Abstract
Tumor microvasculature possesses a high degree of heterogeneity in its structure and function. These features have been demonstrated to be important for disease diagnosis, response assessment, and treatment planning. The exploratory efforts of quantifying tumor vascular heterogeneity with DCE-MRI have led to promising results in a number of studies. However, the methodological implementation in those studies has been highly variable, leading to multiple challenges in data quality and comparability. This paper reviews several heterogeneity quantification methods, with an emphasis on their applications on DCE-MRI pharmacokinetic parametric maps. Important methodological and technological issues in experimental design, data acquisition, and analysis are also discussed, with the current opportunities and efforts for standardization highlighted.
1. Introduction
Development of novel antiangiogenic agents has posed a new challenge to medical imaging technologies. Unlike the systemic cytocidal effect of traditional cytotoxic drugs, these new agents are selectively targeted to the endothelial cells of tumor neovasculature, inducing a rapid shutdown of tumor blood supply without substantially affecting any morphologic metric [1]. Their cytostatic nature makes the conventional method of tumor size measurement inadequate for response evaluation. Several molecular and functional imaging techniques, namely, dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) [2, 3], positron emission tomography (PET) [2, 4], and dynamic contrast enhanced computed tomography (DCE-CT) [2, 5], have been developed in response to this challenge by monitoring pathophysiologic changes in various aspects of tumor vascular structure and functionality.
Among these imaging techniques, DCE-MRI has the unique advantage of not using ionizing radiation. Consequently, its repeated use in response assessment is not limited by patient dose considerations [2, 6]. In fact, daily or weekly repeats of DCE-MRI examinations are common in preclinical and clinical studies, allowing researchers to capture rapid change in tumor vasculature during the short “normalization window” after administration of an antiangiogenic agent [7]. Moreover, the wide availability of MR scanners makes this technique easily accessible to both clinicians and patients. As of September, 2010, American College of Radiology (ACR) has accredited 5498 MR facilities in the United States, compared to only 963 ACR-accredited PET facilities. All these advantages make DCE-MRI a convenient yet flexible modality for noninvasive evaluation of tumor microvasculature and angiogenesis. It has been applied to many preclinical studies and early phase clinical trials of various antiangiogenic and antivascular agents, as well as in nontrial settings for tumor grading and clinical outcome prediction [8, 9]. Quantitative imaging biomarkers derived from pharmacokinetic modeling of DCE-MRI data have been demonstrated to be correlated with various physiologic and pathophysiologic parameters in human and animal models, and found to be predictive of response rate and progression in patients accepting PTK/ZK treatment [10, 11].
Although preclinical and clinical results have shown that DCE-MRI is a powerful technique for the detection of subtle changes in tumor vascular function, many improvements are still needed for this technique to reach its maximum potential. Particularly, the important issue of tumor vascular heterogeneity needs to be addressed. Heterogeneous distribution of neovasculature has been reported in almost all types of solid tumor [12–14]. This phenomenon reflects the more fundamental heterogeneity in the clones of different tumor cell genomic subtypes and the expression of various pro- and antiangiogenic genes [15–17]. Experimental evidence shows that tumor vascular heterogeneity is associated with disease progression and malignancy [18]. Recent studies suggest that cancer relapse and metastasis are likely to be governed by a small population of cancer stem cells in aberrant vascular niches [19–21]. This new understanding strongly implies tumor vascular heterogeneity may have key importance for the understanding of disease mechanism and the discovery of novel treatment strategies. However, so far this critical information was simply ignored or only briefly mentioned as qualitative description in most preclinical and clinical studies. Even in the limited number of cases where quantification efforts have been made, substantial discrepancies exist in quantification method, parameter choice, and result interpretation. This lack of consensus has greatly hindered the investigation of tumor vascular heterogeneity and its influence on treatment response with DCE-MRI. In this paper, we critically review different quantification methods for heterogeneity currently available in the literature, and discuss several methodological and technological issues that are important in assessment of tumor vascular heterogeneity. Although these methods can be applied to any DCE-MRI image or parametric map, we emphasize their application on pharmacokinetic parametric maps as pharmacokinetic modeling is the most advanced DCE-MRI data analysis approach that provides pathophysiologically meaningful quantitative interpretation of DCE-MRI data.
2. Imaging Tumor Vascular Heterogeneity with DCE-MRI
In most solid tumors, the neovasculature developed through angiogenesis has an abnormal structure, with defects on walls that make them more permeable than normal vessels. As blood carrying a low molecular weight contrast agent (such as Gadopentetate Dimeglumine) flows through tumor vasculature, the contrast agent molecule leaks into the interstitial space and accumulates there. Interaction between the paramagnetic Gd3+ ion and adjacent water protons enhances tissue longitudinal (T1) relaxation rate, generating a brighter signal in T1-weighted images. The magnitude and rate of signal enhancement are determined by tissue vascular fraction, perfusion, microvascular permeability, and relative volume of the extracellular extravascular space (EES). The brightness information can be converted to contrast agent concentration with the linear relationship between gadolinium concentration and T1 relaxation rate [27]. The dynamic change of contrast agent concentration can be described by various model-free parameters or fitted to a pharmacokinetic model (Figure 1) [22–26], yielding parametric maps that give an intuitive and quantifiable illustration of tumor vascular heterogeneity. The most commonly used pharmacokinetic and model-free parameters are summarized in Tables 1 and 2, using the standard nomenclature proposed in the consensus paper by Tofts et al. [28]. Details about the use and interpretation of these parameters are beyond the scope of this paper. Interested readers are referred to the in-depth reviews written by Tofts et al. [28] and Evelhoch [29].
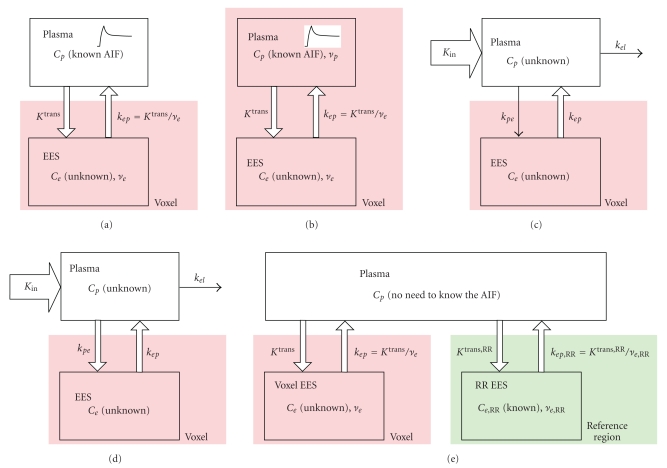
Figure 1.
In the literature, several models are available for DCE-MRI pharmacokinetic analysis. The Tofts' model ((a), also known as Kety's or Larsson's model) [22] models the gadolinium concentration dynamic change in a tissue voxel as the result of convolution between an exponential kernel and a measured or assumed arterial input function (AIF). A plasma term with volume fraction vp is added in the extended Toft's model (b) [23] by the assumption that the influence of the intravascular compartment is nonnegligible. The Brix's model (c) [24] takes the gadolinium concentration in blood plasma as another unknown variable and solves it with the tissue dynamic curve from a set of ordinary differential equations. In the adjusted Brix's model (d) [25], an oversimplified assumption in the original Brix's model is corrected, leading to a more realistic description of the plasma signal. In the reference region model (e) [26], two dynamic curves are simultaneously measured from the target voxel and a nearby reference region with known Ktrans and ve values. The need for an AIF is eliminated by jointly solving two pharmacokinetic equations for target voxel and reference region.
Table 1.
Pharmacokinetic parameters often used in DCE-MRI pharmacokinetic analysis.
| Parameter | Definition | Units | Alternative names | Literature |
|---|---|---|---|---|
| Ktrans | Volume transfer constant between EES and blood plasma | min−1 | EF, KPS | [22, 23, 26] |
| ve | EES volume per unit tissue volume | N/A | Interstitial space | [22, 23, 26] |
| vp | Blood plasma volume per unit tissue volume | N/A | [23] | |
| kep | Rate constant from EES to blood plasma | min−1 | k21 | [22–26] |
| kpe | Rate constant from blood plasma to EES | min−1 | k12 | [25] |
| kel | Elimination rate constant | min−1 | [24, 25] | |
| Amp | Amplitude of the normalized dynamic curve | N/A | A | [24, 25] |
Table 2.
Model-free parameters often used for DCE-MRI dynamic curve description†.
| Parameter | Definition | Units | Alternative names | Literature |
|---|---|---|---|---|
| Area-under-the-curve (AUC) | Area under the signal intensity or gadolinium dynamic curve | a.u·min or mmol·min/L | IAUC, AUGC, IAUGC | [29] |
| Relative signal intensity (RSI) | RSI = S(t)/S0 | N/A | [30] | |
| Initial slope | Maximum or average slope in the initial enhancement | a.u./min | Enhancement slope, upslope, enhancement rate | [31] |
| Washout slope | Maximum or average slope in the washout phase | a.u./min | Downslope, washout rate | [32] |
| Peak enhancement ratio (PER) | PER = (Smax − S0)/S0 | N/A | Maximum signal enhancement ratio (SERmax) | [33] |
| Tmax | Time from contrast agent arrive to peak enhancement | s | Time-to-peak (TTP) | |
| Maximum intensity-time ratio (MITR) | MITR = PER/Tmax | s−1 | [34] |
†Unlike the pharmacokinetic parameters, the nomenclature for model-free parameters has not been standardized.
Notation and abbreviations: S(t): MR signal intensity at time t; S0: precontrast signal intensity; Smax : maximum signal intensity; a.u.: arbitrary unit.
Heterogeneity in tumor vasculature is subject to change over time. During tumor growth, proliferation of tumor cells is faster than angiogenesis, leading to the formation of poorly perfused hypoxic zones near the tumor center. Rim enhancement is frequently observed in DCE-MRI images of various types of primary cancers, metastases, and xenografts. In the breast, it is a strong indicator of cancer in small lesions and malignancy in larger masses [18]. Other more complicated patterns of enhancement are also common. In general, fast progressing diseases and malignancies are associated with highly heterogeneous enhancement patterns in DCE-MRI images. This feature can be used to facilitate cancer diagnosis and staging. In human gliomas, Rose et al. found that low- and high-grade tumors are significantly different in several spatial heterogeneity statistics of DCE-MRI pharmacokinetic parameters [35, 36]. In breast imaging, different internal enhancement patterns have been identified as an important diagnostic characteristic by the ACR Breast Imaging-Reporting and Data System (BI-RADS) MRI lexicon [18].
In cancer treatment, tumor heterogeneity is usually associated with poor response, a higher chance of developing metastasis, and substantially shorter progression-free and overall survival [37, 38]. The mechanism is probably through the selection of more aggressive tumor cells that can better tolerate the hostile acidic, hypoxic environment in poorly perfused low-enhancing regions [19, 39]. Treatment-induced changes in tumor vascular heterogeneity have been reported in several preclinical and clinical settings. Checkley et al. observed that in human PC-3 prostate adenocarcinoma xenografts receiving ZD6474 treatment, the core region in the xenograft tumor revealed a larger reduction in the pharmacokinetic parameter Ktrans than the enhancing rims, suggesting a change in the distribution patterns of perfusion and permeability [40]. De Lussanet et al. found that primary rectal cancer patients receiving radiation therapy have lower intratumoral heterogeneity in the pharmacokinetic parameters Ktrans and vp than patients without radiation therapy [41]. Several groups also explored the possibility of predicting treatment efficacy with tumor vascular heterogeneity and its early change. Johansen et al. investigated the relationship between the distributions of DCE-MRI model-free parameters and patient 5-year survival in a small cohort of 24 breast cancer patients receiving neoadjuvant chemotherapy. They found that patients surviving for more than 5 years have significantly less heterogeneous relative signal intensity (RSI) distribution than nonsurvivors in the baseline DCE-MRI study performed prior to the start of therapy [30]. In a similar study, Padhani et al. correlated early changes in the distribution of DCE-MRI pharmacokinetic parameters with treatment response assessment. Changes in vascular heterogeneity quantified by the range of Ktrans distribution at the end of cycle two were found to be predictive of both clinical and histopathologic responses after three to six cycles of therapy [42]. Yuh et al. analyzed serial DCE-MRI studies of 101 patients during early treatment of cervical cancer. Part of their data has low temporal resolution so a complete pharmacokinetic analysis was not possible. Tumor vascular heterogeneity was thus quantified by the lower percentiles of voxel signal intensities in the plateau phase of the dynamic curves. Low values of the 5th to 20th percentiles were found to be correlated with poor primary tumor control and short disease-free survival. The 5th percentile measured at 2–2.5 weeks of therapy provided the best prediction for 8-year primary tumor control [43].
3. Quantifying Tumor Vascular Heterogeneity
For a long time, radiologists identify intratumoral heterogeneity solely by their visual perception of the image. Even though some level of consensus can be achieved with stringent training of readers and standardization of descriptive vocabularies, this approach is still subjective. Many factors, like the imaging hardware, personal bias, or even the physical condition of the reader, can affect the readers' judgement and lead to intra- and interobserver variations in assessment [44]. This problem is now becoming even more meaningful with the development of novel quantitative imaging techniques like DCE-MRI and perfusion CT. These techniques introduce a new dimension of spatially resolved functional information into the data space, which is usually displayed as pseudocolor overlays on the conventional morphologic images. Human color perception, however, is both nonlinear to luminance and nonuniform for different colors. As demonstrated by Giesel et al., reader's perception of intratumoral homogeneity/heterogeneity can easily be exaggerated or understated by simply choosing a different display range or color scheme (Figure 2) [45, 46]. In order to tackle this problem, several alternative methods have been developed by researchers in different branches of science. All these methods are quantitative, seeking to extract a set of well-defined quantities rather than ambiguous verbal descriptions to summarize some key features of tumor vascular heterogeneity.
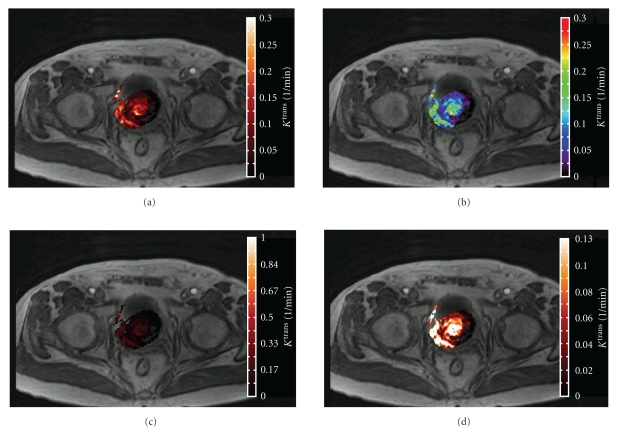
Figure 2.
Visual assessment of tumor vascular heterogeneity on a pseudocolor map is inherently subjective: the same parametric map of the pharmacokinetic parameter Ktrans is visualized as pseudocolor overlays with different display ranges and color schemes on a morphologic image in a bladder cancer patient. Compared to the single color scheme ((a), “blackbody radiation”), the multicolor scheme “rainbow” in (b) exaggerates subtle differences in Ktrans near color boundaries, creating artificial structures in the pseudocolor map. If the display range is chosen too large, the parametric map is perceived as being more “homogeneous” due to underrepresentation of the spatial variation (c). A small display range, on the other hand, tends to exaggerate the size of the “hotspot”, which may lead to an erroneous characterization of the disease (d).
In this section, we review these methods in separate topics. Technological issues and methodological considerations that we deem important in study design and data acquisition and analysis will be discussed in the next section.
3.1. Subregional Region-of-Interests (ROIs)
A simple, straightforward strategy for dealing with intratumoral heterogeneity is to subdivide the tumor into regions with less spatial variability. In the breast, many researchers prefer to analyze a small region with the highest enhancement (i.e., the “hot-spot”) instead of the whole tumor. If the hot-spot ROI is small enough and correctly placed, the hot-spot average is equivalent to the maximum of the whole tumor histogram. The hot-spot analysis can be performed semiautomatically, thus reducing the observer-related variability in ROI placement [34, 47]. In an early study, Mussurakis et al. were unable to detect a statistically significant difference in peak enhancement ratio (PER) between 71 malignant and 37 benign lesions, and concluded that hotspot analysis may not provide any diagnostic advantage over the whole-tumor method [33]. However, their result is probably undermined by an inappropriate selection of parameter: it is now known that in DCE-MRI, the most profound difference between malignant and benign breast lesions is not the maximum signal enhancement, but the shape of the entire dynamic enhancement curve. Malignant tumors have a quick uptake of contrast agent. Their dynamic curves are characterized by a fast increase to the peak followed by a slow washout or a stable “plateau”. Benign lesions usually have a persistently increasing dynamic curve in which the maximum signal intensity is achieved by the end of the observation window [48]. Liney et al. took this factor into consideration by introducing a new model-free parameter, the maximum-intensity-time-ratio (MITR), that corrects PER with the time to peak enhancement (Tmax). Their results from a study of similar size (81 malignant and 36 benign lesions) agreed with Mussurakis et al. conclusion about PER, but also showed that the lesion differentiation ability of the hot-spot analysis is significantly improved over the whole-tumor method when MITR is used [34]. Other parameters that have been demonstrated to be effective in hot-spot analysis include signal enhancement ratio obtained at a fixed time point 60–90 seconds postinjection [49] and the pharmacokinetic parameter Ktrans [50].
Inspired by the commonly seen “rim enhancement” pattern in solid tumors, some researchers also segment a tumor into a poorly enhancing “core” and a strongly enhancing “periphery” or “rim” [40, 51]. Benjaminsen et al. developed this rough segmentation methodology into a quantitative approach to describe radial heterogeneity in tumor perfusion [52]. In this method, a tumor parametric map is divided into several concentric bands with approximately the same width. Median or mean parameter values are calculated for each band and plotted against band number or radius. The Rofstad group in Norway used this method to study the radial distribution of tumor perfusion and EES fraction in a series of preclinical studies with human melanoma xenografts A-07 and R-18 between 2004 and 2008 [52–57]. At about the same time, Jia et al. applied a similar method to the model-free parameter washout slope in colorectal cancer liver metastasis, and gave this approach an intuitive name “onion-peeling” [32]. A major advantage of this method is that the radial distribution curve can be normalized and compared between parameters obtained with different assessment techniques. Therefore, it can be used as a validation approach. Graff et al. used this method to compare the radial distributions of the DCE-MRI pharmacokinetic parameter EF and tumor blood supply measured with a freely diffusible radioactive tracer Na99mTcO4. The close match of radial distribution curves partially validated the authors' hypothesis that EF measures tumor perfusion in these preclinical models [55]. Their results also showed that A-07 and R-18 xenografts are characterized by different radial distributions of EF [55] and λ (a DCE-MRI pharmacokinetic parameter proportional to ve, [53]), suggesting a potential application in tumor characterization.
It should be noted that the success of the “onion-peeling” method in Rofstad's studies is partly due to their relatively simple preclinical model. Growth of in vivo tumors may be restricted or slowed down on certain directions by the existing anatomical structures, leading to an irregular or diffused tumor shape in which a “center” can hardly be defined. Nonradial heterogeneity patterns are also frequently seen in many human tumors (Figure 3). Therefore, this method is probably working best in small scale preclinical studies with well-controlled tumor growth conditions. In large scale clinical studies, other quantification methods that are less specific about the heterogeneity pattern, such as the texture analysis methods, might be more appropriate.
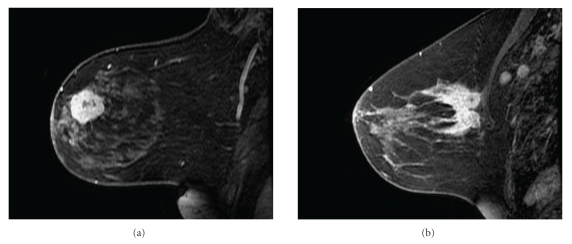
Figure 3.
While the “onion-peeling” method has been demonstrated to be useful in animal xenografts, the specific assumption of radial heterogeneity may not apply to all in vivo human tumors. It can only be used on lesions with regular shape and a radial enhancement pattern (a). The lesion in (b) has an irregular, speculated shape that makes it difficult to define its center. Its enhancement pattern is also more stochastic, without a clear difference between the periphery and the tumor core. Applying the “onion-peeling” method to a tumor like this will not generate much useful information about the underlying tumor biology.
3.2. Histogram Analysis
Intratumoral heterogeneity is also reflected by the shapes of parameter distribution histograms. Tumors are typically characterized by a skewed distribution with a long tail towards strong enhancement or high Ktrans values. Multiple peaks may be observed if the tumor contains a significant fraction of poorly perfused hypoxic mass. Disease progression is usually accompanied by an expansion of the histogram to the right, with the peak broadening its width but decreasing its height. A successful treatment, on the contrary, “suppresses” the histogram leftwards, making the peak narrower and more normal [30, 42]. Functional principal component analysis also revealed that antiangiogenic treatment with ZD6474 causes the disappearance of a second peak with high Ktrans values in mice bearing PC-3 human prostate adenocarcinoma xenografts [58]. A number of studies have demonstrated that incorporating summary statistics describing histogram width or asymmetry leads to substantial improvement in disease diagnosis and response assessment. In primary breast tumors, both the sensitivity and specificity of malignant/benign lesion differentiation can be improved by adding the Ktrans standard deviation information to the conventional analysis approach using Ktrans mean alone [59]. In another study, median Ktrans was demonstrated to be less sensitive than the range of the Ktrans histogram in predicting treatment response in breast cancer patients receiving systemic chemotherapy [42]. Similar results were also reported in cervical cancer patients receiving radiation therapy, where Mayr et al. found that the 10th percentile of RSI is a better predictor for tumor recurrence than the mean or median values [60].
So far, there is no consensus on the optimal method for histogram analysis. Some researchers perform a bin-to-bin comparison between two normalized histograms [61]. Although the rationale for this approach is straightforward to understand, it is in general an inefficient technique: a certain number of bins are needed for the histogram to accurately reflect the shape of the underlying distribution. In clinical research, this number is typically between 10 and 30. When all these bins are simultaneously compared, the need for multicomparison correction makes the whole test very insensitive. For example, if an overall significance level of 0.05 is assigned to a comparison between two normalized histograms with 20 bins each, the individual significance level for every single bin will be only 0.05/20 = 0.0025 with the basic Bonferroni correction [62]. With such a low individual significance level, subtle changes in the histogram are very likely to be masked.
A better strategy for histogram analysis is to summarize only the most relevant information into a few descriptive quantities. Almost all summary statistics have been used in the literature to describe the distribution histogram. A partial list includes the range [42], mean, median, standard deviation [59], skewness [9], kurtosis [63], entropy [63], and various percentiles [30, 41, 43, 60, 64]. There is no decisive conclusion about which statistics is the best. However, the percentiles are probably more robust than the range and moment statistics on pharmacokinetic parametric maps. An “allowed range” is usually set for each pharmacokinetic parameter by the iterative fitting algorithm to accelerate convergence. If the true range of the actual distribution is wider than this “allowed range”, the long tails outside the allowed range will be truncated and replaced by artificial “peaks” on the maximum or minimum allowed values. Unlike the range and moment statistics, the percentiles are insensitive to this artificial distortion of histogram shape because they are not affected by changes in a few extreme values.
3.3. Texture Analysis
The histogram analysis only looks at the distribution of individual voxels within a tumor ROI. The spatial correlation between voxels is ignored. Theoretically, a tumor with an enhancing rim could have an identical histogram with a tumor having many enhancing foci scattered over the entire mass. Spatial distribution information is needed to better differentiate lesions with similar histogram but distinctive enhancement patterns. A number of studies have been conducted along this direction using image texture analysis techniques [31, 65–70].
In DCE-MRI data analysis, the most frequently used texture analysis technique is the cooccurrence matrix method first described by Haralick et al. [71]. Briefly speaking, the cooccurrence matrix element Pd,θ(i, j) measures the probability of starting from any voxel with value i in an image, moving d voxels along direction θ, and arriving at another voxel with value j. Therefore, the cooccurrence matrix is a two dimensional histogram describing a joint distribution of all possible moves with step size d and direction θ on the image. Textural features, such as contrast and correlation, can be exacted from this 2D histogram. They are then averaged over four directions (θ = 0°, 45°, 90°, and 135°) to remove angular dependency. In medical imaging studies, the step size d is usually taken as 1 voxel. The voxel values are usually discretized to reduce cooccurrence matrix dimension. Sinha et al. used this technique to obtain eight textural features that made a reasonably good discrimination (specificity 70%, sensitivity 75%) between 20 malignant and 23 benign breast lesions [65]. Chen et al. extended Haralick et al. 2D technique to 3D, and achieved a significant improvement in classification performance using features obtained from a volumetric texture analysis in a cohort of 77 malignant and 44 benign lesions [66]. Positive results were also reported with various model-free [31] and pharmacokinetic parametric maps [67] as well as on four dimensional dynamic series [68]. Nie et al. further associated the textural features (gray level entropy and sum average) with the visual descriptors in BI-RADS lexicon (enhancement intensity and internal enhancement patterns), thus giving the highly mathematical approach an intuitive interpretation [69].
Currently, the major problem associated with DCE-MRI texture analysis is the high dimensionality of the feature space. A more detailed discussion of this issue will be given in the next section. Researchers are encouraged to adopt a concise study design and dimensionality reduction techniques to improve the reliability of their conclusions.
3.4. Spatial-Parametric Hypervolume Geometry
An interesting approach of heterogeneity quantification was described in the exploratory works of Rose et al. [35, 36]. In their approach, DCE-MRI pharmacokinetic parameters were not treated in the conventional way as voxel values in a 3D imaging volume. Instead, they were interpreted as the height on a fourth dimension in an “extruded” 4D binary hypervolume. Geometric features like (hyper-)surface area, (hyper-)volume, (hyper-)surface to (hyper-)volume ratio, and box-count fractal dimension can be calculated for this spatial-parametric hypervolume, and used as quantitative descriptors of tumor vascular heterogeneity [35]. In a recent study, the Rényi family of fractal dimensions was also investigated [36]. Although the small sample sizes prevented any decisive conclusion about its value to be made at this moment, initial results in glioma grading and liver metastasis treatment response seem to be promising.
This method is conceptually interesting in that spatial and parametric information are treated equally as dimensions in the 4D hypervolume. Geometric features of the hypervolume thereby describe variations in both spatial and parametric domains by nature. BI-RADS lexicon suggests that both morphologic and heterogeneity information are critical in diagnosis of breast cancer. This is perhaps true for all solid tumors. Therefore, geometric features of the spatial-parametric hypervolume compose a pool of potential biomarkers that can simultaneously quantify tumor morphology and heterogeneity. Such a biomarker, if identified, may have important uses in tumor diagnosis and response prediction because it can improve the robustness of the classifier/predictor through reduction of input space dimensionality.
4. Technological and Methodological Considerations
4.1. Avoid Data Sparsity in Study Design
Many descriptive and textural features of imaging data can be extracted from the analysis of tumor heterogeneity. Some researchers tend to use a large number of features in their studies. In an extreme example we found more than 600 features were investigated in only 23 patients [70]. While the researchers' desire of getting the maximum yield from their data is understandable, this practice is fundamentally problematic because as the number of features increases, the problem of data sparsity arises due to Bellman's “curse of dimensionality” [72]. A sparsely-distributed sample set cannot provide adequate information to fully represent the complex behaviors in a large population. Consequently, any prediction made from such a sample set is highly unstable and thus practically useless [73]. This problem is probably responsible for our finding that some complex discriminators are claimed to have (artificially) excellent performance in a relatively small patient population but never develop into real clinical application.
There is no real remedy for the sparse data problem except for thorough consideration and careful planning in the designing stage of a study. The number of features used for classification or prediction must be strictly controlled from the very beginning. A rule of thumb is to keep the ratio between the number of patients in each class and the number of features above 10 [74], that is, for a typical study of about 100 patients divided into two groups, the number of features that can be investigated should not exceed five. This stringent restriction requires the investigators to include only the most promising features into their analysis rather than use all the parameters they can get unselectively. Of course, information about the other less promising features is also interesting and may lead to important discoveries. We recommend to report those as exploratory data. No effort should be made to get any statistical inference from these data. In this way, these data can be preserved without harming the statistical power of the whole study. They may serve as part of the raw data set for future metaanalyses which may provide statistically meaningful information that can never be derived from any single study.
Some researchers chose to classify individual voxels instead of the whole tumor or subject [67, 68, 75]. While this method does help to greatly reduce the stringent restriction on feature set size, caution is needed in the interpretation of the results. Simply stating that a voxel has a “responder-like” curve or “benign” parameters is superficial and sometimes misleading. It is more preferable to characterize voxels with imaging features that have well-recognized association with tumor biology on cellular or tissue levels. Correlation with other established biomarkers is also important when such information is available.
Dimensionality reduction techniques are also powerful tools that may help the investigators to fight the “curse of dimensionality” [74]. It should be noted that dimensionality reduction techniques are only supplements to, but not replacement for, careful study design and successful dimensionality control. They should only be applied to a small feature set carefully selected according to the rule of thumb mentioned above. Applying a dimensionality reduction technique to a poorly designed study would not add any strength to its conclusions.
4.2. Need for Standardization in DCE-MRI Data Acquisition and Analysis
Currently there is no “standard” way of performing DCE-MRI. A number of methodologies have been developed by different groups. Some of them use a T1-weighted fast low-angle shot (FLASH) sequence [76], while the others use saturation recovery or inversion recovery techniques [77]. The measured signal intensities may be converted to gadolinium concentration through the use of a calibration phantom [78], calculation of relative signal enhancement [79], or precontrast T1 [80] or proton density measurements [81]. Data analysis methods are about equally variable, too. Many model-free and pharmacokinetic parameters have been described in the literature (Tables 1 and 2). Most model-free parameters do not have a clear definition that is strictly followed within the DCE-MRI community. For example, many investigators reported the model-free parameter area-under-the-curve (AUC) in their studies, but the way they measured it can be quite different: in some studies it was measured from the relative signal enhancement dynamic curve, while in the others the gadolinium concentration dynamic curve was used. The measurement time can also vary from 30 to 180 seconds postinjection. Even though pharmacokinetic parameters are generally better defined than model-free parameters, considerable variability still exists in the choice of the pharmacokinetic models [22–26] and deconvolution techniques [82, 83].
It is known that many factors in data acquisition and analysis can substantially affect the reliability of results in DCE-MRI studies. The accuracy and precision of the pharmacokinetic parameter estimates, for example, are largely determined by the signal-to-noise ratio (SNR) and temporal resolution of the MR sequence [84]. Systematic errors can be introduced from various sources, such as voluntary or involuntary patient motion, inappropriate use of postprocessing filters, inhomogeneity in B1 excitation field and receiver coil sensitivity, breakdown of linearity in the gadolinium concentration calibration curve [85], and unequal definitions of the same pharmacokinetic parameter (such as Ktrans) in different models [22, 23]. Therefore, the methodological variability described above actually contains a large number of potential error sources. Their complex yet poorly understood interaction makes it extremely difficult to compare DCE-MRI data from different sources. This problem is especially important in the study of tumor vascular heterogeneity. As we have mentioned in the discussions above, an individual study can only investigate a very small fraction of all parameters that characterize intratumoral heterogeneity. The only way to accelerate the investigation process is through meta-analysis that combines results from different studies. Obviously, this can only be achieved with high quality data acquired and analyzed following an optimized standard procedure.
A standardization effort is currently underway driven by the DCE-MRI subcommittee of the Radiological Society of North American (RSNA) Quantitative Imaging Biomarkers Alliance (QIBA) initiative [86]. The most recent progress of their collaborative effort is a comprehensive technical profile that specifies basic standards in DCE-MRI data acquisition, analysis, and quality control on 1.5 Tesla systems. It is recommended that DCE-MRI should be performed on a clinical scanner qualified with a standard phantom, using a 3D FLASH or equivalent sequence without any magnetization preparation or postprocessing filter. The image intensity should be converted to gadolinium concentration through the use of a variable flip angle precontrast T1 map [87] after necessary corrections for patient motion and inhomogeneity in B1 field or coil sensitivity. The pharmacokinetic parameter Ktrans (estimated with the standard Tofts' model) and model-free parameter IAUGC (AUC measured from the gadolinium concentration curve up to 90 seconds postinjection) are recommended as standard quantitative endpoints that should be employed in clinical trials and practice. Guidelines on the minimally acceptable and ideal ranges of various scan parameters (like temporal resolution and slice thickness) are also provided. Investigators in this area are encouraged to follow the progress of the QIBA initiative through journal publications and online updates (http://qibawiki.rsna.org/index.php?title=DCE-MRI_subctte), and adopt the most up-to-date consensus to their own studies to ensure their data have the highest quality standard and the maximum level of comparability.
An important issue that is not addressed in the present version of QIBA DCE-MRI profile is the lack of quality control in the vascular parameter fitting stage. Many researchers only report the “best fit” estimates for pharmacokinetic parameters and ignore the inherent uncertainty in those estimates. This practice leads to a substantial underestimation of the variability in pharmacokinetic parameters. It also raises a philosophical question whether it is appropriate to treat reliable and unreliable parameter estimates equally in the assessment of tumor heterogeneity. Hence, we suggest that DCE-MRI researchers should always report the asymptotic confidence intervals with the “best fit” estimates for pharmacokinetic parameters, and adopt an exclusion criterion or weighting scheme that controls the influence of unreliable estimates from noisy data. We also expect that as the awareness of this issue is raised, a consensus will be developed through the collective effort of the whole DCE-MRI community, and will be included in future standards.
4.3. Validation
It is difficult to validate DCE-MRI parameters because they measure a mixed effect of blood flow, microvascular permeability, and EES volume. A number of studies have demonstrated broad correlations between various DCE-MRI parameters and tissue perfusion [88], microvascular density (MVD) [45, 89], or angiogenic factor expression [90]. However, controversial results are also reported in the literature [91–93]. One reason for this controversy might be tumor heterogeneity at the subvoxel level. In a human cervical carcinoma xenograft study, matched DCE-MRI images and histological sections showed substantial intravoxel heterogeneity in morphology and tissue composition, even though the voxel size (0.23 × 0.47 × 2.0 mm3) is already ten times smaller than those used in human studies [94]. Therefore, DCE-MRI parameters may be better validated against immunohistochemical measurements averaged over several fields randomly picked from the region of interest.
Beside the ROI-averaged parameter values, the distribution of DCE-MRI parameters also needs to be validated in studies of tumor vascular heterogeneity. This task is relatively simple if the “gold standard” is another imaging modality with similar spatial resolution, for which direct comparison can be made between summary statistics [95], histogram shapes, or certain heterogeneity patterns [52, 55]. MVD values or angiogenic factor expression levels need to be compared with DCE-MRI parameters within multiple subregional ROIs or along a line profile. Special techniques, like MRI-guided biopsy [96], may be used to help reduce sampling error caused by mismatch between DCE-MRI images and histological slides.
5. Conclusion
Experimental evidence shows that vascular heterogeneity plays an important role in tumor biology. Several methods have been developed to quantify tumor vascular heterogeneity with DCE-MRI. Initial studies in patients and animal models have shown promising results in disease diagnosis, tumor grading, and response assessment. These results give us additional understanding of tumor biology and justify the necessity of continuous development along this direction. However, the quality of a DCE-MRI heterogeneity study can easily be undermined by various methodological and technological issues, such as poor study design or lack of standardization. This paper may serve as a guideline to help investigators understand and also avoid these possible pitfalls in study design, data acquisition, and analysis in future studies of tumor vascular heterogeneity.
Acknowledgments
The authors are grateful to Dr. Guang Jia for his help in creating Figure 2. This work was supported by grant ODOD AGMT TECH 03-051 and the Wright Center of Innovation in Biomedical Imaging.
References
- 1.Jain RK, Duda DG, Willett CG, et al. Biomarkers of response and resistance to antiangiogenic therapy. Nature Reviews Clinical Oncology. 2009;6(6):327–338. doi: 10.1038/nrclinonc.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rehman S, Jayson GC. Molecular imaging of antiangiogenic agents. Oncologist. 2005;10(2):92–103. doi: 10.1634/theoncologist.10-2-92. [DOI] [PubMed] [Google Scholar]
- 3.Taylor JS, Tofts PS, Port R, et al. MR imaging of tumor microcirculation: promise for the new millenium. Journal of Magnetic Resonance Imaging. 1999;10(6):903–907. doi: 10.1002/(sici)1522-2586(199912)10:6<903::aid-jmri1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 4.de Langen AJ, van den Boogaart VEM, Marcus JT, Lubberink M. Use of H215O-PET and DCE-MRI to measure tumor blood flow. Oncologist. 2008;13(6):631–644. doi: 10.1634/theoncologist.2007-0235. [DOI] [PubMed] [Google Scholar]
- 5.Cao M, Liang Y, Shen C, Miller KD, Stantz KM. Developing DCE-CT to quantify intra-tumor heterogeneity in breast tumors with differing angiogenic phenotype. IEEE Transactions on Medical Imaging. 2009;28(6):861–871. doi: 10.1109/TMI.2008.2012035. [DOI] [PubMed] [Google Scholar]
- 6.Bushberg JT. The Essential Physics of Medical Imaging. 2nd edition. Philadelphia, Pa, USA: Lippincott Williams & Wilkins; 2002. [Google Scholar]
- 7.Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307(5706):58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 8.O’Connor JPB, Jackson A, Parker GJM, Jayson GC. DCE-MRI biomarkers in the clinical evaluation of antiangiogenic and vascular disrupting agents. British Journal of Cancer. 2007;96(2):189–195. doi: 10.1038/sj.bjc.6603515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson A, O’Connor JPB, Parker GJM, Jayson GC. Imaging tumor vascular heterogeneity and angiogenesis using dynamic contrast-enhanced magnetic resonance imaging. Clinical Cancer Research. 2007;13(12):3449–3459. doi: 10.1158/1078-0432.CCR-07-0238. [DOI] [PubMed] [Google Scholar]
- 10.Morgan B, Thomas AL, Drevs J, et al. Dynamic contrast-enhanced magnetic resonance imaging as a biomarker for the pharmacological response of PTK787/ZK 222584, an inhibitor of the vascular endothelial growth factor receptor tyrosine kinases, in patients with advanced colorectal cancer and liver metastases: results from two phase I studies. Journal of Clinical Oncology. 2003;21(21):3955–3964. doi: 10.1200/JCO.2003.08.092. [DOI] [PubMed] [Google Scholar]
- 11.Mross K, Drevs J, Müller M, et al. Phase I clinical and pharmacokinetic study of PTK/ZK, a multiple VEGF receptor inhibitor, in patients with liver metastases from solid tumours. European Journal of Cancer. 2005;41(9):1291–1299. doi: 10.1016/j.ejca.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis—Correlation in invasive breast carcinoma. The New England Journal of Medicine. 1991;324(1):1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 13.Schor AM, Pazouki S, Morris J, Smither RL, Chandrachud LM, Pendleton N. Heterogeneity in microvascular density in lung tumours: comparison with normal bronchus. British Journal of Cancer. 1998;77(6):946–951. doi: 10.1038/bjc.1998.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyss MT, Hofer S, Hefti M, et al. Spatial heterogeneity of low-grade gliomas at the capillary level: a PET study on tumor blood flow and amino acid uptake. Journal of Nuclear Medicine. 2007;48(7):1047–1052. doi: 10.2967/jnumed.106.038489. [DOI] [PubMed] [Google Scholar]
- 15.Heppner GH. Tumor heterogeneity. Cancer Research. 1984;44(6):2259–2265. [PubMed] [Google Scholar]
- 16.Shipitsin M, Campbell LL, Argani P, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal G, Su MY, Nalcioglu O, Feig SA, Chen JH. Significance of breast lesion descriptors in the ACR BI-RADS MRI lexicon. Cancer. 2009;115(7):1363–1380. doi: 10.1002/cncr.24156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuthapisith S, Eremin J, El-Sheemey M, Eremin O. Breast cancer chemoresistance: emerging importance of cancer stem cells. Surgical Oncology. 2010;19(1):27–32. doi: 10.1016/j.suronc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nature Reviews Cancer. 2007;7(10):733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 21.Sneddon JB, Werb Z. Location, location, location: the cancer stem cell niche. Cell Stem Cell. 2007;1(6):607–611. doi: 10.1016/j.stem.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsson HB, Stubgaard M, Søndergaard L, Henriksen O. In vivo quantification of the unidirectional influx constant for Gd-DTPA diffusion across the myocardial capillaries with MR imaging. Journal of Magnetic Resonance Imaging. 1994;4(3):433–440. doi: 10.1002/jmri.1880040332. [DOI] [PubMed] [Google Scholar]
- 23.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. Journal of Magnetic Resonance Imaging. 1997;7(1):91–101. doi: 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 24.Brix G, Semmler W, Port R, Schad LR, Layer G, Lorenz WJ. Pharmacokinetic parameters in CNS Gd-DTPA enhanced MR imaging. Journal of Computer Assisted Tomography. 1991;15(4):621–628. doi: 10.1097/00004728-199107000-00018. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Liang J, Heverhagen JT, et al. Improving the pharmacokinetic parameter measurement in dynamic contrast-enhanced MRI by use of the arterial input function: theory and clinical application. Magnetic Resonance in Medicine. 2008;59(6):1448–1456. doi: 10.1002/mrm.21608. [DOI] [PubMed] [Google Scholar]
- 26.Yankeelov TE, Luci JJ, Lepage M, et al. Quantitative pharmacokinetic analysis of DCE-MRI data without an arterial input function: a reference region model. Magnetic Resonance Imaging. 2005;23(4):519–529. doi: 10.1016/j.mri.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein EJ, Burnett KR, Hansell JR, et al. Gadolinium DTPA (an NMR proton imaging contrast agent): chemical structure, paramagnetic properties and pharmacokinetics. Physiological Chemistry and Physics and Medical NMR. 1984;16(2):97–104. [PubMed] [Google Scholar]
- 28.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T1-weighted MRI of a diffusable tracer: standardized quantities and symbols. Journal of Magnetic Resonance Imaging. 1999;10(3):223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 29.Evelhoch JL. Key factors in the acquisition of contrast kinetic data for oncology. Journal of Magnetic Resonance Imaging. 1999;10(3):254–259. doi: 10.1002/(sici)1522-2586(199909)10:3<254::aid-jmri5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 30.Johansen R, Jensen LR, Rydland J, et al. Predicting survival and early clinical response to primary chemotherapy for patients with locally advanced breast cancer using DCE-MRI. Journal of Magnetic Resonance Imaging. 2009;29(6):1300–1307. doi: 10.1002/jmri.21778. [DOI] [PubMed] [Google Scholar]
- 31.Karahaliou A, Vassiou K, Arikidis NS, Skiadopoulos S, Kanavou T, Costaridou L. Assessing heterogeneity of lesion enhancement kinetics in dynamic contrast-enhanced MRI for breast cancer diagnosis. British Journal of Radiology. 2010;83(988):296–306. doi: 10.1259/bjr/50743919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia G, O’Dell C, Heverhagen JT, et al. Colorectal liver metastases: contrast agent diffusion coefficient for quantification of contrast enhancement heterogeneity at MR imaging. Radiology. 2008;248(3):901–909. doi: 10.1148/radiol.2491071936. [DOI] [PubMed] [Google Scholar]
- 33.Mussurakis S, Gibbs P, Horsman A. Primary breast abnormalities: selective pixel sampling on dynamic gadolinium enhanced MR images. Radiology. 1998;206(2):465–473. doi: 10.1148/radiology.206.2.9457201. [DOI] [PubMed] [Google Scholar]
- 34.Liney GP, Gibbs P, Hayes C, Leach MO, Turnbull LW. Dynamic contrast-enhanced MRI in the differentiation of breast tumors: user-defined versus semi-automated region-of-interest analysis. Journal of Magnetic Resonance Imaging. 1999;10(6):945–949. doi: 10.1002/(sici)1522-2586(199912)10:6<945::aid-jmri6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 35.Rose CJ, Mills S, O’Connor JP, et al. Quantifying heterogeneity in dynamic contrast-enhanced MRI parameter maps. Medical Image Computing and Computer-Assisted Intervention. 2007;10(2):376–384. doi: 10.1007/978-3-540-75759-7_46. [DOI] [PubMed] [Google Scholar]
- 36.Rose CJ, Mills SJ, O’Connor JPB, et al. Quantifying spatial heterogeneity in dynamic contrast-enhanced MRI parameter maps. Magnetic Resonance in Medicine. 2009;62(2):488–499. doi: 10.1002/mrm.22003. [DOI] [PubMed] [Google Scholar]
- 37.Simpson-Herren L, Noker PE, Wagoner SD. Variability of tumor response to chemotherapy II. Contribution of tumor heterogeneity. Cancer Chemotherapy and Pharmacology. 1988;22(2):131–136. doi: 10.1007/BF00257310. [DOI] [PubMed] [Google Scholar]
- 38.Giesel FL, Bischoff H, von Tengg-Kobligk H, et al. Dynamic contrast-enhanced mri of malignant pleural mesothelioma: a feasibility study of noninvasive assessment, therapeutic follow-up, and possible predictor of improved outcome. Chest. 2006;129(6):1570–1576. doi: 10.1378/chest.129.6.1570. [DOI] [PubMed] [Google Scholar]
- 39.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6(19):2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 40.Checkley D, Tessier JJ, Kendrew J, Waterton JC, Wedge SR. Use of dynamic contrast-enhanced MRI to evaluate acute treatment with ZD6474, a VEGF signalling inhibitor, in PC-3 prostate tumours. British Journal of Cancer. 2003;89(10):1889–1895. doi: 10.1038/sj.bjc.6601386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Lussanet QG, Backes WH, Griffioen AW, et al. Dynamic contrast-enhanced magnetic resonance imaging of radiation therapy-induced microcirculation changes in rectal cancer. International Journal of Radiation Oncology Biology Physics. 2005;63(5):1309–1315. doi: 10.1016/j.ijrobp.2005.04.052. [DOI] [PubMed] [Google Scholar]
- 42.Padhani AR, Hayes C, Assersohn L, et al. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced mr imaging: initial clinical results. Radiology. 2006;239(2):361–374. doi: 10.1148/radiol.2392021099. [DOI] [PubMed] [Google Scholar]
- 43.Yuh WTC, Mayr NA, Jarjoura D, et al. Predicting control of primary tumor and survival by DCE MRI during early therapy in cervical cancer. Investigative Radiology. 2009;44(6):343–350. doi: 10.1097/RLI.0b013e3181a64ce9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mussurakis S, Buckley DL, Coady AM, Turnbull LW, Horsman A. Observer variability in the interpretation of contrast enhanced MRI of the breast. British Journal of Radiology. 1996;69(827):1009–1016. doi: 10.1259/0007-1285-69-827-1009. [DOI] [PubMed] [Google Scholar]
- 45.Giesel FL, Choyke PL, Mehndiratta A, et al. Pharmacokinetic analysis of malignant pleural mesothelioma-initial results of tumor microcirculation and its correlation to microvessel density (CD-34) Academic Radiology. 2008;15(5):563–570. doi: 10.1016/j.acra.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Borland D, Taylor RM. Rainbow color map (still) considered harmful. IEEE Computer Graphics and Applications. 2007;27(2):14–17. doi: 10.1109/mcg.2007.323435. [DOI] [PubMed] [Google Scholar]
- 47.Mussurakis S, Buckley DL, Horsman A. Dynamic MRI of invasive breast cancer: assessment of three region-of-interest analysis methods. Journal of Computer Assisted Tomography. 1997;21(3):431–438. doi: 10.1097/00004728-199705000-00017. [DOI] [PubMed] [Google Scholar]
- 48.Turnbull LW. Dynamic contrast-enhanced MRI in the diagnosis and management of breast cancer. NMR in Biomedicine. 2009;22(1):28–39. doi: 10.1002/nbm.1273. [DOI] [PubMed] [Google Scholar]
- 49.Gribbestad IS, Nilsen G, Fjøsne HE, Kvinnsland S, Haugen OA, Rinck PA. Comparative signal intensity measurements in dynamic gadolinium-enhanced MR mammography. Journal of Magnetic Resonance Imaging. 1994;4(3):477–480. doi: 10.1002/jmri.1880040339. [DOI] [PubMed] [Google Scholar]
- 50.Hayes C, Padhani AR, Leach MO. Assessing changes in tumour vascular function using dynamic contrast-enhanced magnetic resonance imaging. NMR in Biomedicine. 2002;15(2):154–163. doi: 10.1002/nbm.756. [DOI] [PubMed] [Google Scholar]
- 51.Jiang L, Zhao D, Constantinescu A, Mason RP. Comparison of BOLD contrast and Gd-DTPA dynamic contrast-enhanced imaging in rat prostate tumor. Magnetic Resonance in Medicine. 2004;51(5):953–960. doi: 10.1002/mrm.20069. [DOI] [PubMed] [Google Scholar]
- 52.Benjaminsen IC, Graff BA, Brurberg KG, Rofstad EK. Assessment of tumor blood perfusion by high-resolution dynamic contrast-enhance MRI: a preclinical study of human melanoma xenografts. Magnetic Resonance in Medicine. 2004;52(2):269–276. doi: 10.1002/mrm.20149. [DOI] [PubMed] [Google Scholar]
- 53.Benjaminsen IC, Brurberg KG, Ruud EBM, Rofstad EK. Assessment of extravascular extracellular space fraction in human melanoma xenografts by DCE-MRI and kinetic modeling. Magnetic Resonance Imaging. 2008;26(2):160–170. doi: 10.1016/j.mri.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 54.Graff BA, Benjaminsen IC, Melås EA, Brurberg KG, Rofstad EK. Changes in intratumor heterogeneity in blood perfusion in intradermal human melanoma xenografts during tumor growth assessed by DCE-MRI. Magnetic Resonance Imaging. 2005;23(9):961–966. doi: 10.1016/j.mri.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 55.Graff BA, Benjaminsen IC, Brurberg KG, Ruud EBM, Rofstad EK. Comparison of tumor blood by perfusion assessed by dynamic contrast-enhanced MRI with tumor blood supply assessed by invasive imaging. Journal of Magnetic Resonance Imaging. 2005;21(3):272–281. doi: 10.1002/jmri.20265. [DOI] [PubMed] [Google Scholar]
- 56.Gaustad JV, Benjaminsen IC, Graff BA, Brurberg KG, Ruud EBM, Rofstad EK. Intratumor heterogeneity in blood perfusion in orthotopic human melanoma xenografts assessed by dynamic contrast-enhanced magnetic resonance imaging. Journal of Magnetic Resonance Imaging. 2005;21(6):792–800. doi: 10.1002/jmri.20321. [DOI] [PubMed] [Google Scholar]
- 57.Brurberg KG, Benjaminsen IC, Dørum LMR, Rofstad EK. Fluctuations in tumor blood perfusion assessed by dynamic contrast-enhanced MRI. Magnetic Resonance in Medicine. 2007;58(3):473–481. doi: 10.1002/mrm.21367. [DOI] [PubMed] [Google Scholar]
- 58.O’Connor E, Fieller N, Holmes A, Waterton J. How to analyse dynamic MRI in oncology: advanced histogram analysis gives better statistical power & insight than simple averaging. In: Proceedings of the 13th Scientific Meeting & Exhibition (ISMRM '05); May 2005; Miami, Fla, USA. [Google Scholar]
- 59.Issa B, Buckley DL, Turnbull LW. Heterogeneity analysis of Gd-DTPA uptake: improvement in breast lesion differentiation. Journal of Computer Assisted Tomography. 1999;23(4):615–621. doi: 10.1097/00004728-199907000-00024. [DOI] [PubMed] [Google Scholar]
- 60.Mayr NA, Yuh WTC, Arnholt JC, et al. Pixel analysis of MR perfusion imaging in predicting radiation therapy outcome in cervical cancer. Journal of Magnetic Resonance Imaging. 2000;12(6):1027–1033. doi: 10.1002/1522-2586(200012)12:6<1027::aid-jmri31>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 61.Watson Y, Cheung S, Roberts C, et al. Prognostic power of DCE-MRI heterogeneity analysis in patient with advanced solid tumours. In: Proceedings of the 14th Scientific Meeting & Exhibition (ISMRM '06); August 2006; Seattle, Wash, USA. [Google Scholar]
- 62.Armitage P, Berry G. Statistical Methods in Medical Research. 3rd edition. Cambridge, UK: Blackwell Science; 1994. [Google Scholar]
- 63.Ertaş G, Gülçür HÖ, Tunaci M. Improved lesion detection in MR mammography: three-dimensional segmentation, moving voxel sampling, and normalized maximum intensity-time ratio entropy. Academic Radiology. 2007;14(2):151–161. doi: 10.1016/j.acra.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 64.Mayr NA, Wang JZ, Zhang D, et al. Synergistic effects of hemoglobin and tumor perfusion on tumor control and survival in cervical cancer. International Journal of Radiation Oncology Biology Physics. 2009;74(5):1513–1521. doi: 10.1016/j.ijrobp.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 65.Sinha S, Lucas-Quesada FA, DeBruhl ND, et al. Multifeature analysis of Gd-enhanced MR images of breast lesions. Journal of Magnetic Resonance Imaging. 1997;7(6):1016–1026. doi: 10.1002/jmri.1880070613. [DOI] [PubMed] [Google Scholar]
- 66.Chen W, Giger ML, Li H, Bick U, Newstead GM. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magnetic Resonance in Medicine. 2007;58(3):562–571. doi: 10.1002/mrm.21347. [DOI] [PubMed] [Google Scholar]
- 67.Kale MC, Clymer BD, Koch RM, et al. Multispectral co-occurrence with three random variables in dynamic contrast enhanced magnetic resonance imaging of breast cancer. IEEE Transactions on Medical Imaging. 2008;27(10):1425–1431. doi: 10.1109/TMI.2008.922181. [DOI] [PubMed] [Google Scholar]
- 68.Woods BJ, Clymer BD, Kurc T, et al. Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. Journal of Magnetic Resonance Imaging. 2007;25(3):495–501. doi: 10.1002/jmri.20837. [DOI] [PubMed] [Google Scholar]
- 69.Nie K, Chen JH, Yu HJ, Chu Y, Nalcioglu O, Su MY. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Academic Radiology. 2008;15(12):1513–1525. doi: 10.1016/j.acra.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prescott JW, Zhang D, Wang JZ, et al. Temporal analysis of tumor heterogeneity and volume for cervical cancer treatment outcome prediction: preliminary evaluation. Journal of Digital Imaging. 2010;23(3):342–357. doi: 10.1007/s10278-009-9179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Transactions on Systems, Man and Cybernetics. 1973;3(6):610–621. [Google Scholar]
- 72.Bellman R. Dynamic Programming. Princeton, NJ, USA: Princeton University Press; 1957. [Google Scholar]
- 73.Hastie T, Tibshirani R, Friedman JH. The Elements of Statistical Learning Data Mining, Inference, and Prediction. New York, NY, USA: Springer; 2001. [Google Scholar]
- 74.Jain AK, Duin RPW, Mao J. Statistical pattern recognition: a review. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2000;22(1):4–37. [Google Scholar]
- 75.Lee SH, Kim JH, Kim KG, Park JS, Park SJ, Moon WK. Optimal clustering of kinetic patterns on malignant breast lesions: comparison between K-means clustering and three-time-points method in dynamic contrast-enhanced MRI. In: Proceedings of the 29th Annual International Conference of IEEE Engineering in Medicine and Biology Society (EMBS ’07); August 2007; Lyon, France. pp. 2089–2093. [DOI] [PubMed] [Google Scholar]
- 76.Galbraith SM, Lodge MA, Taylor NJ, et al. Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis. NMR in Biomedicine. 2002;15(2):132–142. doi: 10.1002/nbm.731. [DOI] [PubMed] [Google Scholar]
- 77.Makkat S, Luypaert R, Sourbron S, Stadnik T, De Mey J. Quantification of perfusion and permeability in breast tumors with a deconvolution-based analysis of second-bolus T1-DCE data. Journal of Magnetic Resonance Imaging. 2007;25(6):1159–1167. doi: 10.1002/jmri.20937. [DOI] [PubMed] [Google Scholar]
- 78.McGrath DM, Bradley DP, Tessier JL, Lacey T, Taylor CJ, Parker GJM. Comparison of model-based arterial input functions for dynamic contrast-enhanced MRI in tumor bearing rats. Magnetic Resonance in Medicine. 2009;61(5):1173–1184. doi: 10.1002/mrm.21959. [DOI] [PubMed] [Google Scholar]
- 79.Yang C, Stadler WM, Karczmar GS, Milosevic M, Yeung I, Haider MA. Comparison of quantitative parameters in cervix cancer measured by dynamic contrast-enhanced MRI and CT. Magnetic Resonance in Medicine. 2010;63(6):1601–1609. doi: 10.1002/mrm.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Priest AN, Gill AB, Kataoka M, et al. Dynamic contrast-enhanced MRI in ovarian cancer: initial experience at 3 tesla in primary and metastatic disease. Magnetic Resonance in Medicine. 2010;63(4):1044–1049. doi: 10.1002/mrm.22291. [DOI] [PubMed] [Google Scholar]
- 81.Meng R, Chang SD, Jones EC, Goldenberg SL, Kozlowski P. Comparison between population average and experimentally measured arterial input function in predicting biopsy results in prostate cancer. Academic Radiology. 2010;17(4):520–525. doi: 10.1016/j.acra.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Makkat S, Luypaert R, Sourbron S, Stadnik T, De Mey J. Assessment of tumor blood flow in breast tumors with T1-dynamic contrast-enhanced MR imaging: impact of dose reduction and the use of a prebolus technique on diagnostic efficacy. Journal of Magnetic Resonance Imaging. 2010;31(3):556–561. doi: 10.1002/jmri.22077. [DOI] [PubMed] [Google Scholar]
- 83.Fluckiger JU, Schabel MC, DiBella EVR. Model-based blind estimation of kinetic parameters in dynamic contrast enhanced (DCE)-MRI. Magnetic Resonance in Medicine. 2009;62(6):1477–1486. doi: 10.1002/mrm.22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henderson E, Rutt BK, Lee TY. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magnetic Resonance Imaging. 1998;16(9):1057–1073. doi: 10.1016/s0730-725x(98)00130-1. [DOI] [PubMed] [Google Scholar]
- 85.Jackson E, Gupta S, Rosen M, et al. QIBA DCE-MRI technical committee update: phantom studies and first DCE-MRI profile. In: Proceedings of the 96th Scientific Assembly and Annual Meeting of the Radiological Society of North America (RSNA '10); December 2010; Chicago, Ill, USA. [Google Scholar]
- 86.Jackson E, Ashton E, Evelhoch JL, et al. Multivendor, multisite DCE-MRI phantom validation study. In: Proceedings of the 95th Scientific Assembly and Annual Meeting of the Radiological Society of North America (RSNA '10); December 2009; Chicago, Ill, USA. [Google Scholar]
- 87.Gupta RK. A new look at the method of variable nutation angle for the measurement of spin-lattice relaxation times using fourier transform NMR. Journal of Magnetic Resonance. 1977;25(1):231–235. [Google Scholar]
- 88.Pärkkä JP, Niemi P, Saraste A, et al. Comparison of MRI and positron emission tomography for measuring myocardial perfusion reserve in healthy humans. Magnetic Resonance in Medicine. 2006;55(4):772–779. doi: 10.1002/mrm.20833. [DOI] [PubMed] [Google Scholar]
- 89.Buckley DL, Drew PJ, Mussurakis S, Monson JRT, Horsman A. Microvessel density in invasive breast cancer assessed by dynamic Gd-DTPA enhanced MRI. Journal of Magnetic Resonance Imaging. 1997;7(3):461–464. doi: 10.1002/jmri.1880070302. [DOI] [PubMed] [Google Scholar]
- 90.Knopp MV, Weiss E, Sinn HP, et al. Pathophysiologic basis of contrast enhancement in breast tumors. Journal of Magnetic Resonance Imaging. 1999;10(3):260–266. doi: 10.1002/(sici)1522-2586(199909)10:3<260::aid-jmri6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 91.Hulka CA, Edmister WB, Smith BL, et al. Dynamic echo-planar imaging of the breast: experience in diagnosing breast carcinoma and correlation with tumor angiogenesis. Radiology. 1997;205(3):837–842. doi: 10.1148/radiology.205.3.9393545. [DOI] [PubMed] [Google Scholar]
- 92.Su MY, Cheung YC, Fruehauf JP, et al. Correlation of dynamic contrast enhancement MRI parameters with microvessel density and VEGF for assessment of angiogenesis in breast cancer. Journal of Magnetic Resonance Imaging. 2003;18(4):467–477. doi: 10.1002/jmri.10380. [DOI] [PubMed] [Google Scholar]
- 93.Costouros NG, Lorang D, Zhang Y, et al. Microarray gene expression analysis of murine tumor heterogeneity defined by dynamic contrast-enhanced MRI. Molecular Imaging. 2002;1(3):301–308. doi: 10.1162/15353500200202124. [DOI] [PubMed] [Google Scholar]
- 94.Ellingsen C, Egeland TAM, Galappathi K, Rofstad EK. Dynamic contrast-enhanced magnetic resonance imaging of human cervical carcinoma xenografts: pharmacokinetic analysis and correlation to tumor histomorphology. Radiotherapy and Oncology. 2010;97(2):217–224. doi: 10.1016/j.radonc.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 95.Kierkels RGJ, Backes WH, Janssen MHM, et al. Comparison between perfusion computed tomography and dynamic contrast-enhanced magnetic resonance imaging in rectal cancer. International Journal of Radiation Oncology Biology Physics. 2010;77(2):400–408. doi: 10.1016/j.ijrobp.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 96.Ménard C, Susil RC, Choyke P, et al. An interventional magnetic resonance imaging technique for the molecular characterization of intraprostatic dynamic contrast enhancement. Molecular Imaging. 2005;4(1):63–66. doi: 10.1162/15353500200504181. [DOI] [PMC free article] [PubMed] [Google Scholar]