Abstract
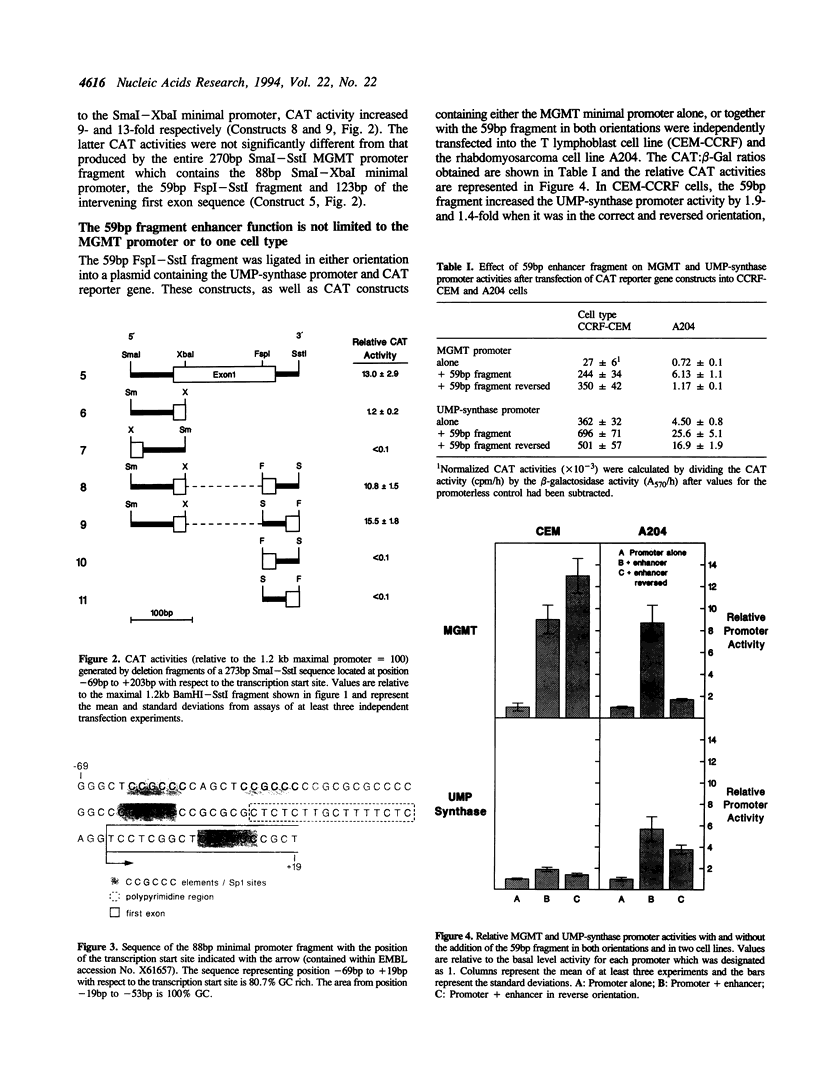
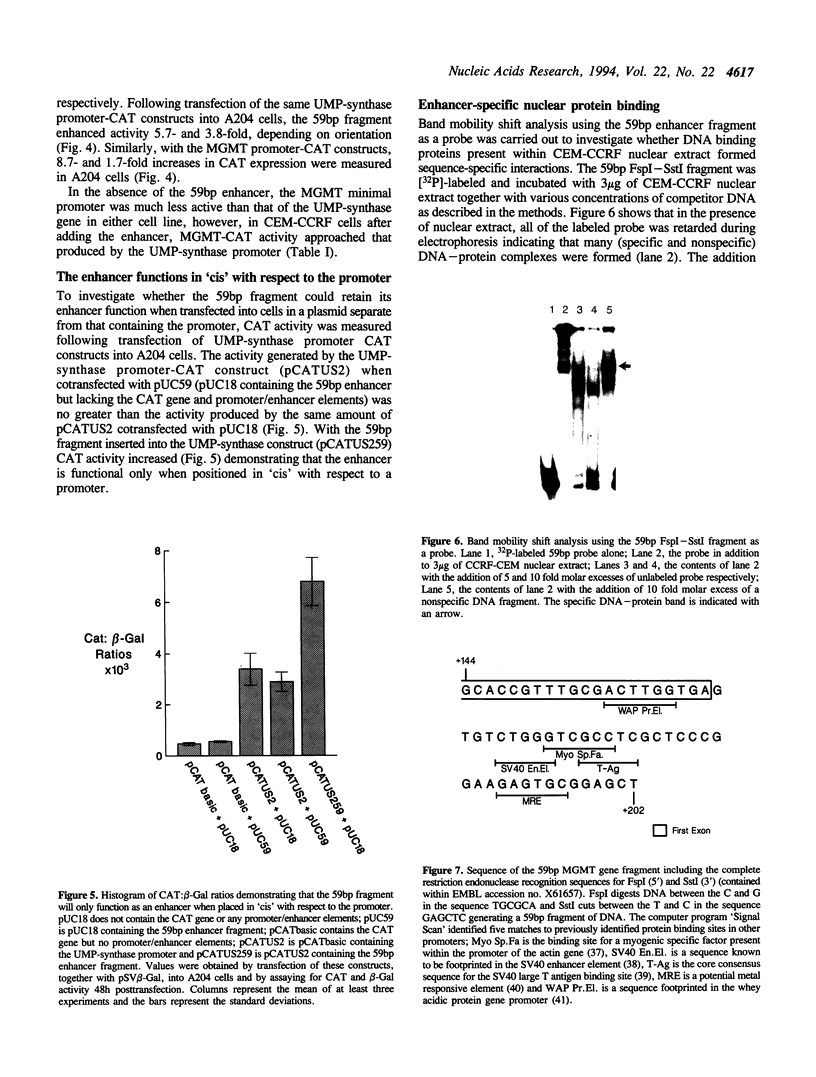
The DNA repair enzyme, O6-methylguanine DNA methyltransferase (MGMT) is responsible for repair of damage induced by alkylating agents that produce adducts at O6-guanine in DNA. Although the MGMT gene promoter has housekeeping gene promoter characteristics, unlike these genes which are expressed at a constant level, MGMT transcriptional activity varies between cell types. During an attempt to identify regions of the MGMT regulatory sequence sensitive to variations in transcription factors between cell types, we have identified a 59 bp enhancer which is required for efficient MGMT promoter function. This fragment produced increased transcriptional activity in reporter gene constructs containing either the MGMT or UMP-synthase promoter when transfected into either of two cell lines; it seems therefore that this enhancer may interact with relatively common trans-acting factors. Functional activity is only detected when the enhancer is in 'cis' with respect to the promoter, suggesting that complexes are formed between proteins bound to the enhancer and promoter sequences. We propose that the enhancer-binding protein may be a novel transcription factor since there are no obvious consensus sequences within the 59 bp sequence for known DNA-binding proteins.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen R. D., Taplitz S. J., Wong S., Bristol G., Larkin B., Herschman H. R. Metal-dependent binding of a factor in vivo to the metal-responsive elements of the metallothionein 1 gene promoter. Mol Cell Biol. 1987 Oct;7(10):3574–3581. doi: 10.1128/mcb.7.10.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison M. L. Enhancers: mechanisms of action and cell specificity. Annu Rev Cell Biol. 1988;4:127–153. doi: 10.1146/annurev.cb.04.110188.001015. [DOI] [PubMed] [Google Scholar]
- Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994 Apr 8;77(1):1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- Citron M., Decker R., Chen S., Schneider S., Graver M., Kleynerman L., Kahn L. B., White A., Schoenhaus M., Yarosh D. O6-methylguanine-DNA methyltransferase in human normal and tumor tissue from brain, lung, and ovary. Cancer Res. 1991 Aug 15;51(16):4131–4134. [PubMed] [Google Scholar]
- Davidson I., Fromental C., Augereau P., Wildeman A., Zenke M., Chambon P. Cell-type specific protein binding to the enhancer of simian virus 40 in nuclear extracts. Nature. 1986 Oct 9;323(6088):544–548. doi: 10.1038/323544a0. [DOI] [PubMed] [Google Scholar]
- DeLucia A. L., Lewton B. A., Tjian R., Tegtmeyer P. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J Virol. 1983 Apr;46(1):143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döffinger R., Pawlita M., Sczakiel G. Electrotransfection of human lymphoid and myeloid cell lines. Nucleic Acids Res. 1988 Dec 23;16(24):11840–11840. doi: 10.1093/nar/16.24.11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice D. C., Feldman P. A., Colberg-Poley A. M., Buckery R. M., Neubauer R. H. A sensitive method for the detection of beta-galactosidase in transfected mammalian cells. Biotechniques. 1991 Dec;11(6):739-40, 742-3. [PubMed] [Google Scholar]
- Foote R. S., Mitra S., Pal B. C. Demethylation of O6-methylguanine in a synthetic DNA polymer by an inducible activity in Escherichia coli. Biochem Biophys Res Commun. 1980 Nov 28;97(2):654–659. doi: 10.1016/0006-291x(80)90314-9. [DOI] [PubMed] [Google Scholar]
- Fornace A. J., Jr, Papathanasiou M. A., Hollander M. C., Yarosh D. B. Expression of the O6-methylguanine-DNA methyltransferase gene MGMT in MER+ and MER- human tumor cells. Cancer Res. 1990 Dec 15;50(24):7908–7911. [PubMed] [Google Scholar]
- Friedberg E. C. The molecular biology of nucleotide excision repair of DNA: recent progress. J Cell Sci Suppl. 1987;6:1–23. doi: 10.1242/jcs.1984.supplement_6.1. [DOI] [PubMed] [Google Scholar]
- Harris L. C., Potter P. M., Remack J. S., Brent T. P. A comparison of human O6-methylguanine-DNA methyltransferase promoter activity in Mer+ and Mer- cells. Cancer Res. 1992 Nov 15;52(22):6404–6406. [PubMed] [Google Scholar]
- Harris L. C., Potter P. M., Tano K., Shiota S., Mitra S., Brent T. P. Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene. Nucleic Acids Res. 1991 Nov 25;19(22):6163–6167. doi: 10.1093/nar/19.22.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. C., Remack J. S., Brent T. P. In vitro methylation of the human O6-methylguanine-DNA methyltransferase promoter reduces transcription. Biochim Biophys Acta. 1994 Mar 1;1217(2):141–146. doi: 10.1016/0167-4781(94)90027-2. [DOI] [PubMed] [Google Scholar]
- He X. M., Ostrowski L. E., von Wronski M. A., Friedman H. S., Wikstrand C. J., Bigner S. H., Rasheed A., Batra S. K., Mitra S., Brent T. P. Expression of O6-methylguanine-DNA methyltransferase in six human medulloblastoma cell lines. Cancer Res. 1992 Mar 1;52(5):1144–1148. [PubMed] [Google Scholar]
- Lewin B. Commitment and activation at pol II promoters: a tail of protein-protein interactions. Cell. 1990 Jun 29;61(7):1161–1164. doi: 10.1016/0092-8674(90)90675-5. [DOI] [PubMed] [Google Scholar]
- Lubon H., Hennighausen L. Nuclear proteins from lactating mammary glands bind to the promoter of a milk protein gene. Nucleic Acids Res. 1987 Mar 11;15(5):2103–2121. doi: 10.1093/nar/15.5.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermelstein F. H., Flores O., Reinberg D. Initiation of transcription by RNA polymerase II. Biochim Biophys Acta. 1989 Sep 21;1009(1):1–10. doi: 10.1016/0167-4781(89)90071-7. [DOI] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Myrnes B., Norstrand K., Giercksky K. E., Sjunneskog C., Krokan H. A simplified assay for O6-methylguanine-DNA methyltransferase activity and its application to human neoplastic and non-neoplastic tissues. Carcinogenesis. 1984 Aug;5(8):1061–1064. doi: 10.1093/carcin/5.8.1061. [DOI] [PubMed] [Google Scholar]
- Nakatsu Y., Hattori K., Hayakawa H., Shimizu K., Sekiguchi M. Organization and expression of the human gene for O6-methylguanine-DNA methyltransferase. Mutat Res. 1993 Jan;293(2):119–132. doi: 10.1016/0921-8777(93)90063-m. [DOI] [PubMed] [Google Scholar]
- Nicosia A., Monaci P., Tomei L., De Francesco R., Nuzzo M., Stunnenberg H., Cortese R. A myosin-like dimerization helix and an extra-large homeodomain are essential elements of the tripartite DNA binding structure of LFB1. Cell. 1990 Jun 29;61(7):1225–1236. doi: 10.1016/0092-8674(90)90687-a. [DOI] [PubMed] [Google Scholar]
- Olsson M., Lindahl T. Repair of alkylated DNA in Escherichia coli. Methyl group transfer from O6-methylguanine to a protein cysteine residue. J Biol Chem. 1980 Nov 25;255(22):10569–10571. [PubMed] [Google Scholar]
- Ostrowski L. E., von Wronski M. A., Bigner S. H., Rasheed A., Schold S. C., Jr, Brent T. P., Mitra S., Bigner D. D. Expression of O6-methylguanine-DNA methyltransferase in malignant human glioma cell lines. Carcinogenesis. 1991 Sep;12(9):1739–1744. doi: 10.1093/carcin/12.9.1739. [DOI] [PubMed] [Google Scholar]
- Pegg A. E., Byers T. L. Repair of DNA containing O6-alkylguanine. FASEB J. 1992 Mar;6(6):2302–2310. doi: 10.1096/fasebj.6.6.1544541. [DOI] [PubMed] [Google Scholar]
- Pieper R. O., Costello J. F., Kroes R. A., Futscher B. W., Marathi U., Erickson L. C. Direct correlation between methylation status and expression of the human O-6-methylguanine DNA methyltransferase gene. Cancer Commun. 1991 Aug;3(8):241–253. doi: 10.3727/095535491820873092. [DOI] [PubMed] [Google Scholar]
- Pieper R. O., Futscher B. W., Dong Q., Ellis T. M., Erickson L. C. Comparison of O-6-methylguanine DNA methyltransferase (MGMT) mRNA levels in Mer+ and Mer- human tumor cell lines containing the MGMT gene by the polymerase chain reaction technique. Cancer Commun. 1990;2(1):13–20. doi: 10.3727/095535490820874812. [DOI] [PubMed] [Google Scholar]
- Prestridge D. S. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci. 1991 Apr;7(2):203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Prost E., Moore D. D. CAT vectors for analysis of eukaryotic promoters and enhancers. Gene. 1986;45(1):107–111. doi: 10.1016/0378-1119(86)90138-1. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991 Nov;5(11):1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- Rhodes S. J., Chen R., DiMattia G. E., Scully K. M., Kalla K. A., Lin S. C., Yu V. C., Rosenfeld M. G. A tissue-specific enhancer confers Pit-1-dependent morphogen inducibility and autoregulation on the pit-1 gene. Genes Dev. 1993 Jun;7(6):913–932. doi: 10.1101/gad.7.6.913. [DOI] [PubMed] [Google Scholar]
- Saffhill R., Margison G. P., O'Connor P. J. Mechanisms of carcinogenesis induced by alkylating agents. Biochim Biophys Acta. 1985 Dec 17;823(2):111–145. doi: 10.1016/0304-419x(85)90009-5. [DOI] [PubMed] [Google Scholar]
- Stapleton G., Somma M. P., Lavia P. Cell type-specific interactions of transcription factors with a housekeeping promoter in vivo. Nucleic Acids Res. 1993 May 25;21(10):2465–2471. doi: 10.1093/nar/21.10.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Walsh K., Schimmel P. Two nuclear factors compete for the skeletal muscle actin promoter. J Biol Chem. 1987 Jul 15;262(20):9429–9432. [PubMed] [Google Scholar]
- Wang Y., Kato T., Ayaki H., Ishizaki K., Tano K., Mitra S., Ikenaga M. Correlation between DNA methylation and expression of O6-methylguanine-DNA methyltransferase gene in cultured human tumor cells. Mutat Res. 1992 Mar;273(2):221–230. doi: 10.1016/0921-8777(92)90083-f. [DOI] [PubMed] [Google Scholar]
- Zerivitz K., Akusjärvi G. An improved nuclear extract preparation method. Gene Anal Tech. 1989 Sep-Oct;6(5):101–109. doi: 10.1016/0735-0651(89)90016-2. [DOI] [PubMed] [Google Scholar]
- Zhu Q. S., Heisterkamp N., Groffen J. Unique organization of the human BCR gene promoter. Nucleic Acids Res. 1990 Dec 11;18(23):7119–7125. doi: 10.1093/nar/18.23.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wronski M. A., Brent T. P. Effect of 5-azacytidine on expression of the human DNA repair enzyme O6-methylguanine-DNA methyltransferase. Carcinogenesis. 1994 Apr;15(4):577–582. doi: 10.1093/carcin/15.4.577. [DOI] [PubMed] [Google Scholar]
- von Wronski M. A., Harris L. C., Tano K., Mitra S., Bigner D. D., Brent T. P. Cytosine methylation and suppression of O6-methylguanine-DNA methyltransferase expression in human rhabdomyosarcoma cell lines and xenografts. Oncol Res. 1992;4(4-5):167–174. [PubMed] [Google Scholar]