.
Co-overexpression and purification of the S. oneidensis RNAP core
The pYK201 plasmid was transformed into BL21 Star (DE3) cells (Invitrogen). An overnight culture (10 ml) from a single colony was subcultured into 1 L of LB media supplemented with 100 μg/ml of ampicillin at 37 °C and grown to an OD600 of 0.3–0.5; the culture was cooled on ice to room temperature before induction by the addition of a final concentration of 0.5 mM IPTG. The cells were grown for additional 5 h at room temperature, then harvested by centrifugation for 15 min at 5000g at 4 °C. The cell pellet was resuspended in 35 ml of nickel binding buffer (NBB: 50 mM sodium phosphate, 10 mM Tris–HCl, 0.5 M NaCl, 5 mM imidazole, 4 mM 2-mercaptoethanol [2-ME], 5% glycerol, pH 8.0), disrupted by sonication, and the lysate was centrifuged for 30 min at 15,000 rpm at 4 °C; subsequent steps were carried out at 4 °C.
One milliliter of Ni-NTA agarose (Qiagen) was equilibrated with 10 ml of NBB in a 20-ml disposable column (Econo-Pac, Bio-Rad), and the cleared lysate was passed through the column by gravity flow, followed by a 10 ml NBB (plus 10 mM imidazole) wash. Proteins were eluted with 3 column volume (CV) of NBB containing 20 mM, 40 mM, and 150 mM imidazole, respectively, and fractions were collected at 1 CV each. The fractions were analyzed by SDS–PAGE, and peak fractions containing RNAP were pooled, and diluted with TGED buffer (20 mM Tris–HCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 5% glycerol) to a final NaCl concentration of 0.15 M.
The resulting sample was loaded onto a MonoQ 10/10 (GE Healthcare) column pre-equilibrated with TGED with 0.2 M NaCl, using Aktä purifier (GE Healthcare). Purification using the anion exchange column was carried out as described [29]. Fractions containing pure RNAP were pooled and dialyzed against storage buffer (10 mM Tris–HCl, pH 7.9, 50% glycerol, 0.1 mM EDTA, 0.1 mM DTT, 0.1 M NaCl). The typical yield was about 20–30 mg of purified RNAP from a 1L culture.
Overexpression and purification of the S. oneidensis σ subunits
Soluble σ70 (pSH001) were expressed in TOP10 cells (Invitrogen). The cells were grown in LB media supplemented with 100 μg/ml ampicillin at 37 °C until OD600 0.7–0.9, cooled on ice to room temperature, induced with 0.5 mM IPTG, then grew for another 5 h at room temperature. The purification procedure was the same as the steps described above. Insoluble σ38 (pNG079), σ32 (pNG076), and σ70N399C (pNG103) were produced in BL21-Gold (DE3) cells (Stratagene), growth conditions, and purification steps were described previously [30]. Typical yields for σ70, σ70N399C, σ38 and σ32 are 2 mg/l, 10 mg/l, 20 mg/l, and 15 mg/l, respectively.
The rpoE gene (pNG083) was expressed in soluble form by BL21-Gold (DE3) as previously described [31]. The cell pellet was resuspended in 35 ml of buffer A (50 mM Tris–HCl, pH 7.4, 0.5 M NaCl, 10 mM MgCl2, 5 mM 2-ME, 10% glycerol), and lysed by sonication. The lysate was centrifuged at 19,000g for 30 min at 4 °C. The soluble proteins in the supernatant were applied onto a 1 ml Ni-NTA agarose column, and washed with 20 CV of buffer B (buffer A at pH 8.0) plus 10 mM imidazole. The column was washed with 3 CV of buffer B plus 20 mM, then 40 mM imidazole, and the protein was eluted with 8 ml of buffer B with 150 mM imidazole. The eluted protein was diluted to ~1 mg/ml and dialyzed in TGED with 0.15 MNaCl, then applied to a Resource Q column (GE Healthcare) pre-equilibrated with TGED plus 0.15 M NaCl. A linear gradient of 0.15–1 M NaCl was run over 120 min at a flow rate of 0.25 ml/min. Fractions of purified σ24, identified by SDS–PAGE, were pooled and stored as an ~65% ammonium sulfate precipitate [32] at 4 °C. Yields were typically 1 mg/L.
5′-Rapid amplification of cDNA ends (RACE)
The 5′-RACE method [33] was used to extend partial cDNA clones by amplifying the 5′ sequences of S. oneidensis mRNAs. Total RNA was extracted from S. oneidensis cells grown at 35 °C using the RiboPure-Bacteria kit (Ambion) with genomic DNA contamination removed by DNase I. First strand cDNA synthesis was performed using 5 μg of total RNA and 50 ng of gene specific primer GSP1 (Supplementary Table 2). The RNA template was then degraded by the RNase H, and the first strand cDNAs were separated from unincorporated nucleotides and primers using S.N.A.P. columns (Invitrogen) or QIAquick spin filters (Qiagen), dependent on size of transcript. A homopolymeric dC- or dA-tail was added (via terminal transferase) to the 3′ termini of the cDNAs. Tailed cDNAs were subsequently amplified with Taq polymerase (Invitrogen) using a nested gene specific primer, GSP2, and the oligo anchor primer (Supplementary Table 2). Resulting PCR products were diluted 1:100 and subjected to a second PCR using the second nested gene specific primer PM and an abridged oligo anchor primer (Supplementary Table 2). The products of these reactions were sequenced.
Formation of RNAP holoenzyme
Micromolar concentrations of RNAP holoenzymes were prepared in a 4:1 molar ratio of σ (σ70, σ38, σ32, or σ24) to RNAP core in a transcription buffer (TB: 50 mM Tris–HCl, 3 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 100 mM KCl, 100 μg/ml BSA); incubated on ice for several hours, then at 30 °C for 20 min (except in the case of σ32 where all steps were done at 42 °C).
Electrophoretic mobility shift assay
Promoter regions of interest in the S. oneidensis genome were PCR-amplified with Pfu polymerase and primers listed in Supplementary Table 3. Open complexes were formed by incubating 50 nM holoenzyme (50 nM RNAP core: 200 nM σ) with 20 nM of promoter DNA fragments in 20 μl of buffer TB at 30 °C for 10 min. Open complexes were then challenged with a final concentration of 40 μg/mL heparin for 1 min at 30 °C, and loaded onto a 5% non-denaturing polyacrylamide gel (37.5:1 acrylamide/bisacrylamide, 16.5 × 14.5 cm), and ran for 90 min at 150 V. The resulting gel was stained with SYBR® Green (Invitrogen) according to the manufacturer protocol, and imaged on a Molecular Imager ®FX (Bio-Rad).
Single-round radioactive transcription assay
Promoter sequences were PCR-amplified with primers listed in Supplementary Table 3 and inserted between the BamHI and HindIII restriction sites of the pTH8 vector (containing a T7 early terminator, ~300 bp downstream of the HindIII site) [34]. One hundred nanomolar of holoenzymes was incubated with buffer TB containing 13 nM of DNA template in a final volume of 10 μl. After 15 min incubation at 30 °C (or 42 °C for σ32), a 1 μl of a NTP mixture (1.5 mM ATP, CTP, GTP, 75 μM UTP, 3 μCi [α-32P]UTP, and 1 μg heparin) was added into the pre-formed complex. Eight microliters of stop solution (98% deionized formamide, 10 mM EDTA, 0.025% xylene cyanol FF, 0.025% bromophenol blue) was added after 7 min, then samples were loaded onto a pre-run 6% denaturing PAGE gel (19:1 acrylamide/bisacrylamide, 6 M Urea, 0.5 × TBE, 22 × 20 cm), and ran for 90 min at 21 W. RNA products were visualized using PhosphorImager (Molecular Dynamics, Inc.) and ImageQuant Software (GE Healthcare).
Site-specific labeling of σ70 and DNA with fluorophores
Site-specific labeling of σ70,N399C was performed by the incorporation of Cy3B maleimide using a solid state-based labeling (SSL) [35]. σ70,N399C incorporated the Cy3B maleimide with 65–70% efficiency, while non-specific labeling of the σ70ΔCys derivative was 1% using the same dye. DNA fragments of the lacCONS promoter derivatives [18] (Fig. 4A), end labeled with Alexa647, (Invitrogen) were prepared as described [30].
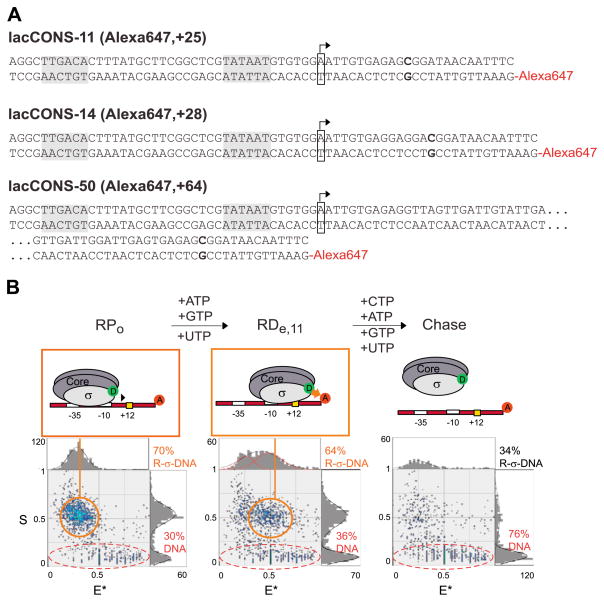
Fig. 4.
smFRET measurement to monitor subunit composition of S. oneidensis RNAP during early and mature elongation. (A) Derivatives of lacCONS promoter used in this study. The promoter −35 and −10 elements are boxed in each construct, as is the start site, which is boxed and indicated with an arrow. The first guanine residue on the template strand at position +12, +15, or +51 is bolded, indicating the halt site in the elongation complex, and the downstream position of the Alexa647 fluorescent label is indicated. (B) Top, overview of ALEX-FRET. ALEX-FRET assesses the σ70 content and translocational position of the transcription complex as it transitions from open complex (RPo, left) to the elongation complex (RDe,11, middle) after the addition of the nucleotide subset, and finally, to the dissociated chase after the addition of all four nucleotides (right). Bottom, ALEX histograms of FRET experiments with RPo, RDe,11, and chased RDe,11.
Transcription complex formation for single-molecule biophysical studies
Reaction mixtures of 80 nM RNAP core and 100 nM wild-type or Cy3B-labeled σ70 derivative in TB were used for preparation of open complexes, similar to previously described [21]. Samples were incubated 20 min at 30 °C; 20 nM dual-labeled or Alexa647-labeled promoter dsDNA was then added and samples were further incubated 10 min at 30 °C. Heparin-Sepharose (GE Healthsciences, 0.8 μl of 1 mg/ml) was added to disrupt non-specific RNAP-promoter complexes and to remove free RNAP [30]. After addition, reactions were gently mixed for 15 s, and then centrifuged on a bench top centrifuge at 2000g for 10 s. Twenty microliters of aliquots were transferred to tubes containing 1 μL of 10 mM ApA (Ribomed Biotechnologies), pre-warmed at 30 °C, and incubated for 5 min at 30 °C.
Abortive products were prepared by adding 0.8 μl of 1.25 mM GTP and UTP to the prepared RPo complex, while halted elongation complexes were prepared by adding 2 μl of 1.25 mM ATP, GTP, and UTP in TB and incubating for an additional 5 min. Ability of halted complexes to resume transcription elongation was tested with the addition of 2 μl of 1.25 mM ATP, GTP, and UTP with 6.25 mM CTP (chase reaction), and samples were incubated a further 5 min at 30 °C.
Single-molecule fluorescence spectroscopy
Sample preparation, alternating-laser excitation-microscopy, data acquisition, and analysis were as described in [18,21,36,37]. Alternating-laser excitation was achieved by using 532 and 638 nm laser light, with an alternation period of 100 μs, and duty cycle of 50%. Excitation intensities were 150–200 μW for 532 nm excitation (donor excitation) and 120 μW for 638 nm excitation (acceptor excitation). Data collection occurred over 15 or 30 min periods at 25 °C.
Determination of σ70 retention
Fractional DNA occupancy, θ, was calculated for RPo, RDe, and chased RDe as described [36]. θ = [donor–acceptor species]/([acceptor-only species] + [donor–acceptor species]). RPo was measured first for 15 min, then RDe was measured for 30 min, and finally, the chased RDe was measured for 15 min. The concentration of donor–acceptor and acceptor-only species were determined by the molecule count using S thresholds [36].
Results
Co-overexpression and purification of in vivo assembled RNAP
The co-overexpression construct, pYK201, was designed for facile genetic manipulations. As shown in Fig. 1A, the expression is driven by a single T7 promoter and produces a long polycistronic mRNA encoding all of the core subunits (α, β, β′, and ω). Each RNAP core subunit sequence was inserted as a cassette into the co-expression plasmid via unique restriction sites (Fig. 1A). Therefore, each subunit sequence can be mutagenized before substituting into the co-overexpression plasmid, providing numerous combinations for mutational studies on RNAP. Above all, this co-overexpression construct was designed to ensure high yield of the RNAP core. First, the translational enhancer element upstream of the ribosome-binding site (rbs) is maintained such that all subunits are translated at high levels [38,39]. Second, we introduced an ATG start codon in front of the wild-type rpoC gene because the wild-type start codon (GTG) is not preferable for high level expression in E. coli [40]. Third, a T7 expression vector and the BL21 Star (DE3) E. coli strain were selected as the expression plasmid and host, respectively, since the T7 RNA polymerase has excellent processivity, generating long (>10 kb) transcripts, and the BL21 Star (DE3) strain has been reported to stabilize long mRNAs by suppressing endogenous RNase activity [41].
A hexahistidine tag fused to the C-terminus of the α subunit allowed complexes containing α subunit(s) to be purified by immobilized metal ion affinity chromatography (IMAC), then further purified by anion exchange chromatography. The subsequent fractions, resolved in SDS–PAGE, showed that anion exchange chromatography can provide highly pure RNAP core complexes (Fig. 1B). The typical yield of the fully assembled RNAP core was about 20–30 mg/L of E. coli culture, 10 times higher than immunoaffinity purification from S. oneidensis [11].
Beyond ensuring high yield, the co-overexpression allows us to produce homogeneous S. oneidensis RNAP core assembled by species- specific subunit interactions. Since S. oneidensis and E. coli RNAP core subunits share ~80% identity in amino acid sequences (Table 2), there may be inter-species subunit–subunit interactions in the E. coli host cell. We tested subunit cross-reactivity between the two species by overexpressing C-terminal hexahistidine tagged S. oneidensis α (pSH101 in Table 1) in E. coli. SDS–PAGE after IMAC showed no detectable E. coli subunits associated with the overexpressed S. oneidensis α (Fig. 1C). In addition, mass spectroscopy data through nanoLC/MS/MS of β and β′ subunits from the purified S. oneidensis RNAP core confirmed that there is no detectable cross-reactivity (analyzed by Midwest Bio-Services, Supplementary Material). Thus, we conclude that the co-overexpression system produces large quantities of highly pure S. oneidensis RNAP.
Table 2.
Amino acid sequence comparison of S. oneidensis and E. coli RNAP subunits.
| RNAP subunit | Amino acid percentage identity (%) |
|---|---|
| α | 88 |
| β | 80 |
| β′ | 80 |
| ω | 72 |
| σ70 | 79 |
| σ38 | 80 |
| σ32 | 69 |
| σ24 | 73 |
Expression and stability of S. oneidensis σ factors
To generate S. oneidensis RNAP holoenzymes, we created expression constructs of the S. oneidensis σ factors, σ70 (rpoD), σ38 (rpoS), σ32 (rpoH), and σ24 (rpoE). Among these factors, σ70 is the primary initiation factor responsible for the expression of housekeeping genes [42]. In this study, we solubly expressed σ70 under a trc-promoter expression system [43] since it was reported that the robust T7 promoter expression system drives the overexpressed E. coli σ70 into formation of insoluble inclusion bodies [16]. Insoluble σ70 requires refolding steps [44], reducing the yield of active protein [45]. Soluble S. oneidensis σ70 was produced using the milder trc-promoter expression system, induced at room temperature. The resulting purified S. oneidensis σ70 after anion exchange chromatography is shown in Fig. 1D.
Soluble production of the other σ factors, including the mutant σ70 described later, using the trc promoter was not possible because the expression level and purity were much lower than that of wild-type σ70 under the same conditions. In order to produce large quantity of highly pure mutant σ70, σ38, σ32, and σ24, we reverted to the T7 expression system. Previous reports have established that soluble overexpression of E. coli σ24 by the T7 promoter can be achieved by high concentration isopropyl-β-D-thiogalactopyranoside (IPTG) induction of expression at low cell density in E. coli culture [31]. Using this method, we were able to overexpress soluble, highly pure S. oneidensis σ24 (Fig. 1D). No detectable amounts of soluble protein could be produced for the other σ’s, mutant σ70, σ38, and σ32, using these conditions. The overexpression of these proteins under the T7 promoter resulted in their localization to inclusion bodies; therefore, these insoluble σ factors were purified and refolded from insoluble inclusion bodies as described in [30].
Identification of σ specific promoters and characterization of transcriptional activity
Putative promoter sequences for σ70, σ32, and σ24 were found in the S. oneidensis genome by examining available S. oneidensis microarray data [8,9,46–48], and by comparing known promoters from E. coli [49,50] and Salmonella enterica [51]. Upstream sequences of predicted genes were analyzed with AlignACE [52] to find potential regulatory motifs with E. coli sequences as a reference. Conserved motifs for both σ32 and σ24 were found to be nearly identical to the E. coli binding sites (Supplementary Material), and comparison of predicted sites for S. oneidensis σ32 were found to be in good agreement with previously predicted sites [9]. Upstream regions of dnaK for σ32 and rpoE for σ24, which have the strongest correlation to known E. coli promoters, were selected for further characterization. The dmsAB-1 promoter was selected for the study of σ70 due to its significance in the reductive function of S. oneidensis [53]. Since σ38 can recognize some σ70 promoters [54], we utilized the dmsAB-1 promoter for the study of both σ70 and σ38.
The transcription start site of dmsAB-1, dnaK, and rpoE were determined using 5′-rapid amplification of cDNA ends (RACE) [33]. A combination of homopolymeric dC- or dA-tailing was utilized to accurately locate the transcription start site. The mapped transcriptional start sites for dmsAB-1, dnaK, and rpoE are shown in Fig. 2A; the upstream −35 and −10 promoter elements were found to be in good agreement with our predicted motifs.
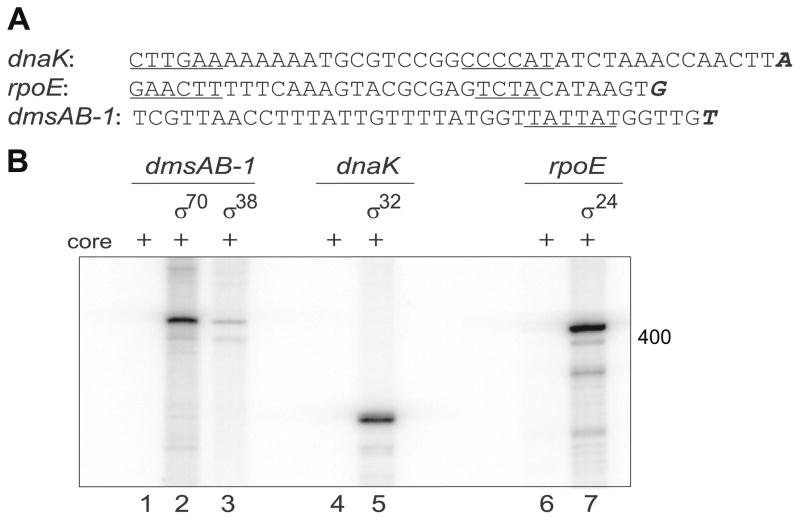
Fig. 2.
In vitro transcription assay. (A) Transcriptional start site mapped by 5′-RACE; start sites are in bold and italic font, predicted −35/−10 elements are underlined. (B) Single-round in vitro transcription reactions with different holoenzymes were performed with specific promoter sequences (inserted into the pTH8 vector). Transcription products using dmsAB-1 DNA are shown in lanes 1–3 (325 bps); dnaK DNA in lanes 4–5 (316 bps); and rpoE DNA in lanes 6–7 (354 bps); transcription products using core only are shown in lanes 1, 4, and 6. Gels were visualized by PhosphorImager.
The promoters of dmsAB-1, dnaK, and rpoE were then amplified by PCR, and their interaction with holoenzyme containing σ70/σ38, σ32, and σ24, respectively, was tested by electrophoretic mobility shift assay (EMSA, Supplementary Material). For each promoter, a specific interaction of the holoenzyme and its predicted promoter was observed and this interaction survived a challenge with heparin [55], which reduces non-specific interactions (Supplementary Material). Additionally, our EMSA result validated that σ38 can recognize the σ70 promoter, dmsAB-1.
To further evaluate the functionality of the recombinant S. oneidensis RNAP holoenzymes, we examined their ability to transcribe specific mRNA products by performing single-round in vitro transcription on plasmid templates containing the promoters described above. The RNAP of each σ factor was shown to yield specific transcription products from these templates (Fig. 2B). Control experiments of RNAP core only transcriptions were performed on each promoter with no detectable specific transcription product, and we also observed a decrease in σ70 transcription activity when rifampicin [56], a known transcription inhibitor, was added (Supplementary Material).
Single-molecule characterization of transcription initiation by σ70-RNAP
Transcription initiation requires the presence of the RNAP core enzyme and its initiation factor, σ70. In the E. coli system, this complex binds to promoter DNA and unwinds ~14 base pairs (bp) surrounding the transcription start site to yield a catalytically competent RNAP-promoter open complex (RPo) [57–59]. In subsequent steps of transcription initiation, RNAP enters into initial synthesis of RNA as an RNAP-promoter initial transcribing complex (RPitc), typically engaging in abortive cycles of synthesis and release of short RNA products, and, upon the synthesis of an RNA product of 9–11 nucleotides (nt), it breaks its interactions with promoter DNA, leaves the promoter, and enters into processive synthesis of RNA as an RNAP-DNA elongation complex (RDe) [57–60].
To determine if S. oneidensis σ70-RNAP complex, like its E. coli counterpart, “scrunches” DNA during transcription initiation, we monitored the fluorescence resonance energy transfer (FRET) between a fluorescent donor incorporated at a site in the −10/−35 spacer DNA (position −15) and a fluorescent acceptor incorporated at a site in downstream DNA (position +15), similar to previous E. coli studies [18,19] (Fig. 3A). As in those studies, we utilized confocal optical microscopy with alternating-laser excitation (ALEX) [18,21,36,37] to detect and quantify the fluorescence from single molecules as they diffuse through the detection volume. For each single molecule, we extract two parameters: a donor–acceptor stoichiometry parameter, S, which reports on the association state of the molecule, and an observed efficiency of donor–acceptor energy transfer, E*, which reports on the proximity of the fluorescent probes [36,37]. With these two parameters, we are able to detect the colocalization of the two fluorophores on the DNA and to monitor the distance changes within single molecules of RPo and RPitc.
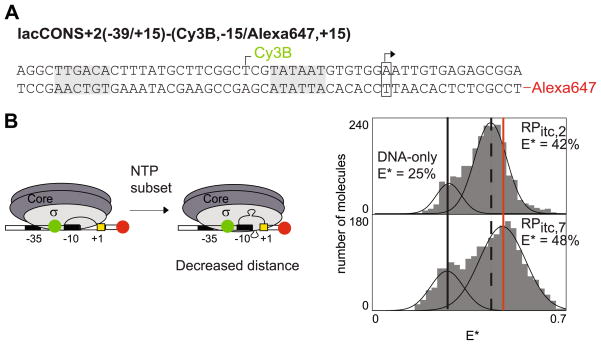
Fig. 3.
Initial transcription by S. oneidensis RNAP involves scrunching of promoter DNA. (A) Derivative of lacCONS promoter utilized to monitor the contraction of DNA between positions −15 (Cy3B, as donor) and +15 (Alexa647, as acceptor). The promoter −35 and −10 elements are grey-colored, as is the start site, which is boxed and indicated with an arrow. (B) Left, schematic of DNA scrunching by RNAP. Right, two representative E* histograms compromising of the free DNA (lowest E* value, solid black line), RPo (dashed black line) or RPitc,≤ 7 (higher E* values, corresponding to DNA bending by the RNAP, solid red line).
We first attempted to monitor the transcription initiation of S. oneidensis RNAP on its native promoter, dmsAB-1, identified in experiments described above; however, we found that the binding affinity of this promoter was lower than that required by singlemolecule measurements [61]. We have not been able to overcome this limitation to date, and the requirement for strong S. oneidensis promoters combined with the lack of mapped S. oneidensis makes the selection of appropriate S. oneidensis promoters very difficult at this time. However, we observed significant cross-reactivity of the S. oneidensis σ70-RNAP with E. coli promoters during the ensemble characterization described above (Supplementary Material), and specifically, the S. oneidensis holoenzyme was shown to bind with high affinity to an E. coli promoter, lacCONS, which has been utilized for previous single-molecule studies of E. coli transcription [18,21,30].
Using this promoter, we performed measurements that monitored the formation of the open complex, RPo, and then upon the addition of a subset of nucleotides, UTP and GTP, to RPo, monitored the structural changes of DNA during iterative abortive synthesis of RNA products up to 7 nt in length (RPitc≤7, Fig. 3B, left). The results indicated that, upon transition from RPo to RPitc,≤ 7, mean E* significantly increases (Fig. 3B, right), which implies that the mean donor–acceptor distance, mean R, significantly decreases. This distance changes are consistent with previous E. coli experiments and verifies that S. oneidensis does indeed “scrunch” the DNA segment between −10/−35 spacer DNA and downstream DNA (Fig. 3B, left).
Single-molecule characterization of σ70 retention during the transition from early to mature elongation
Beyond examining the RNAP complex during transcription initiation, we can also examine the fate of the σ70-RNAP complex as it transitions into elongation. The retention of σ70 during elongation challenges proposals hypothesizing that there are distinct subunit composition states for initiation and elongation and indicates that σ factors may influence elongation or regulate other stages of the transcription cycle [24].
In order to monitor the subunit composition of the σ70-RNAP complex during the transition from initiation to elongation, we needed to fluorescently label σ70 and the promoter DNA. We exploited the high sequence identity between S. oneidensis and E. coli σ70 (~80% overall, and ~98% in the region of interest, amino acids 362–613) to select an amino acid residue for labeling that has the highest probability of being solvent-accessible and compatible with FRET, based on known E. coli labeling sites. Given the extensive amount of fluorescent work done with E. coli σ70, selection of an appropriate position was relatively straight-forward and the S. oneidensis σ70 asparagine residue at 399 (residue 396 in E. coli σ70) was chosen, and a single-cysteine derivative, N399C was created, expressed, and purified. The shift in the S. oneidensis σ70 labeling position relative to its E. coli counterpart is due largely to the insertion of three unique amino acids at positions 198–200 in the S. oneidensis σ70 sequence, which lie in the linker region between the evolutionary conserved regions, 1.2 and 2, of σ70 [62] (Supplementary Material). The DNA-binding and transcriptional activity of this single-cysteine derivative was examined, and the σ70,N399C had DNA-binding and transcriptional activity comparable to the wild-type σ70, consistent with E. coli mutant σ70,N396C [63] (determined by abortive initiation studies, data not shown), making it appropriate for single-molecule studies.
For these experiments, we incorporated the FRET donor, Cy3B, at σ70 position 399, which has a high probability of being at the leading edge of the RNAP, and we incorporated a FRET acceptor on the downstream DNA (Fig. 4B). In a single experiment using ALEX-FRET, we are able to detect the interaction of fluorescently labeled σ70-RNAP complex with fluorescently labeled DNA by extracting the donor–acceptor S (the stoichiometry term defining the presence of one or both fluorophores as previous described [64,65]), which allows us to simultaneously define the σ70 content of the elongation complex, and we are also able to define the translocational position of a fluorescently labeled transcription complex, by monitoring efficiency of donor–acceptor energy transfer, E*, which reports on the proximity of the transcription complex and the downstream DNA.
Stable elongation complexes were formed using three derivatives of the lacCONS promoter as in the previous experiment. Each derivative allows for a distinct static elongation complex to be probed, two in the early transition from initiation to early elongation (+11 and +14) and one in mature elongation at +50. The static elongation points are reached by halting elongation with nucleotide starvation. CTP is omitted from the nucleotide mixture, and the elongation halts at the first template-strand guanine at position +12, +15, or +51 (Fig. 4A) [18,21,30,66].
Fig. 4 and Table 3 present the representative ALEX results for the open complex (RPo) and the static elongation complex (RDe,11) of the lacCONS promoter. For RPo, two observable species corresponding to the donor–acceptor complex (σ70-RNAP-DNA, S ≈ 0.55, and E* ≈ 0.3) and acceptor-only species (DNA-only, S < 0.3) are indicated. The addition of the NTP subset (ATP, GTP, UTP) permits elongation and the formation of the stable elongation complex at +11, RDe,11. Approximately 80% of the complexes are converted to a species that exhibits the same S value but a higher FRET efficiency (S ≈ 0.52 and E* ≈ 0.49), which can be chased with the addition of all four NTPs. These results are consistent with previous studies in E. coli and indicate that a majority of the complexes are functional and competent to undergo the transition from initiation to elongation under these conditions.
Table 3.
Summary of the σ70 retentions and half-lives for the three elongation complexes.
| Halted elongation point | Initial σ70 retention (%) | σ70 Retention half-life (min) |
|---|---|---|
| RDe,11 | 100 ± 7 | 80 ± 16 |
| RDe,14 | 59 ± 4 | 75 ± 23 |
| RDe,50 | 45 ± 9 | 43 ± 13 |
The kinetics and stability of the RDe,11 complex were also examined and the results are shown in Fig. 5A and Table 3. The extent of σ70 retention as a function of time is presented, and the initial retention and half-life of the complex are determined. σ70 retention was monitored by examining the fractional DNA occupancy ratios as a function of time for the open complex, after NTP addition, and during chase. The retention ratios for the open complex and chase experiments are virtually constant as a function of time (data not shown) and can be scaled to 100% for the open complex and 0% or baseline for the chase complex. Using these corrections, the results indicate the initial extent of retention in RDe,11 is ~100% with a half-life of σ70 retention of 80 min.
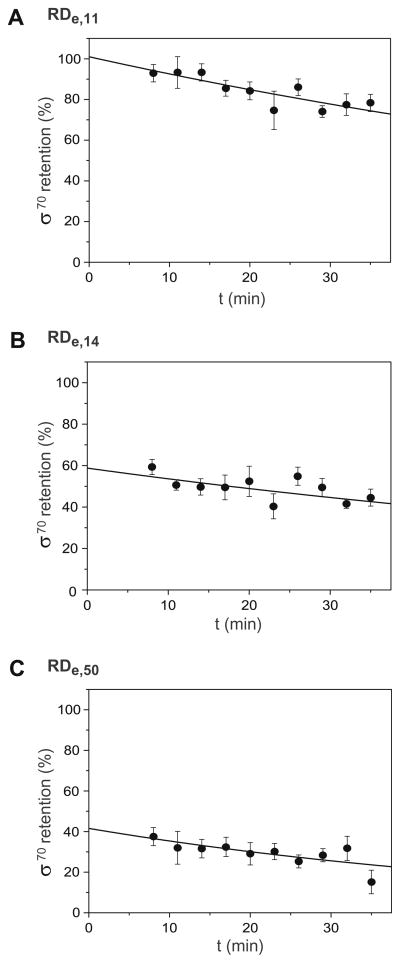
Fig. 5.
Kinetics of σ release during early and mature elongation. (A) Fractional occupancy θ (t) for RPo on lacCONS-11 template DNA. Five independent measurements are fitted with a single-exponential (solid line) and errors bar reflect the standard error of mean (SEM). (B) σ70 retention in early elongation complex, RDe,11. The same as (A), with the y intercept corresponding to 5 min after the addition of the NTP subset (ATP, GTP, UTP) to RPo. (B) Early elongation complex with filled RNA exit channel, RDe,14. (C) Mature elongation complex, RDe,50.
Two other elongation points were examined in this manner, RDe,14 and RDe,50. At RDe,14, the RNA exit channel becomes completely filled [67–71], and the interactions between the exiting nascent RNA chain and σ70 as elongation continues may disrupt contacts between the σ70 and the RNAP, contributing to the σ release [72,73]. The initial retention of σ70 in RDe,14 is ~59% with a half-life of ~75 min (Fig. 5B and Table 3). These results indicate that in a portion of the transcription complexes σ70 remains associated with RNAP upon the formation of RDe,14; however, a significant portion of the σ70 was released at a time-scale faster than our measurements resolution (less than 10 min). Finally, RDe,50 represents a mature elongation complex, and the initial retention of σ70 in RDe,50 is ~45% with a half-life of ~43 min (Fig. 5C and Table 3). These results indicate that in approximately half of the transcription complexes, σ70 remains associated with and translocated with RNAP upon the formation of RDe,50, consistent with RDe,14 experiments and previous studies in E. coli [21].
Discussion
We have described the establishment of a co-overexpression system of the S. oneidensis core RNAP in E. coli, its activity with four σ factors, and the single-molecule characterization of its transcription initiation and subunit composition during elongation. The design of the multi-subunit expression system yields milligrams of active RNAP core enzyme, a 10-fold increase from previously reported S. oneidensis RNAP core purification [11]. Mass spectrometry and S. oneidensis α-only overexpression experiment shows no detectable cross-reactivity between S. oneidensis and E. coli RNAP core subunits, under co-overexpression condition even though there is a strong sequence similarity between S. oneidensis and E. coli (72–88% for RNAP core subunits, as shown in Table 2). Additionally, each subunit was inserted into the co-overexpression construct via unique restriction sites, this cassette design simplifies genetic manipulations of subunits, e.g., deletions, insertions, point-mutations, etc., thus facilitating the creation of RNAP mutants that may otherwise be difficult to generate. Our approach provides a reliable source of RNAP for biological studies of S. oneidensis transcription machinery.
Existing reports on global transcriptome response to various stimuli have provided important insights into the complex nature of regulatory mechanisms in S. oneidensis. Most relevant to the utilization of S. oneidensis as a bioremediation tool is the response of its transcriptome to different terminal electron acceptors [8]. In the presence of metals and thiosulfate, an upregulation of σ factors σ32 and σ24 was observed, while σ54 and σ38 were downregulated [8]. Elucidation of the role of these σ factors in S. oneidensis could provide new insights into regulation schemes involved with respiration. To date, in vitro characterization of S. oneidensis σ factors has not been reported. In this study, we have produced recombinant S. oneidensis σ factors (σ70, σ38, σ32, and σ24) with high purity and yield, which allows us to characterize the interaction of these holoenzymes with their specific S. oneidensis promoters.
The selection of individual promoters (dmsAB-1, dnaK, and rpoE) for each σ factor (σ70, σ32, and σ24, respectively) was based on S. oneidensis microarray data. We verified these promoters through bioinformatics and biochemical assays. Previous work by Saffarini et al. [53] indicated the presence of a binding site for the transcription regulator, CRP, upstream of the DMSO reductase operon. Given the interest in alternative electron acceptor utilization by S. oneidensis, we chose this promoter for ourσ70 studies, although strongerσ70 promoters with higher similarity to E. coli promoters may exist. Additionally, in our assays, we have utilized the same promoter, dmsAB-1, for both σ70 and σ38. Overlap in σ70 and σ38 promoters has been noted previously in E. coli through the recognition of a canonical −10 hexamer sequence [54]. While identification of a set of unique σ38 promoters is desirable, overlap at this promoter provided the most expedient means for examining the specificity and activity of the RNAP holoenzyme containing σ38. While strength and specificity of a promoter consensus sequence can vary throughout the genome, strong promoters such as dnaK and rpoE, demonstrate the similarity between the S. oneidensis and E. coli σ factors. Elucidation of the individual regulons associated with the S. oneidensis σ factors, particularly σ32 and σ24, could illuminate the unique functions of these σ factors in respiration and anaerobic growth of this organism, and provide important evolutionary clues to the development of robust metabolisms in S. oneidensis.
Finally, we have reported the application of existing single-molecule fluorescence assays to the characterization of the newly reconstituted transcription machinery from S. oneidensis. Using smFRET, we were able determine that “scrunching” was the underlying mechanism by which S. oneidensis RNAP translocates relative to DNA during abortive initiation. This mechanism has been observed in both E. coli and T7 RNAP [18–20], and the conservation of this mechanism by S. oneidensis indicates that is may be a general mechanism for transcription initiation. In another smFRET assays, we were also able to determine the fate of the S. oneidensis σ70 during early and mature elongation, and verify that σ70 release is non-obligatory in S. oneidensis, again providing evidence for the generality of this mechanism to other bacterial species. These assays have not only provided insight into the underlying transcription mechanism of S. oneidensis, but also indicate that singlemolecule characterization of other bacterial organism and more importantly other transcription components, like the alternative σ factors produced here, could also be achieved using a similar expression and purification strategy.
We have demonstrated that the reconstitution of functional holoenzymes from S. oneidensis provides an excellent path for biochemical and biophysical characterizations, and that we can extend transcriptional studies beyond well-characterized systems into new transcription components and/or new bacterial organisms, and that with continued efforts, comparative studies between E. coli and S. oneidensis could reveal important information about how environmental stress and evolutionary processes effect transcription in these organisms and help to elucidate the role RNAP plays in gene regulation under stress conditions.
Supplementary Material
Acknowledgments
We thank Dr. M. Uljana Mayer Dr. Liang Shi for providing the S. oneidensis RNAP subunits clones, and Dr. Mayer, Devdoot Majumdar, and Yuval Ebenstein for critical reading of the article; the Dr. Jay D. Gralla group for help with the radioactive transcription assays; Irina Sorokina for helpful discussion of the MALDI-MS data. We also acknowledge the Shewanella Federation for helpful discussions. This work was supported by Department of Energy Grant FG03-02ER63339 and NIH Grant GM069709-01 to S.W.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.pep.2008.11.013.
Footnotes
Abbreviations used: RNAP, RNA polymerase; EMSA, electrophoretic mobility shift assay; DMSO, dimethyl sulfoxide; TMAO, trimethylamine N-oxide; rbs, ribosome binding site; IMAC, immobilized metal ion affinity chromatography; SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; MALDI-MS, matrix-assisted laser desorption/ionization mass spectrometry; IPTG, isopropyl β-D-1-thiogalactopyranoside; RACE, 5′-rapid amplification of cDNA ends; ORF, open reading frame; 2-ME, 2-mercaptoethanol; CV, column volume; EDTA, ethylenediaminetetra-acetic acid; DTT, dithiothreitol; BSA, bovine serum albumin, smFRET, single-molecule fluorescence resonance energy transfer; LE, leading edge; ALEX, alternating-laser excitation; nt, nucleotide; SEM, standard error of mean.
References
- 1.Myers CR, Nealson KH. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science. 1988;240:1319–1321. doi: 10.1126/science.240.4857.1319. [DOI] [PubMed] [Google Scholar]
- 2.Tiedje JM. Shewanella-the environmentally versatile genome. Nature Biotechnology. 2002;20:1093–1094. doi: 10.1038/nbt1102-1093. [DOI] [PubMed] [Google Scholar]
- 3.Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, MacGregor BJ, Ringelberg DB, White DC, Nishijima M, Sano H, Burghardt J, Stackebrandt E, Nealson KH. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. International Journal of Systematic Bacteriology. 1999;49:705–724. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- 4.Marshall MJ, Beliaev AS, Dohnalkova AC, Kennedy DW, Shi L, Wang Z, Boyanov MI, Lai B, Kemner KM, McLean JS, Reed SB, Culley DE, Bailey VL, Simonson CJ, Saffarini DA, Romine MF, Zachara JM, Fredrickson JK. C-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biology. 2006;4:e268. doi: 10.1371/journal.pbio.0040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nature Biotechnology. 2002;20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 6.Daraselia N, Dernovoy D, Tian Y, Borodovsky M, Tatusov R, Tatusova T. Reannotation of Shewanella oneidensis genome. OMICS. 2003;7:171–175. doi: 10.1089/153623103322246566. [DOI] [PubMed] [Google Scholar]
- 7.Leaphart AB, Thompson DK, Huang K, Alm E, Wan XF, Arkin A, Brown SD, Yan L, Liu X, Wickman GS, Zhou J. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. Journal of Bacteriology. 2006;188:1633–1642. doi: 10.1128/JB.188.4.1633-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beliaev AS, Klingeman DM, Klappenbach JA, Wu L, Romine MF, Tiedje JM, Nealson KH, Fredrickson JK, Zhou J. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. Journal of Bacteriology. 2005;187:7138–7145. doi: 10.1128/JB.187.20.7138-7145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao H, Wang Y, Liu X, Yan T, Wu L, Alm E, Arkin A, Thompson DK, Zhou J. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. Journal of Bacteriology. 2004;186:7796–7803. doi: 10.1128/JB.186.22.7796-7803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symposia Quantitative Biology. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 11.Probasco MD, Thompson NE, Burgess RR. Immunoaffinity purification and characterization of RNA polymerase from Shewanella oneidensis. Protein Expression and Purification. 2007;55:23–30. doi: 10.1016/j.pep.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Mayer MU, Shi L, Squier TC. One-step, non-denaturing isolation of an RNA polymerase enzyme complex using an improved multi-use affinity probe resin. Molecular BioSystems. 2005;1:53–56. doi: 10.1039/b500950b. [DOI] [PubMed] [Google Scholar]
- 13.Artsimovitch I, Svetlor V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. Journal of Biological Chemistry. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 14.Kuznedelov K, Minakhin L, Severinov K. Preparation and characterization of recombinant Thermus aquaticus RNA polymerase. Methods Enzymology. 2003;370:94–108. doi: 10.1016/S0076-6879(03)70009-3. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, Lewis PJ. Overproduction and purification of recombinant Bacillus subtilis RNA polymerase. Protein Expression Purification. 2008;59:86–93. doi: 10.1016/j.pep.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Fujita N, Ishihama A. Reconstitution of RNA polymerase. Methods in Enzymology. 1996;273:121–130. doi: 10.1016/s0076-6879(96)73011-2. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Severinov K, Goldfarb A, Ebright RH. Rapid RNA polymerase genetics: one-day, no-column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proceedings of the National Academy of Sciences of the United States America. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang GQ, Roy R, Ha T, Patel SS. Transcription initiation in a single-subunit RNA polymerase proceeds through DNA scrunching and rotation of the N-terminal subdomains. Molecular Cell. 2008;30:567–577. doi: 10.1016/j.molcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapanidis AN, Margeat E, Laurence TA, Doose S, Ho SO, Mukhopadhyay J, Kortkhonjia E, Mekler V, Ebright RH, Weiss S. Retention of transcription initiation factor [sigma]70 in transcription elongation: single-molecule analysis. Molecular Cell. 2005;20:347–356. doi: 10.1016/j.molcel.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay J, Kapanidis AN, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH. Translocation of sigma(70) with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell. 2001;106:453–463. doi: 10.1016/s0092-8674(01)00464-0. [DOI] [PubMed] [Google Scholar]
- 23.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Molecular Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Shimamoto N, Kamigochi T, Utiyama H. Release of the sigma subunit of Escherichia coli DNA-dependent RNA polymerase depends mainly on time elapsed after the start of initiation, not on length of product RNA. Journal of Biological Chemistry. 1986;261:11859–11865. [PubMed] [Google Scholar]
- 26.Dummler A, Lawrence AM, de Marco A. Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microbial Cell Factories. 2005;4:34. doi: 10.1186/1475-2859-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callaci S, Heyduk E, Heyduk T. Conformational changes of Escherichia coli RNA polymerase sigma70 factor induced by binding to the core enzyme. Journal of Biological Chemistry. 1998;273:32995–33001. doi: 10.1074/jbc.273.49.32995. [DOI] [PubMed] [Google Scholar]
- 28.Owens JT, Miyake R, Murakami K, Chmura AJ, Fujita N, Ishihama A, Meares CF. Mapping the sigma 70 subunit contact sites on Escherichia coli RNA polymerase with a sigma 70-conjugated chemical protease. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6021–6026. doi: 10.1073/pnas.95.11.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hager DA, Jin DJ, Burgess RR. Use of mono Q high resolution ion exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- 30.Mukhopadhyay J, Mekler V, Kortkhonjia E, Kapanidis AN, Ebright YW, Ebright RH, Sankar LAaSG. Fluorescence resonance energy transfer (FRET) in analysis of transcription-complex structure and function. Methods in Enzymology. 2003;371:144–159. doi: 10.1016/S0076-6879(03)71010-6. [DOI] [PubMed] [Google Scholar]
- 31.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the σE (σ24) heat shock sigma factor of Escherichia coli. EMBO Journal. 1995;14:1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Englard S, Seifter S. Precipitation techniques. Methods in Enzymology. 1990;182:285–300. doi: 10.1016/0076-6879(90)82024-v. [DOI] [PubMed] [Google Scholar]
- 33.Scotto-Lavino E, Du G, Forhman MA. 5′ end cDNA amplification using classic RACE. Nature Protocols. 2006;1:2555–2562. doi: 10.1038/nprot.2006.480. [DOI] [PubMed] [Google Scholar]
- 34.Hunt TP, Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proceedings of the National Academy of Sciences of the United States America. 1985;82:8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim Y, Ho SO, Gassman NR, Korlann Y, Landorf EV, Collart FR, Weiss S. Efficient site-specific labeling of proteins via cysteines. Bioconjugate Chemistry. 2008;19:786–791. doi: 10.1021/bc7002499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J. Accurate FRET measurements within single diffusing biomolecules using alternatinglaser excitation. Biophysical Journal. 2005;88:2939. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olins PO, Rangwala SH. A novel sequence element derived from bacteriophage T7 mRNA acts as an enhancer of translation of the lacZ gene in Escherichia coli. Journal of Biological Chemistry. 1989;264:16973–16976. [PubMed] [Google Scholar]
- 39.Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expression and Purification. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 40.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 41.Lopez PJ, Marchand I, Joyce SA, Dreyfus M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Molecular Microbiology. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- 42.Helmann JD, Chamberlin MJ. Structure and function of bacterial sigma factors. Annual Review of Biochemistry. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 43.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 44.Burgess RR, Sankar A. Purification of overproduced Escherichia coli RNA polymerase [sigma] factors by solubilizing inclusion bodies and refolding from Sarkosyl. Methods in Enzymology. 1996;273:145–149. doi: 10.1016/s0076-6879(96)73014-8. [DOI] [PubMed] [Google Scholar]
- 45.Zhi H, Jin DJ. Purification of highly-active and soluble Escherichia coli [sigma]70 polypeptide overproduced at low temperature. Methods in Enzymology. 2003;370:174–180. doi: 10.1016/S0076-6879(03)70015-9. [DOI] [PubMed] [Google Scholar]
- 46.Beliaev AS, Thompson DK, Fields MW, Wu L, Lies DP, Nealson KH, Zhou J. Microarray transcription profiling of a Shewanella oneidensis etrA mutant. Journal of Bacteriology. 2002;184:4612–4616. doi: 10.1128/JB.184.16.4612-4616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beliaev AS, Thompson DK, Khare T, Lim H, Brandt CC, Li G, Murray AE, Heidelberg JF, Giometti CS, Yates J, III, Nealson KH, Tiedje JM, Zhou J. Gene and protein expression profiles of Shewanella oneidensis during anaerobic growth with different electron acceptors. OMICS. 2002;6:39–60. doi: 10.1089/15362310252780834. [DOI] [PubMed] [Google Scholar]
- 48.Thompson DK, Beliaev AS, Giometti CS, Tollaksen SL, Khare T, Lies DP, Nealson KH, Lim H, Yates J, III, Brandt CC, Tiedje JM, Zhou J. Transcriptional and proteomic analysis of a ferric uptake regulator (Fur) mutant of Shewanella oneidensis: possible involvement of Fur in energy metabolism, transcriptional regulation, and oxidative stress. Applied and Environmental Microbiology. 2002;68:881–892. doi: 10.1128/AEM.68.2.881-892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rezuchova B, Miticka H, Homerova D, Roberts M, Kormanec J. New members of the Escherichia coli sigma E regulon identified by a two-plasmid system. FEMS Microbiology Letters. 2003;225:1–7. doi: 10.1016/S0378-1097(03)00480-4. [DOI] [PubMed] [Google Scholar]
- 50.Ramirez-Santos J, Collado-Vides J, Garcia-Varela M, Gomez-Eichelmann MC. Conserved regulatory elements of the promoter sequence of the gene rpoH of enteric bacteria. Nucleic Acids Research. 2001;29:380–386. doi: 10.1093/nar/29.2.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skovierova H, Rezuchova B, Homerova D, Roberts M, Kormanec J. Characterization of the σE-dependent rpoEp3 promoter of Salmonella enterica serovar Typhimurium. FEMS Microbiology Letters. 2006;261:53–59. doi: 10.1111/j.1574-6968.2006.00325.x. [DOI] [PubMed] [Google Scholar]
- 52.Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nature Biotechnology. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- 53.Saffarini DA, Schultz R, Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. Journal of Bacteriology. 2003;185:3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka K, Takayanagi Y, Fujita N, Ishihama A, Takahashi H. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, {sigma}38, is a second principal {sigma} factor of RNA polymerase in stationary-phase Escherichia coli. Proceedings of the National Academy of Sciences of the United States America. 1993;90:3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfeffer SR, Stahl SJ, Chamberlin MJ. Binding of Escherichia coli RNA polymerase to T7 DNA. Displacement of holoenzyme from promoter complexes by heparin. Journal of Biological Chemistry. 1977;252:5403–5407. [PubMed] [Google Scholar]
- 56.Hartmann G, Honikel KO, Knusel F, Nuesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochimica et Biophysica Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 57.Murakami KS, Darst SA. Bacterial RNA polymerases: the whole story. Current Opinion in Structural Biology. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- 58.Record MT, Jr, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. [Google Scholar]
- 59.Young BA, Gruber TM, Gross CA. Views of transcription initiation. Cell. 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 60.Hsu LM. Promoter clearance and escape in prokaryotes. Biochimica et Biophysica Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 61.Laurence TA, Weiss S. Analytical chemistry. How to detect weak pairs. Science. 2003;299:667–668. doi: 10.1126/science.1081025. [DOI] [PubMed] [Google Scholar]
- 62.Malhotra A, Severinova E, Darst SA. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 63.Owens JT, Miyake R, Murakami K, Chmura AJ, Fujita N, Ishihama A, Meares CF. Mapping the sigma70 subunit contact sites on Escherichia coli RNA polymerase with a sigma70-conjugated chemical protease. Proceedings of the National Academy of Sciences of the United States America. 1998;95:6021–6026. doi: 10.1073/pnas.95.11.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proceedings of the National Academy of Sciences of the United States America. 2004;101:8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee NK, Kapanidis AN, Wang Y, Michalet X, Mukhopadhyay J, Ebright RH, Weiss S. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophysical Journal. 2005;88:2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carpousis AJ, Gralla JD. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. Journal of Molecular Biology. 1985;183:165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- 67.Borukhov S, Severinov K. Role of the RNA polymerase sigma subunit in transcription initiation. Research in Microbiology. 2002;153:557–562. doi: 10.1016/s0923-2508(02)01368-2. [DOI] [PubMed] [Google Scholar]
- 68.Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. Journal of Molecular Biology. 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 69.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Current Opinion in Structural Biology. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 71.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 72.Murakami KS, Masuda S, Darst SA. Crystallographic analysis of Thermus aquaticus RNA polymerase holoenzyme and a holoenzyme/promoter DNA complex. Methods Enzymology. 2003;370:42–53. doi: 10.1016/S0076-6879(03)70004-4. [DOI] [PubMed] [Google Scholar]
- 73.Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A. The sigma 70 subunit of RNA polymerase mediates a promoter–proximal pause at the lac promoter. Nature Structural & Molecular Biology. 2004;11:544–550. doi: 10.1038/nsmb757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.