Abstract
The expression of genes that reside near telomeres is attenuated through telomere position-effect variegation (TPEV). Using a URA3 reporter located at TEL-VIIL of S. cerevisiae, it was demonstrated that the disruptor of telomeric silencing-1 (Dot1) regulates TPEV by catalyzing the methylation of H3K79. URA3 was also used as a reporter to demonstrate that H3K79 methylation is required for HM silencing. Surprisingly, a genome-wide expression analysis of mutants defective in H3K79 methylation patterns indicated that only a few telomeric genes, such as the COS12 located at TEL-VIIL, are subject to H3K79 methylation-dependent natural silencing. Consistently, loss of Dot1 did not globally alter Sir2/3 occupancy in subtelomeric regions, but did lead to some telomere-specific changes. Furthermore, we demonstrated that H3K79 methylation by Dot1 does not play a role in the maintenance of natural HML silencing. Our results show that the commonly used URA3 reporter located at TEL-VIIL or at the mating loci may not report on natural PEV and that studies concerning the epigenetic mechanism of silencing in yeast should employ assays that report on the natural pattern of gene expression.
INTRODUCTION
Eighty years ago, H.J. Muller described the phenomenon of position-effect variegation (PEV) of gene expression (which he called “eversporting displacement”), in which the expression of genes in Drosophila that are brought near heterochromatin is silenced (Muller and Altenburg, 1930). Following the identification of ADH4 as the most distal gene in the S. cerevisiae genome on TEL-VIIL (Walton et al., 1986), Gottschling and colleagues replaced ADH4 with a URA3 reporter at this location and established that expression of URA3 is silenced in a semi-stable, but heritable manner (Aparicio et al., 1991; Gottschling et al., 1990; Rusche et al., 2003). This process has been referred to as telomeric position effect variegation (TPEV) and is thought to be due to the heterochromatic nature of chromosome ends in budding yeast. Indeed, when URA3 is moved into presumptive euchromatin 20 Kb or more away from the telomere, its transcription increases (Aparicio and Gottschling, 1994; Gottschling et al., 1990). A similar reporter-based assay has also been developed by placing the ADE2 gene adjacent to the telomere sequence (a TG1-3 tract) near the right end of chromosome V (TEL-VR) (Singer et al., 1998). The finding that both TEL-VIIL and TEL-VR seem to be subject to TPEV of gene expression has been inferred to represent telomeric silencing genome-wide, and these two reporter systems have been widely used for the past twenty years in studies of telomeric gene silencing (for example: Aparicio et al., 1991; Cubizolles et al., 2006; Gardner et al., 2005; Gottschling et al., 1990; Krogan et al., 2002; Miller et al., 2001; Murphy et al., 2003; Ng et al., 2002; Nislow et al., 1997; Xu et al., 2007).
Transcriptional silencing of genes near telomeres is associated with specific chromatin structures (Rusche et al., 2003). Histone modifications such as acetylation, methylation, and monoubiquitination have been linked to transcriptional silencing (Grewal and Elgin, 2007; Shahbazian and Grunstein, 2007; Shilatifard, 2006). In budding yeast, several proteins involved in adding or removing histone modifications are involved in silencing a URA3 reporter when it is placed near telomeres or at the cryptic mating type loci, HML and HMR. Reporter-based silencing at telomeres and silencing at the mating type loci is mediated by the silent information regulators, Sir3, Sir4 and the conserved Sir2 NAD+-dependent histone deacetylase, which assemble along the nucleosome fiber (Gartenberg, 2000; Rine et al., 1979; Rusche et al., 2003). Mating-type silencing also requires the Sir1 silencing protein, while telomeric silencing involves yKu70/yKu80, Rap1 and other factors involved in maintenance of the chromosome end.
Dot1, the methyltransferase that catalyzes mono-, di- and trimethylation of histone H3 lysine 79 (H3K79), has been shown to be involved in the regulation of telomeric and HM silencing using the URA3 reporter (Lacoste et al., 2002; Ng et al., 2002; van Leeuwen et al., 2002). Dot1 can interact with the histone H4 N-terminal tail acetylated on lysine 16, a mark regulated by the Sas2 acetyltransferase and the Sir2 histone deacetylase (Altaf et al., 2007). In addition, the removal of H3K79 methylation as well as H3K4 methylation are important for heterochromatin formation at the silent mating-type loci (Osborne et al., 2009). Furthermore, the mammalian and Drosophila Dot1 complex, DotCom, was recently purified and it was demonstrated that its specific H3K79 trimethylation functions in the Wnt signaling pathway (Mohan et al., 2010).
Based on the observation that H3K79 is hypermethylated in euchromatin and hypomethylated in heterochromatin, it has been proposed that methylation of H3K79 plays an indirect role in telomeric silencing, perhaps by restricting the association of the Sir proteins with euchromatin, thereby enhancing their assembly on telomeric heterochromatin (Ng et al., 2002; van Leeuwen et al., 2002). Consistent with a function such as a boundary factor, it has been proposed that the loss of Dot1, or mutations changing K79 of H3 to another residue, results in spreading of the Sir proteins beyond the heterochromatic regions of the few regions tested (Ng et al., 2002; Shilatifard, 2006; van Leeuwen et al., 2002). Conversely, it has been shown that the loss of the Sir proteins results in the intrusion of H3K79 methylation into heterochromatin, suggesting that the Sir proteins assembled on heterochromatin in turn restrict access of Dot1 to these telomeric regions (Ng et al., 2002; van Leeuwen et al., 2002).
To better understand the role of H3K79 methylation via Dot1 in TPEV, we identified and tested a series of mutants that differentially effect H3K79 di- and/or tri-methylation. As described below, our study suggests that the loss of Dot1 does not globally alter Sir2/3 occupancy in the subtelomeric regions, indicating that other factors in addition to H3K79 methylation are required for the regulation of Sir2/3 occupancy in the subtelomeric regions. Furthermore, our studies show that H3K79 methylation established by Dot1 does not play a role in the maintenance of natural silencing of telomeric and HM loci.
RESULTS
Ard1 was required for proper normal levels of H3K79 trimethylation
A systematic genome-wide screen of the yeast deletion collection for mutants that affect H3K79 methylation previously revealed that Swi4 and Swi6 are required for H3K79 di-, but not trimethylation (Schulze et al., 2009). Employing the same methods, ARD1 was identified as a factor that is required for maintaining the proper levels of trimethylated H3K79, with no global effect on dimethylated H3K79 (Supplementary Figure 1A–D). In addition, strains deleted for ARD1 also had reduced levels of histone H2B monoubiquitination (Supplementary Figure 2A). However, Ard1 was not required for the proper expression of several of the known genes required for H2B monoubiquitination, as its deletion did not grossly change the steady-state level of the mRNAs tested (Supplementary Figure 2B). Ard1 and Nat1 are in a complex, however, our GPS analysis demonstrated that H3K79 trimethylation levels are a bit higher in nat1 null strains than in ard1 null background (data not shown). More detailed molecular analysis is required to understand the links between Ard1 and Dot1 and the Ard1/Nat1 complex and silencing. However, our finding suggests that Ard1 regulates the establishment of H3K79 trimethylation by regulating the H2B monoubiquitination levels either directly or indirectly.
Di- and trimethylation of H3K79 were required for URA3 reporter expression on TEL-VII-L
Histone H3K79 methylation catalyzed by Dot1 is required for silencing of the URA3 reporter gene located near the end of chromosome 7 (TEL-VIIL) (Ng et al., 2002; van Leeuwen et al., 2002). To determine whether di- and trimethylated forms of H3K79 are specifically required for URA3 silencing, we scored the phenotype of cells whose only copy of URA3 is at this location (TELVII::URA3) (Gottschling et al., 1990). Wild-type cells were resistant to 5-FOA (Supplementary Figure 2C), indicating that URA3 expression is silenced. In contrast, cells lacking RAD6, which are unable to monoubiquitinate H2BK123, a pre-requisite for H3K79 trimethylation, were sensitive to 5-FOA, and therefore, defective for silencing of URA3 expression. Consistent with the requirement for H3K79 trimethylation in gene silencing, ard1Δ mutants were defective in the silencing of reporters at the telomeres and at the silent mating-type loci (Aparicio et al., 1991; Mullen et al., 1989; Whiteway et al., 1987). To determine the requirement for H3K79 dimethylation in telomeric silencing, we tested the sensitivity of TEL-VIIL::URA3 strains deleted for SWI4 to 5-FOA, and also observed reduced growth similar to rad6Δ cells (Supplementary Figure 2C).
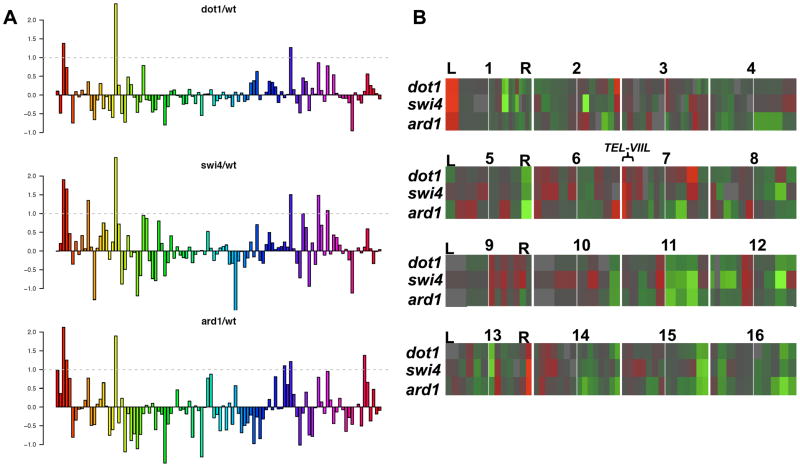
We confirmed the results of the reporter-based silencing assay with gene expression data using Affymetrix Gene Chip Yeast Genome 2.0 Arrays (see M&M) (Figure 1A). The pattern of gene expression upon the loss of ARD1 was very similar to the loss of either DOT1 or SWI4, resulting in derepression of a small number of genes lying within 20 kb of the chromosome ends (Figure 1A). This result suggests that both di- and trimethylation of H3K79 are required for a minor telomere-associated silencing of gene expression at these loci.
Figure 1. Histone H3K79 methylation and telomeric silencing.
(A) Barplots indicating gene expression ratios of mutant/wt in log2 format for 106 genes within 20kb of the chromosome ends. The genes are ordered left to right in terms of increasing distance from the end of the chromosome. (B) Heatmap of gene expression within 20 kb of the end of the 16 yeast chromosomes in swi4Δ, ard1Δ, and dot1Δ strains, which are specifically defective in di- and trimethylation of H3K79 respectively, are compared to a wild-type strain. Gene expression ratios are indicated by colored rectangles with red indicating a higher expression in the mutant, and green indicating a higher expression in the wild-type cells. Gray indicates equal expression in both strains. The color scale is plus or minus 3-fold. The numbered rectangles indicate the ends of each yeast chromosome. The genes within 20 kb of the end of the left arm of each chromosome are indicated to the left of the gray line in each rectangle, whereas genes within 20 kb of the end of the right arm of the chromosome are indicated to the right of the gray line. “L” and “R” indicate the left and right arm of each chromosome.
H3K79-mediated telomere-associated gene silencing was limited and uneven
A more detailed analysis of our gene expression data on a chromosome-by-chromosome basis revealed several unexpected findings that challenge the prevailing view of Dot1 as a general regulator of TPEV. Although we did observe loss of derepression at a small number of telomeres, including near TEL-IL, TEL-IIR, TEL-VIIL and TEL-XIIIR, genes near the 28 other telomere ends in budding yeast showed no substantial change in natural expression in the absence of H3K79 methylation or in cells lacking Dot1 (Figure 1B).
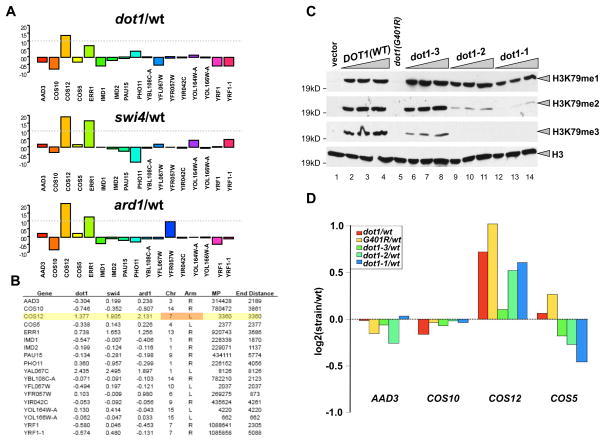
Because of the degree to which the URA3 reporter is silenced is proportional to its proximity to the chromosome end (Aparicio and Gottschling, 1994), we therefore limited further analysis of our expression data to genes within 10 kb of telomeres. Based on these criteria, we found that only a handful of genes increased their natural expression in dot1Δ, swi4Δ and ard1Δ mutants (Figure 2A). The genes closest to a chromosome end – YFR057W (TEL-VIR) and AAD3 (TEL-IIIR) and YOL166W-A)–were not derepressed in the absence of H3K79 methylation (Figure 2A–B). Interestingly, the gene whose silencing was most affected by loss of H3K79 methylation, COS12, resides near the site of the URA3::TEL-VIIL that is commonly used as a reporter of telomere-associated gene silencing (Gottschling et al., 1990; Singer et al., 1998; van Leeuwen et al., 2002) (Figures 1B and 2A-B).
Figure 2. Histone H3K79 methylation effects on natural telomere-associated gene expression.
(A) The log2 ratio of gene expression for mutant/wt is shown for genes within 5kb of chromosomal ends. The dashed grey line indicates two-fold enrichment in the mutant over wt. (B) Table of gene expression values. The log2 ratio of gene expression for mutant/wt is shown for genes within 5kb of chromosomal ends for the mutants indicated in the table, along with the chromosomal position of the gene, the gene midpoint, and the distance between the midpoint and the end of the chromosome. (C) H3K79 methylation status in cells expressing dot1-1, dot1-2, and dot1-3 alleles were examined by Western blotting using anti-H3K79 mono-, di- and trimethylation specific antibodies. Dot1 alleles were cloned in pRS315 vector as described under Materials and Methods. Dot1 (G401R) is a catalytically dead mutant (van Leeuwen et al., 2002). dot1-3 exhibits slightly less accumulation of H3K79 trimethylation, but comparable levels of dimethylation than wild-type; while dot1-2 and -1 lost most of their H3K79 tri- and dimethylation with an observed slight loss in the H3K79 monomethylation signal in dot1-1. (C) Gene expression analysis using dot1-1, -2, and -3 strains. The ratio of gene expression in log2 format for various Dot1 mutants relative to wt is shown for four genes within 5 kb of the chromosomal ends.
To alleviate the possibility that our observations are indirect due to the deletion of SWI4, SWI6 and ARD1, we also generated a series of DOT1 alleles by mutating several conserved lysine residues within Dot1. The resulting mutants were marginally defective in H3K79 trimethylation (dot1-3), tri and dimethylation (dot1-2), and tri-, di- and a bit of monomethylation (dot1-1) (Figure 2C). When we analyzed the expression level of several telomeric genes in these mutants, we found that that COS12 expression on TEL-VIIL requires proper H3K79 di- and trimethylation (Figure 2D). Taken together, our results suggest that only a few genes on specific telomeres are derepressed upon loss of H3K79 methylation.
Genome-wide localization pattern of Sir2/Sir3 and H3K79 methylation
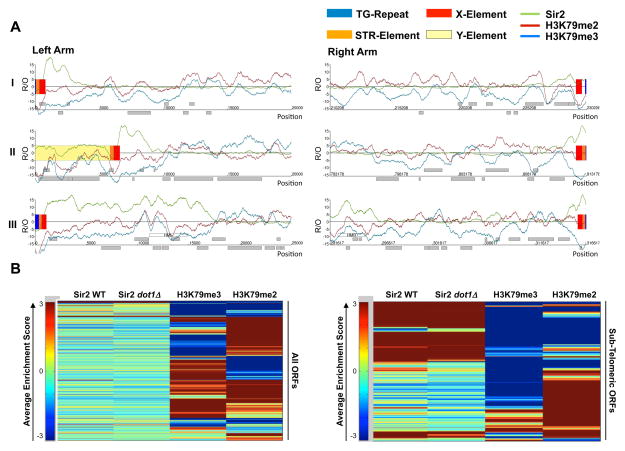
Using ChIP-on-chip, we compiled detailed genome-wide maps of the Sir proteins, Sir2 and Sir3, and compared them to previously published data for H3K79 di- and trimethylation (Schulze, et. al 2009) primarily focusing on telomere-proximal regions which we define here as being within 20 kB from the chromosome ends (Figure 3A, Supp Figure 3 and 5). As previously shown, Sir2 and Sir3 bound to all 32 telomere proximal regions and the silent mating-type loci HML/HMR (Sperling and Grunstein, 2009; Tsankov et al., 2006), whereas H3K79 di- and trimethylation had low levels at almost all of the telomere ends. Consistent with each chromosome end having a distinct composition of telomeric and subtelomeric elements (Pryde and Louis, 1999), each telomere-proximal region had a unique pattern of Sir protein binding and H3K79 methylation. In general, regions bound by Sir proteins were mostly devoid of H3K79 di- or trimethylation supporting the idea of Sir proteins binding and H3K79 methylation being mutually exclusive (Figure 3A, Supplemental Figure 3). To explore the spatial relationship between the Sir proteins and H3K79 methylation more closely, we compared the patterns of Sir2 and H3K79 di- and trimethylation at all genes (Figure 3B). The average enrichment scores for Sir2 as well as H3K79 di- and trimethylation were calculated for all open reading frames (ORFs) and hierarchically clustered. Genome-wide, Sir2, H3K79me2 and H3K79me3 were associated with different groups of ORFs; and within the first 20 kB of all telomere ends, the algorithm clearly separated ORFs bound by Sir2 from ORFs enriched for H3K79 methylation (Figure 3B).
Figure 3. High-Resolution profiles of Sir2, H3K79me2 and H3K79me3.
A) 20 kb of chromosome ends I to III were plotted along the x axis against the relative occupancy (R/O) of Sir2, H3K79 di- and trimethylation in WT cells. Superimposed ChIP-on-chip profiles indicated the distinct distribution of Sir2 and H3K79 methylation. Each chromosome end was unique for its Sir2, H3K79me2 and H3K79me3 occupancy. B) ORFs bound by Sir2 separated from H3K79 di- and trimethylated ORFs. Heatmap visualizing the average enrichment of Sir2 in WT and dot1Δ cells as well as H3K79me2 and H3K79me3 for all known yeast ORFs (left panel) and for all ORFs within 20kB from the chromosome ends (right panel). Each row color-codes the average enrichment score for a particular ORF on the spectrum from red indicating enrichment to blue for depletion. The distribution patterns were hierarchically clustered and plotted.
Other factors in addition to the H3K79 methylation pattern could regulate Sir2/3 occupancy at telomeres
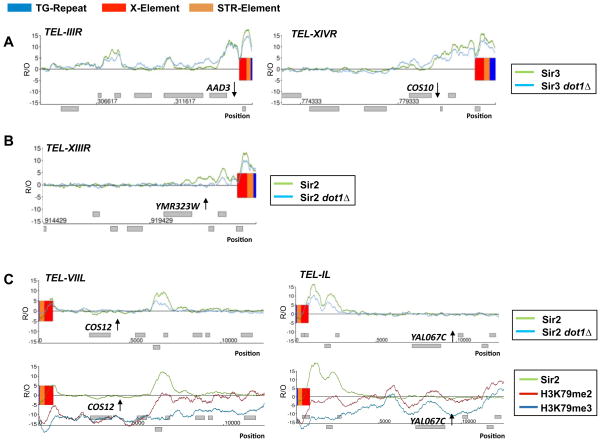
To explore the previously proposed role of H3K79 methylation in regulating the binding of Sir proteins at telomeres, we assessed Sir protein binding in the absence of H3K79 methylation. Using ChIP-on-Chip, we mapped Sir2/Sir3 binding across the genome in cells lacking DOT1 to determine if the binding patterns were altered compared to wild-type cells (Figure 3B, 4 and Supplemental Figure 4 and 5). Only at a few of the telomeres did Sir protein binding notably change upon loss of Dot1, while at the majority of the telomeres, the Sir2/Sir3 binding profiles in the absence of Dot1 appear to be similar to those of wild-type cells (Figure 4, Figure 3B and Supplemental Figure 4 and 5). The few changes observed were fully consistent with our gene expression data. Loss of Dot1 resulted in some increased Sir3 occupancy at the subtelomeric regions of chromosomes IIIR and XIVR (Figure 4A), which was associated with an increased in silencing and lower expression of the genes AAD3 and COS10 found in these regions. At other subtelomeric regions, such as XIIIR, loss of Dot1 caused a marginal decrease in Sir2 binding and coincided with a derepression of YMR323W, a gene located in that region (Figure 4B). Strikingly, the few genes such as COS12 or YAL067C, which were derepressed in cells lacking DOT1, were not bound by Sir proteins in the presence or absence of Dot1, indicating that the effect on gene expression at these loci is independent of Sir2/3 (Figure 4C). Instead, some of these genes are marked moderately by both H3K79 di- and trimethylation and changes in their expression could likely be due to the elimination of H3K79 methylation itself as the result of the deletion of DOT1 (Figure 4C). The region around COS12 on the left arm of chromosome VII is challenging to analyze because it shows a high level of unspecific background enrichment when compared to mock-IP’s, similar to a few other subtelomeric regions (JMS and MSK, unpublished data). Therefore, to meet this challenge, we normalized our data against mock controls, thus providing the most stringent assessment of protein binding and histone modifications in this repetitive region.
Figure 4. Minor alterations of Sir2 and Sir3 binding at subtelomeric regions corresponded to observed gene expression changes.
A) The right arms of chromosome III and XIVR were plotted along the x axis against the relative occupancy (R/O) of Sir3 in a wild-type and dot1 deletion strain. Increased Sir3 occupancy coincided with a lower expression of AAD3 and COS10. B) The right arm of chromosome XIII was plotted along the x axis against the relative occupancy of Sir2 in a wild-type and dot1 deletion strain. Decreased Sir2 occupancy coincided with a higher expression of YMR323W. C) The left arms of chromosome VII and chromosome I were plotted along the x axis against the relative occupancy of Sir2 in a wild-type and dot1 deletion strain as well as H3K79 di- and trimethylation. Only minor changes in Sir2 occupancy were observed at the subtelomere of ChrVIIL and ChrIL in the absence of H3K79 methylation. The superimposed profiles with H3K79me2 and H3K79me3 indicate that COS12 and YAL067C were moderately enriched for H3K79 methylation and not Sir2.
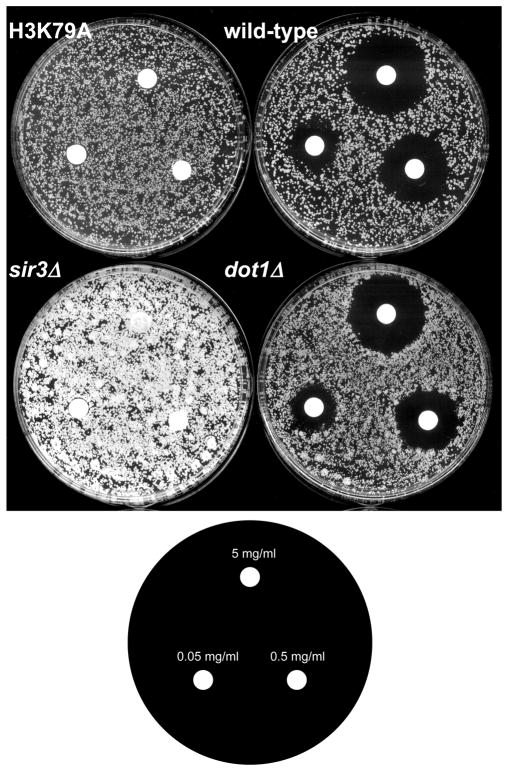
Histone H3K79 methylation by Dot1 is not required for proper HM silencing
Based on URA3 reporter studies, it has been proposed that histone H3K79 methylation by Dot1 is required for proper HML and HMR silencing, in addition to its role in the regulation of telomeric silencing (van Leeuwen et al., 2002). In this study, we re-evaluated the role of H3K79 methylation in silencing at HML silencing by measuring natural silencing at this loci. Therefore, we analyzed the sensitivity of cells to different concentrations of the mating pheromone alpha factor. In wild-type haploid MATa cells, a-specific genes such as the alpha factor receptor STE2 are expressed, allowing for arrest in the G1 phase of the cell cycle in the presence of increasing amounts of the alpha factor peptide. When HML and HMR silencing is disrupted, as occurs in sir2Δ and sir3Δ mutants, genes at the cryptic mating loci are expressed, including the alpha2 gene at HML, which represses the expression of a-specific genes such as STE2. Thus, these mutants are insensitive to the presence of alpha factor and no effect on growth is observed, even at the highest concentrations of alpha factor (Figure 5). Using the same assay, we found that the loss of Dot1 had no effect on growth resistance to increasing concentrations of alpha factor compared to an isogenic wild-type strain, suggesting that H3K79 methylation is not required for proper silencing of mating type loci (Figure 5). Surprisingly though, a single point mutation in H3K79 itself displayed a similar phenotype to that of one with a deletion of Sir2 and/or Sir3 (Figure 5). In addition, our microarray studies in strains bearing an H3K79A mutation demonstrated that the expression mating pheromone alpha2 is up-regulated whereas alpha2 expression is unchanged in dot1 null strains (data not shown). Thus, H3K79A, but not methylation by Dot1 affects HML silencing. In contrast, both strains (H3K79A and dot1 null) show deprepression of COS12 on TEL-VIIL (data not shown).
Figure 5. Histone H3K79 residue is required for proper mating type silencing, but Dot1 and H3K79 methylation does not play a role in this process.
Wild-type, dot1Δ, sir3Δ, or H3K79A strains were plated on YPD and tested for growth inhibition in the presence of the indicated concentration of alpha factor in a halo assay test. Both sir3Δ and H3K79A mutants were resistant to alpha factor arrest. However, strains lacking H3K79 methylation due to deletion of DOT1 are inhibited to a similar extent as the wild-type strain. This indicates that H3K79 methylation in itself is not required for proper HML silencing.
DISCUSSION
Our data on Dot1 suggests that its role in heterochromatic transcriptional regulation is context-dependent and limited to a few telomeres. Our microarray studies demonstrated that the natural expression pattern of only 2 of the 31 unique genes located within 10 kb of a chromosome end were derepressed 2-fold or more in dot1Δ, or other mutants altering the pattern of H3K79 methylation. Of all of the unique genes within 10 kb of a chromosome end, only COS12 (YGL263W) and SEO1 (YAL067C) were derepressed in mutants missing H3K79 methylation. It seems significant that the gene nearest the telomere whose expression is most affected by H3K79 methylation, COS12/YGL263W, is also near the location of the TEL-VIIL::URA3 gene that has been extensively used to report the status of telomere-associated gene silencing and to identify the DOT genes involved in telomeric gene silencing.
In the generation of strains for TPEV studies, the ADH4 gene, which naturally lies within 15 kb from the chromosome end, was replaced with a URA3 reporter with the concerted loss of the subtelomeric repeats and installation of a new tract of telomere sequences (TG1-3) (Gottschling et al., 1990). In our hands, unlike COS12, the natural expression pattern of ADH4 was marginally altered by the loss of H3K79 methylation (data not shown). These findings indicate that the replacement of URA3 in the ADH4 loci and the deletion of the rest of the TEL-VIIL along with the installation of a new tract of telomere sequences (TG1-3) following the URA3 spatially localized URA3 where COS12 naturally resides in the genome. Therefore, URA3 in its artificial location at TEL-VIIL behaves just like COS12 would in its natural location. Consistent with this observation, Rossman et al., (in this issue) demonstrated that in dot1 null cells, URA3 expression is also up-regulated in a strain that deletes COS12 and replaces it with ADH4::URA3.
One possible explanation for our observations regarding the natural derepression of the telomeric-associated genes in the absence of H3K79 methylation might be explained by the fact that these genes are expressed at low levels, and therefore, are not detected via microarray studies. To address this possibility, we have analyzed the intensity of expression for many of the unique telomeric genes and have found them either to be repressed or not highly expressed at all. It is hard to distinguish between these two modes of transcriptional regulation unless one has mutant(s) that result in their derepression. In the case of TEL-VII-L, we detect low intensity of expression for COS12 in the wild-type cells. However, in the absence of H3K79 methylation, the intensity of expression for COS12 is increased, indicating that H3K79 methylation plays an essential role in COS12 transcriptional regulation. Given the fact that H3K79 methylation is associated within COS12 loci, derepression of COS12 in the absence of Dot1 can be explained by a direct role for H3K79 methylation in transcriptional regulation and not TPEV.
Additionally, variable and limited silencing of expression via insertion of reporters near chromosome ends were previously reported (Pryde and Louis, 1999). Silencing of URA3 inserted at different positions near the ends of several chromosomes varied from chromsome to chromsome and from location to location within the same chromosome. Those results are similar to what we observed with direct assays for the natural expression of genes in their native locations near chromosome ends. Another study that used DNA microarrays to probe for the expression of telomere-associated genes found that deletion of YAF9 and SAS5, which encode YEATS-domain proteins involved in telomere-associated gene silencing, only affects the expression of genes near certain chromosome ends (Zhang et al., 2004). Our findings that the loss of Dot1 and H3K79 methylation has no detectable effect on maintenance of HML silencing (Figure 5) questions the generality of the role of Dot1 and H3K79 methylation in heterochromatic silencing in yeast.
In the work presented here, we confirm and substantially extend previous observations about mutually exclusive occupancy of subtelomeric regions by either the Sir proteins or H3K79 methylation, which was derived from one chromosome end (Ng et al., 2002; van Leeuwen et al., 2002). We show here that across the entire genome, loci enriched for the Sir2 and Sir3 proteins are mostly depleted of H3K79 methylation. One possible mechanism for the opposing activities of H3K79 methylation and Sir protein binding might be related to the competition between Dot1 and Sir3 for a binding site on histone H4 (Altaf et al., 2007; Fingerman et al., 2007). However, the negative correlation between H3K79 methylation and Sir binding does not necessarily reflect a causal relationship between their occupancies across all telomeres. As such, our data on the effects of Dot1 on the distribution of Sir proteins at natural telomeric gene loci are consistent with previously published data. Specifically, we confirmed that the loss of Dot1 resulted in a slightly reduced binding of Sir2 and Sir3 to the right end of chromosome VI (Ng et al., 2002; van Leeuwen et al., 2002). Extending these data from a single chromosome end to a genome-wide scale, we observed a similar reduction in Sir2 and Sir3 binding at several subtelomeric regions. However, this likely constitutes a telomere-specific and thus context-dependent effect of H3K79 methylation on Sir protein binding, as Sir2 and Sir3 binding to other subtelomeric regions was not affected by the loss of Dot1. In addition, and consistent with prevailing models, we found that Dot1 prevented the spread of heterochromatin at a limited number of telomeres, as judged by increased binding of Sir3 at loci more distal to the telomere ends. Consistent with this explanation, Sir3 and Sir4 have been shown to bind regions distant from silent domains and repress genes more than 100 kB from heterochromatic loci only upon the simultaneous loss of HTZ1 and SET1 as determined by DamID (Venkatasubrahmanyam et al., 2007).
The role of Dot1 is not restricted to heterochromatin at telomeres, but might also extend to the silent mating type loci HML and HMR. A recent study has elegantly revealed the crucial steps during heterochromatin formation at the HML locus at single-cell resolution (Osborne et al., 2009), supporting earlier kinetic studies of heterochromatin establishment derived from bulk cultures (Katan-Khaykovich and Struhl, 2005). At the native HML locus, loss of Dot1 accelerates the establishment of silencing presumably through exerting an effect on Sir binding, therefore indicating that Dot1 activity is involved in a specific step of the process (Osborne et al., 2009). However, the altered kinetics of the heterochromatin establishment caused by loss of Dot1 has minimal effects on gene silencing at the natural HML (Osborne et al., 2009). This is further supported by studies using bulk yeast cultures showing that silencing of both native silent mating type loci is independent of Dot1 (Yang et al., 2008). Thus our data measuring steady-state expression of native HM loci are consistent with the lack of requirement for Dot1 under those conditions. In contrast, artificially compromising HM silencing by either genetic or DNA sequence-based manipulations necessitates a requirement for Dot1 in this process. Only in combination with a deletion of SIR1 (Osborne et al., 2009; van Welsem et al., 2008) or at a synthetic HML-E silencer (Weber and Ehrenhofer-Murray, 2010) does loss of Dot1 cause derepression of the HM loci. Taken together, our study shows that the role of Dot1 and H3K79 methylation on telomeric and HM silencing is limited, in contrast to previous conclusions from reporter assays. This questions the use of URA3 as a reporter assays for measuring natural silencing. In support of this conclusion, an accompanying study by Rossmann et al. (published in this issue) independently demonstrated that the URA3 reporter assay at telomeres does not reflect on heterochromatin formation, but rather reports on an imbalance in ribonucleotide reductase levels, indicating that metabolic changes caused by 5-FOA are incompatible with the use of URA3 as a reporter for TPEV.
Materials and Methods
Global Proteomic Analysis of Histone Modifications
GPS analyses were carried out as described previously (Schneider et al., 2004) with the use of antibodies specific for dimethylated (Upstate/Millipore) or trimethylated Lys 79 (Abcam) and for dimethylated Lys 4 of histone H3 (Abcam).
Microarray analysis of gene expression
Yeast cells were grown in a rich liquid media and harvested at an OD600 of 0.7 for RNA purification. Biotinylated cRNA was prepared from 1 ug Total RNA using the MessageAmp II one-cycle linear amplification protocol from Ambion (Austin, Tx) and used to probe Affymetrix (Santa Clara, CA) GeneChip. Data from this study were analyzed as described in the Supplemental Methods.
Chromatin Immunoprecipitation and Genome-wide ChIP-on-Chip
Genome-wide ChIP-Chips were performed and analyzed as previously described (Schulze et al., 2009). A detailed description can also be found in the Supplemental Methods.
Generation of dot1 alleles
The mutations of the dot1-1, dot1-2, and dot1-3 are multiple point mutations of lysine to arginine. The dot1-3 has substitutions of lysines ranging from K106 through K443, in which all lysines were substituted with arginine (K106-443R). The dot1-2 contains further substitutions of lysines (K106-508R) in addition to mutations of dot1-3. In the dot1-1, all the lysines from K105 to the C-terminus of Dot1 were substituted to arginine including lysines (K106-579R) in addition to the ones substituted in the dot1-2. The dot1 alleles were cloned into pRS315 in the form of SpeI restriction site-5′UTR (450bp)-dot1 ORF-3′UTR (230bp)-SmaI restriction site by using SpeI and SmaI. The H3K79 methylation states and gene expression profiles were examined in the dot1-(1–3) mutant strains transformed with those dot1 plasmids.
Supplementary Material
Acknowledgments
We are grateful to Dr. Edwin Smith and Mark Johnston for discussions, Laura Shilatifard for editorial assistance and Amy Lindley for technical assistance. Work in MSK’s laboratory is supported by CIHR grant MOP-79442. MSK is a Scholar of the Canadian Institute for Advanced Research, and JMS was supported by a fellowship from the Child and Family Research Institute. SLJ and ASH are supported by funds from the Stowers Institute. The yeast studies in the Shilatifard Laboratory are supported in part by a grant from the National Institute of Health, R01GM069905.
Footnotes
The microarray data files can be accessed at NCBI with GEO accession number GSE27234. The ChIP-on-Chip data files can be accessed online with ArrayExpress accession number E-MEXP-3108.
Supplemental data for this article include extended figures supporting the conclusions of the paper and extended procedures/materials and methods.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Cote J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Gottschling DE. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- Cubizolles F, Martino F, Perrod S, Gasser SM. A homotrimer-heterotrimer switch in Sir2 structure differentiates rDNA and telomeric silencing. Mol Cell. 2006;21:825–836. doi: 10.1016/j.molcel.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Fingerman IM, Li H-C, Briggs SD. A charge-based interaction between histone H4 and Dot1 is required for H3K79 methylation and telomere silencing: identification of a new trans-histone pathway. Genes Dev. 2007:2018–2029. doi: 10.1101/gad.1560607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin. Molecular and cellular biology. 2005;25:6123–6139. doi: 10.1128/MCB.25.14.6123-6139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartenberg MR. The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more. Curr Opin Microbiol. 2000;3:132–137. doi: 10.1016/s1369-5274(00)00064-3. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J. 2005:2138–2149. doi: 10.1038/sj.emboj.7600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A. COMPASS, a histone H3 (Lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem. 2002;277:10753–10755. doi: 10.1074/jbc.C200023200. [DOI] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Cote J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem. 2002;277:30421–30424. doi: 10.1074/jbc.C200366200. [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y, Washburn MP, Florens L, Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kayne PS, Moerschell RP, Tsunasawa S, Gribskov M, Colavito-Shepanski M, Grunstein M, Sherman F, Sternglanz R. Identification and characterization of genes and mutants for an N-terminal acetyltransferase from yeast. Embo J. 1989;8:2067–2075. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ, Altenburg E. The Frequency of Translocations Produced by X-Rays in Drosophila. Genetics. 1930;15:283–311. doi: 10.1093/genetics/15.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GA, Spedale EJ, Powell ST, Pillus L, Schultz SC, Chen L. The Sir4 C-terminal coiled coil is required for telomeric and mating type silencing in Saccharomyces cerevisiae. J Mol Biol. 2003;334:769–780. doi: 10.1016/j.jmb.2003.09.066. [DOI] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne EA, Dudoit S, Rine J. The establishment of gene silencing at single-cell resolution. Nat Genet. 2009;41:800–806. doi: 10.1038/ng.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. Embo J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Strathern JN, Hicks JB, Herskowitz I. A suppressor of mating-type locus mutations in Saccharomyces cerevisiae: evidence for and identification of cryptic mating-type loci. Genetics. 1979;93:877–901. doi: 10.1093/genetics/93.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Schulze JM, Jackson J, Nakanishi S, Gardner JM, Hentrich T, Haug J, Johnston M, Jaspersen SL, Kobor MS, Shilatifard A. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–641. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of Site-Specific Histone Acetylation and Deacetylation. Annu Rev Biochem. 2007 doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C, Mahowald M, Gottschling DE. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling AS, Grunstein M. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc Natl Acad Sci U S A. 2009;106:13153–13159. doi: 10.1073/pnas.0906866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Brown CR, Yu MC, Win MZ, Silver PA, Casolari JM. Communication between levels of transcriptional control improves robustness and adaptivity. Mol Syst Biol. 2006;2:65. doi: 10.1038/msb4100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- van Welsem T, Frederiks F, Verzijlbergen KF, Faber AW, Nelson ZW, Egan DA, Gottschling DE, van Leeuwen F. Synthetic lethal screens identify gene silencing processes in yeast and implicate the acetylated amino terminus of Sir3 in recognition of the nucleosome core. Mol Cell Biol. 2008:3861–3872. doi: 10.1128/MCB.02050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A. 2007;104:16609–16614. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton JD, Paquin CE, Kaneko K, Williamson VM. Resistance to antimycin A in yeast by amplification of ADH4 on a linear, 42 kb palindromic plasmid. Cell. 1986;46:857–863. doi: 10.1016/0092-8674(86)90067-x. [DOI] [PubMed] [Google Scholar]
- Weber JM, Ehrenhofer-Murray AE. Design of a minimal silencer for the silent mating-type locus HML of Saccharomyces cerevisiae. Nucleic acids research. 2010:7991–8000. doi: 10.1093/nar/gkq689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Freedman R, Van Arsdell S, Szostak JW, Thorner J. The yeast ARD1 gene product is required for repression of cryptic mating-type information at the HML locus. Mol Cell Biol. 1987;7:3713–3722. doi: 10.1128/mcb.7.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Molecular cell. 2007;27:890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Britton J, Kirchmaier AL. Insights into the impact of histone acetylation and methylation on Sir protein recruitment, spreading, and silencing in Saccharomyces cerevisiae. J Mol Biol. 2008:826–844. doi: 10.1016/j.jmb.2008.06.059. [DOI] [PubMed] [Google Scholar]
- Zhang H, Richardson DO, Roberts DN, Utley R, Erdjument-Bromage H, Tempst P, Cote J, Cairns BR. The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol Cell Biol. 2004;24:9424–9436. doi: 10.1128/MCB.24.21.9424-9436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.