Abstract
Supercoiled DNA plasmids were exposed in the frozen state to high energy electrons. Surviving supercoiled molecules were separated from their degradation products (e.g., open circle and linear forms) by agarose gel electrophoresis and subsequently quantified by staining and image analysis. Complex survival curves were analyzed using radiation target theory, yielding the radiation-sensitive mass of each form. One of the irradiated plasmids was transfected into cells, permitting radiation analysis of gene expression. Loss of this function was associated with a mass much smaller than the entire plasmid molecule, indicating a lack of energy transfer in amounts sufficient to cause structural damage along the DNA polynucleotide. The method of radiation target analysis can be applied to study both structure and function of DNA.
Keywords: DNA, plasmid, radiation, target analysis
Interactions of ionizing radiation directly with macromolecules results in breakage of covalent bonds. The damage to protein molecules has been examined in great detail [1]; damage was found throughout polypeptides regardless of the site of the primary ionization. In contrast, studies with RNA have shown that radiation damage does not spread throughout the polynucleotide [2]. These findings elicited questions concerning the nature of radiation damage in DNA.
Among the thousands of studies of DNA, several concerned radiation effects on DNA in cells and in various purified states. Effects of ionizing radiation on DNA have been described in detail [3], but have been concerned mostly with indirect radiation effects. The direct effects of radiation on DNA and analysis by target theory were initially reported before advanced analytical techniques were available and only limited interpretations were possible. Single strand breaks (ssbs) were observed twenty times more frequently than double strand breaks (dsbs). This led to considerable speculation and modeling concerning the location of radiation damages in DNA. This study presents the first direct evidence for the restriction of radiation damage to the site of the primary ionization.
The present studies employed highly-purified DNA in the form of supercoiled plasmids. It is well-known that breakage of a covalent bond in the polymer backbone (a ‘nick’) of either of the two DNA strands causes the supercoils to relax, and the plasmid assumes the open circular form. If a second break occurs in the opposing DNA strand near the location of the first break, the resulting double-strand break will cause the circular form to be converted to a linear form. Although these three forms of DNA have the same mass, they differ markedly in hydrodynamic properties, permitting separation by electrophoresis.
In addition to serving as a highly-purified source of DNA with known degradation products, plasmid DNA molecules used in transfection experiments typically encode at least one eukaryotic gene (“reporter gene”). Upon introduction of the plasmid to cultured cells (with the help of agents to facilitate delivery), expression of an encoded gene can be monitored. In order to assess effects of radiation on biological activity, some experiments employed a plasmid encoding green fluorescent protein (GFP), which permitted application of target theory to probe the relationship between radiation damage and reporter gene expression.
Previous reports of irradiated plasmid DNA have appeared; only a few studies concerned the direct effects of ionizing radiation on DNA. Using monochromatic X-rays on and off the K-alpha absorption edge of phosphorus, Hieda et al. [4] showed that radiation interactions directly with the phosphorus atom were not unusually effective in radiation damage; and Boudaiffa et al. [5] showed that single-, double- and multiple-strand breaks were formed in DNA by electrons with energies typical of secondary electrons. It should be noted that both of these studies, as well as that of Purkayastha et al [6] used very low-energy ionizing radiations, (100-2150 eV). In contrast, this study utilized high-energy electrons (13 MeV) employed under conditions (i.e., −135°C) that minimized indirect radiation effects. In the present study, the radiation target method has not only been applied to structural studies of DNA, but the technique has been extended to functional properties as well.
MATERIALS AND METHODS
A 9.9 kbp plasmid, pMB340, was provided by Gary Koe (Valentis Inc, Burlingame CA). The 5 kbp plasmid encoding GFP (pGreen Lantern-1) was originally obtained from Gibco-BRL (Carlsbad CA), propagated in E. coli and purified by Aldevron Custom Plasmid Purification (Fargo ND).
Plasmids were dissolved at a concentration of 3 mg/mL in sterile 2.5 mM Tris-HCl (pH 8.5). Plasmid samples (20 μL) were frozen on dry ice in 2 mL glass ampoules and sealed with an oxygen-gas flame. Sealed, frozen samples were shipped and held at −80°C before and after irradiation at −135°C [7] with 13 MeV electrons from a linear accelerator at the Armed Forces Radiobiology Research Institute (Bethesda MD) or at the National Institute of Standards and Technology (Gaithersburg MD).
To determine the effects of intercalation, some experiments utilized plasmid DNA that had been incubated prior to irradiation with ethidium bromide (1 ethidum bromide molecule per base pair).
Radiation dosimetry was determined by thermoluminescent dosimeters (TLDs) or by EPR analysis of irradiated alanine. Both methods agreed in cross-checks.
Samples of irradiated plasmids were electrophoresed on 0.9% agarose gels [8] that resolved the supercoiled, open circle and linear forms. Included on each gel with irradiated samples was a series of concentrations (from 100 to 1000 ng) of unirradiated plasmid DNA that was used for calibration. After migration, gels were stained with ethidium bromide and quantified by fluorometric image analysis using a Fluor-S MultiImager and Quantity One software (Biorad, Hercules CA). Ethidium staining of the supercoiled form was adjusted by a factor of 1.4 to account for the diminished binding [9].
Two microgram aliquots of the irradiated GFP plasmid were diluted in water and mixed with 8.4 μL of polyethylenimine (10 mM nitrogen, pH 7) to facilitate delivery to African green monkey kidney cells (COS-7). The resulting DNA-polyethylenimine complexes were diluted in cell culture media and incubated with COS-7 cells as previously described [10]. After 4 h, the media was replaced with fresh media, and the cells were grown at 37°C for approximately 40 h. After harvesting and washing, cells were analyzed for GFP expression using a Coulter Epics XL flow cytometer (Hialeah FL). Fluorescence from 5000 cells was excited at 488 nm and monitored at 525 nm; triplicate transfections were measured for each sample.
Radiation target analysis
Target theory is based on the fact that gamma rays and high-energy electrons interact randomly with atoms along their trajectory, transferring an average of 60 electron volts in each interaction. The absorption of this large amount of energy results in breakage of covalent bonds causing severe structural damage. While most radiation damage is indirect, this can be eliminated by use of dry or frozen samples. Under these conditions, the only intact structures surviving are those that had no interactions with the applied radiation.
Random interactions are described by the Poisson equation. If a single random event affects the observed property, then the probability of escape (i.e,, no interactions) is e−x, where x is dependent on the radiation dose, D, the mass of the molecule, m, and a constant, q [11]. The number of surviving molecules, N = N0 e−qmD. If each molecule can express a function, the surviving activity will be A0 e−qmD.
When irradiation is performed at −135°C and the radiation dose is given in megarads (1 Gy = 100 rads), q = (1761)−1; the mass will be expressed in kDa. The radiation inactivation curve will be a straight line with slope k, and 1761 k = m.
Target theory has also been developed for more complex situations where radiation effects required multiple primary ionizations [12]. In these cases, the inactivation curve displays an initial ‘shoulder’. These were not observed in the present studies.
In the present experiments, frozen samples of supercoiled plasmid DNA were exposed to a wide range of radiation doses. The initial supercoiled forms are of very large mass and disappear rapidly after only small radiation exposures. Thus only a few radiation points are available to establish the inactivation curve, resulting in considerable error in determination of the slope and therefore of the target mass.
After irradiation, the open circle and linear forms of the plasmids show a rapid rise and slower decay. The decay was analyzed as a single exponential involving only some of the experimental points. This restriction leads to greater error in mass determinations than would be found from a single exponential followed over the entire dose range. In order to compare results from repeated experiments, target analyses were performed on data normalized to the total amount of DNA in the original samples of that experiment.
A comparable analysis exists for molecules possessing biological activity, such as the GFP gene in the present study. The appearance of green fluorescent protein in transfected cells decreases as a single exponential with radiation dose. Using the same mathematical relationship, the masses of the structures responsible for that activity were determined.
RESULTS
Two different supercoiled plasmids were studied: a ’10 kilobase’ plasmid of ~6.5 MDa, and a ~3.3 MDa plasmid containing the GFP gene whose biological activity was measured in transfection experiments.
’10 kbase plasmid’
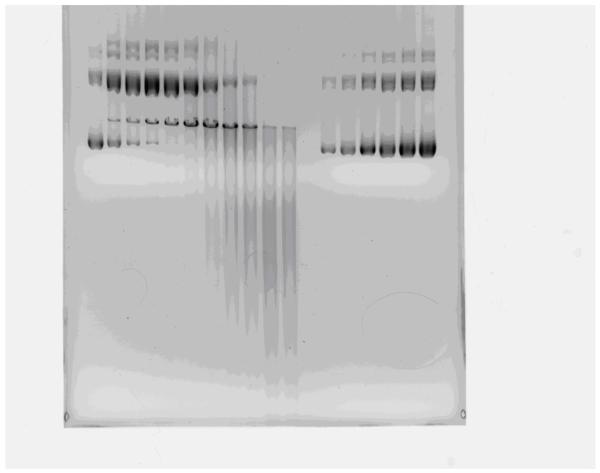
This plasmid of 9.9 kilobase pairs was irradiated and analyzed by electrophoresis for the three molecular forms. Dark bands of supercoiled and open circle DNA are seen in the control samples while the band of linear forms appears only after radiation exposure (Fig. 1). Lower molecular weight forms accumulate as a streak lower on the gel only after exposure to high radiation doses.
Fig. 1.
Gel electrophoresis of 10 kb plasmid. Lanes on the right used for calibration show increasing concentrations (100-1000 ng) of unirradiated plasmid; the lower band shows the predominant supercoiled form; smaller amounts of the open circle form are seen. Lanes on the left show the samples exposed to 0 to 24 Mrads of radiation.
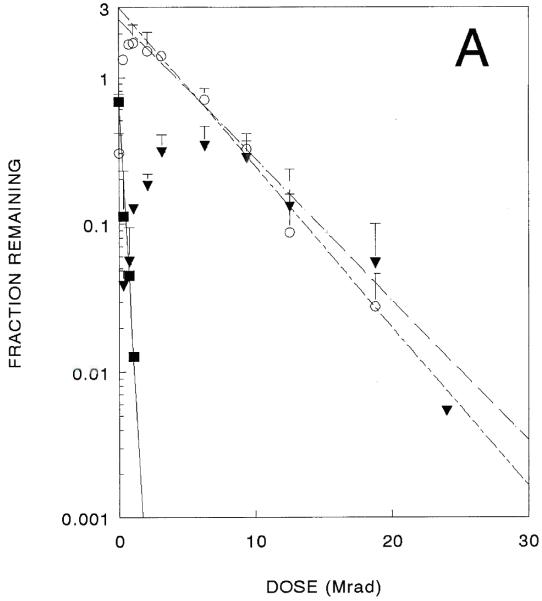
The supercoiled molecules decreased exponentially after only very small doses of radiation. Radiation target sizes calculated from three independent experiments averaged 6900 kDa [10 kbp]. The other forms exhibited more complex radiation responses. Amounts of both open circle and linear forms initially increased after radiation exposure, and both eventually decreased after higher doses (Fig. 2A). Assuming exponential decays, radiation target sizes calculated from three independent experiments are 580 kDa [0.88 kbp] for open circles and 440 kDa [0.66 kbp] for linear forms, as given in Table 1.
Fig. 2.
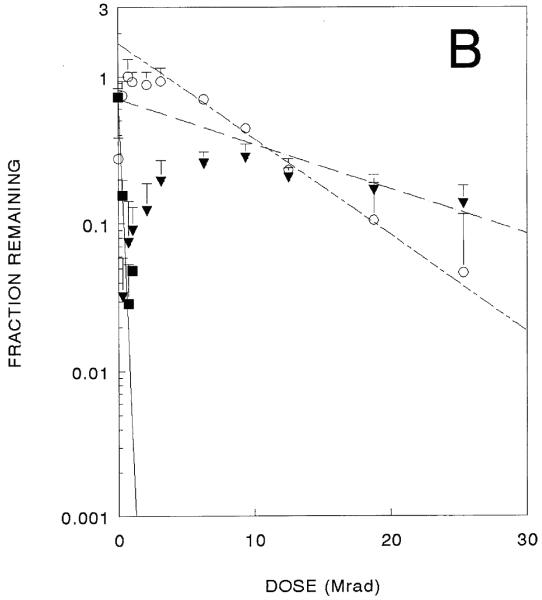
A 10 kbase plasmid inactivation curve. Surviving superccoiled (filled squares). Open circle (circles), and linear forms (filled triangles) are shown (data from three independent experiments). 2B. Identical experiments in the presence of ethidium bromide.
Table 1.
6.5 MDa (’10 kbase’) plasmid Target sizes (in kDa)
| Supercoiled Destruction |
Open Circle Destruction |
Linear form Destruction |
|
|---|---|---|---|
| 6900 ± 600 | 580 ± 140 | 440 ± 51 | |
| +Ethidum Bromide | 9300 ± 5300 | 390 ± 189 | 130 ± 90 |
Parallel experiments were performed in which this plasmid was incubated with ethidium bromide before radiation exposure. Although supercoiled forms show diminished ethidium bromide binding, these studies resulted in similar-shaped curves for all three forms (Fig. 2B) that yielded somewhat smaller target sizes (Table 1). The large errors make it difficult to determine if ethidium bromide intercalation has any effect on target sizes, however the difference in target sizes for destruction of the linear form is statistically significant, suggesting a protective effect on chain scission due to intercalation of ethidium bromide.
‘GFP plasmid’
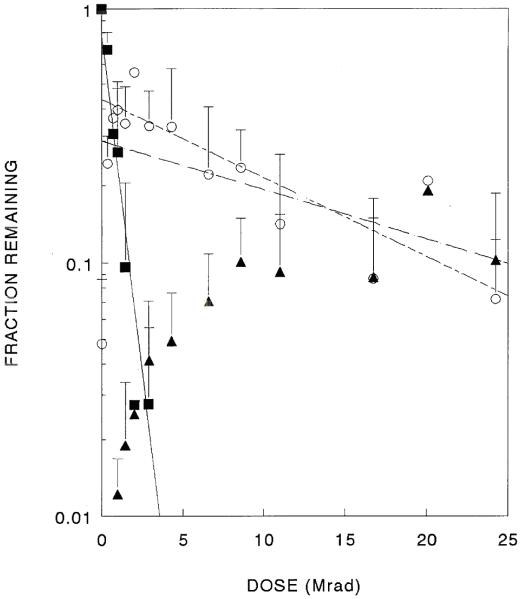
The supercoiled form of the GFP plasmid was rapidly lost by small doses of ionizing radiation (Fig. 3). The amount of the open circle forms rose simultaneously, reached a maximum and then slowly decayed with increasing radiation exposure. Linear forms did not appear until higher doses were obtained; the amounts of these forms also rose and reached a plateau. As with the 10 kbase plasmid, a streak was seen on the gels in samples exposed to large radiation doses.
Fig. 3.
GFP plasmid inactivation curve. Surviving supercoiled (filled squares), open circle (circles) and linear forms (filled triangles) are shown (data from three independent experiments).
In three independent experiments, the amount of the supercoiled form of the 3300 kDa GFP plasmid was observed to decay exponentially with radiation dose, yielding target sizes of 4700 ± 2400 kDa [7.1 kbp] (Table 2), indicating that a single radiation interaction anywhere in the plasmid results in the loss of that molecule from the supercoiled population.
Table 2.
3.3 MDa GFP plasmid Target Sizes (in kDa)
| Supercoiled | Open Circle | Linear Form | Transfection |
|---|---|---|---|
| 4700 ± 2400 | 260 ± 130 | 79 | 1067 ± 155 |
Plasmid preparations contain some plasmid in the open circle form (~20%) prior to irradiation and the levels of open circles increased after low doses of radiation due to the destruction of supercoiled molecules. Further exposure caused an exponential decrease in open circle forms (Fig. 2). Combining data from several identical experiments enabled better target determinations, leading to a target size of 260 kDa [0.790 kpb], for the destruction of open circles (Table 2). This indicates that the disappearance of the open circle form requires an ionization somewhere in a mass of 260 kDa compared to a total mass of 3300 kDa.
The linear form is expected to arise when open circle molecules suffer a polymer cleavage in the intact strand, ‘close to’ the preexisting break in the opposite strand. It could also arise directly from the supercoiled form provided the initial radiation interaction caused nearby cleavages in both strands. Two experiments showed a rise of linear forms to a plateau; no decrease in amount of linear forms was seen in this range of radiation exposures. This suggests that the target for destruction of the linear form was very small. One experiment did show a slight decrease at the highest doses. This decrease in the latter experiment was related to radiation damage in a mass of 79 kDa [0.12 kbp] (Table 2).
These structural analyses are complemented by transfection analysis of GFP signal from the same irradiated samples. In each experiment, the amount of transfection was reduced by radiation exposure to levels of 1-9% of the control. Further exposure did not reduce this level. Independent of radiation damage, the transfection assay has a ‘background’ value due to intrinsic cell fluorescence. Similar phenomena have been seen in irradiation of enzymes in which there was a slight non-enzymatic reaction producing the same product. As in those cases, this component could be subtracted from all the samples, giving a simple exponential decrease in transfection activity (Fig. 4). Fitting a single exponential function to many experimental points led to decreased error in target size determinations. In three experiments, the target size associated with GFP expression was 1067 ± 155 kDa [1.61 kbp], considerably smaller than the 3300 kDa total mass (Table 2).
Fig. 4.
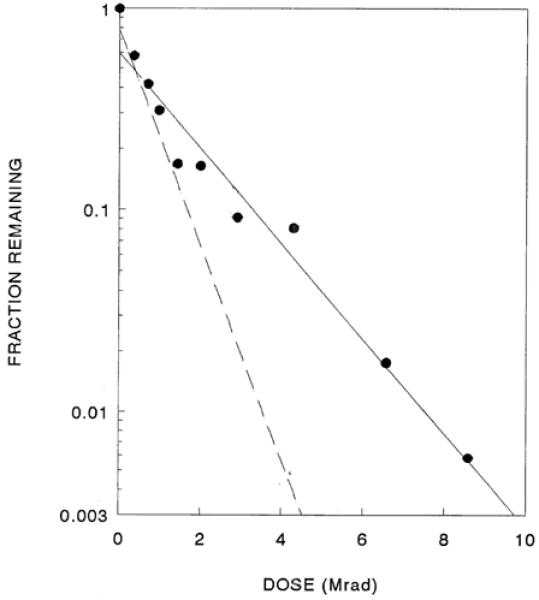
Loss of biological activity (as assessed by transfection) in irradiated GFP plasmids. The data are from the same three experiments shown in Fig. 3; note that the scale on the x-axis is enlarged. The dashed line shows the surviving supercoil content from Figure 3.
Preliminary experiments (data not shown) with lyophilized GFP plasmid with extremely low water content (<0.5%, <0.2 mol water/mol nucleotide) yielded similar results. This suggests that water does not play a role in our observations with high energy electron irradiations, in contrast with reports of irradiation with soft (70 kvp) X-rays [6].
DISCUSSION
High-energy electrons ionize covalent bonds randomly in exposed matter. In the frozen state virtually all damage is due to the action of radiations directly on macromolecules. Under these conditions the random interactions can be analyzed by radiation target theory. In the simplest case, radiation damage due to a single primary ionization leads to an exponential loss of a property with radiation dose at a rate proportional to the mass of the structures involved.
In the present studies, frozen supercoiled molecules were irradiated and surviving properties were analyzed by radiation target theory. In samples of both plasmids exposed to increasing doses of ionizing radiation, the supercoiled forms rapidly disappeared with concomitant increases in the amount of open circle and linear forms. As the supercoiled forms disappeared, the amount of open circle forms reached a maximum and then decreased with radiation dose. The linear forms increased more slowly, reaching a maximum as the open circles diminished.
A simple exponential decrease of measured properties as a function of radiation dose indicates that the effect is due to a single radiation interaction. Thus, the disappearance of supercoiled plasmid is caused by a single radiation interaction anywhere within the mass of the entire molecule. This result does not indicate the fate(s) of the supercoiled forms that are destroyed.
Assays of the open circle and linear forms showed a complex relationship with radiation exposure. These results were similar to observations in another plasmid after exposure to extremely low-energy X-rays [4], i.e., there was an initial dramatic increase in both forms up to a maximum (in open circles) that then decreased.
A ‘nick’ (breakage of a covalent bond in the polymer backbone of a single strand) in supercoiled DNA causes the molecule to unwind into an open circle form [13]. Further radiation exposure causes a decrease in open circle molecules at a rate expected from a much smaller structure. As with the supercoiled form, this result only means that a radiation interaction in this mass alters the structure so that the molecule no longer behaves (electrophoretically) as an open circle plasmid. This electrophoretic mobility will change only when radiation-induced scissions occur within a few bases of a scission already existing in the opposite strand (creating a double-strand break, or dsb). This predicts a target size of the order of 1 kDa (approximately three bases); if multiple single-strand breaks exist in an open circle plasmid, the loss of open circles will occur at a rate of that many multiples. This effect contributes to the target size measured for the destruction of open circles (260-580 kDa) and the present results indicate that single strand breaks in open circles occur about every 15-20 base pairs on average.
Multiple dsbs will fragment an open circle molecule into a variety of fragments which will appear as a smear lower on the gel. Linear forms might also appear directly from irradiated supercoiled plasmids. However, irradiation of a very small DNA structure composed of a 10 kDa and a 5 kDa strand yielded a 15 kDa target size for the loss of each strand [14]. In this small DNA complex, a radiation interaction with one strand resulted in scission of that strand as well as scission somewhere in the opposite chain.
In supercoiled plasmids a single radiation-induced cleavage anywhere will result in loss of the supercoiled structure; it will only be converted to the linear form if a second chain cleavage occurs within a few base pairs (i.e., a double-strand break). However, an increase in linear DNA was only observed after the creation and subsequent destruction of open circle forms. Thus our observations indicate that radiation-induced double-strand breaks in the original supercoiled plasmids must be relatively rare, consistent with the expected probabilities of random events.
Determination of the radiation sensitivity of biological function, transfection, adds a useful dimension to these results. However, it is anticipated that DNA repair in COS-7 cells could resuscitate radiation damage in GFP plasmids, resulting in increased GFP production and diminished target size. The extent of COS-7 repair of photoadducts of plasmids was determined to be 1% [15]. If a similar degree of repair occurred in the GFP system used here, the effect would be too small to significantly alter the estimated target size.
Transfection decreased exponentially in irradiated samples, but at a slower rate than the loss of the supercoiled form. This permitted reliable determination of surviving function over a wide range of radiation exposures, resulting in much smaller error in target size estimates. The transfection target size is considerably smaller than the size of the DNA molecule, only about one-third the mass. The average target size observed for transfection, 1067 kDa, is extremely close to 1068 kDa, the mass of the segment of the plasmid involved in transfection activity (which includes the CMV promoter, GFP gene, SV40 intron and polyadenylation signal; Fig, 4). This result has important ramifications; i.e., radiation damage elsewhere in the plasmid DNA does not affect transfection activity, so clearly there cannot have been any radiation damage (sufficient to cause covalent bond breakage and activity loss) to the active portion. These data indicate that there is negligible transfer of radiation energy along the DNA. This conclusion is consistent with our finding that the intercalation of ethidium bromide in supercoiled DNA (an event that would likely alter energy transfer along the polymer chain) had minimal effects on target sizes (Table 1).
Radiation cleavage of a polynucleotide chain requires a break in the sugar-phosphate polymer backbone, although it has been shown [4] that ionization of the phosphorus atom is not involved. In RNA [2] and in the present results radiation damage did not spread along the polymer backbone, but was restricted to a single area. The observations are consistent with observations in irradiated oligosaccharides [16], glycoproteins [17], and DNA [6]. Radiation damage was localized in oligosaccharides, primarily to a single sugar ring; in glycoproteins, energy was not transferred to the attached polypeptide. Since phosphate is not present in oligosaccharides and glycoproteins, it is clear that radiation damage to the sugar ring is necessary and sufficient to account for all these observations.
Previous studies of ribozyme enzymatic activity [2] reported that any energy transfer along the RNA polynucleotide could not be more than 15 bases. A similar approach in the present study of DNA makes it likely that in DNA, as in RNA, a primary ionization must occur directly in the active sequence. This conclusion suggests that energy transfer along DNA chain is minimal, if it occurs at all. This finding was not previously anticipated and offers new insight into the nature and mechanism of direct radiation damage in DNA.
The direct action of ionizing radiation can be used as a method to analyze both structural and functional mass of DNA. Additional uses for this radiation target analysis could include characterizing genetic elements in the viral genome responsible for DNA packaging, cell binding, and infectivity. For synthetic vectors, this technique could potentially be utilized to identify the size of vector components (e.g., polymers) that result in maximum DNA stability, cellular uptake and transfection activity.
ACKNOWLEDGEMENTS
The authors thank Dr. Martin Gellert for many helpful discussions. Support for these studies was derived in part from the National Institutes of Health through NIBIB grant #1 RO1 EB005476-01 to TJA and partially from the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health.
Footnotes
A preliminary report of some of these studies of one plasmid was presented at the Pacifichem 2005 meeting, December 2005.
REFERENCES
- [1].Kempner ES. Effects of High-Energy Electrons and Gamma Rays Directly on Protein Molecules. J. Pharmaceut. Sci. 2001;90:1637–1646. doi: 10.1002/jps.1114. [DOI] [PubMed] [Google Scholar]
- [2].Bernstein SL, Kempner E. Radiation Target Analysis of RNA. Proc. Nat’l. Acad. Sci. US. 1996;93:6410–6414. doi: 10.1073/pnas.93.13.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hutterman J, Kohnlein W, Teoule R, editors. Effects of Ionizing Radiation on DNA. Springer-Verlag; Berlin: 1978. [Google Scholar]
- [4].Hieda K, Hirono T, Azami A, Suzuki M, Furusawa Y, Maezawa H, Usami N, Yokoya A, Kobayashi K. Single- and Double-strand breaks in pBR322 plasmid DNA by monochromatic X-rays on and off the K-absorption edge of phosphorus. Internat. J. Radiat. Biol. 1996;70:437–445. doi: 10.1080/095530096144914. [DOI] [PubMed] [Google Scholar]
- [5].Boudaiffa B, Hunting D, Cloutier P, Huels MA, Sanche L. Induction of single- and double-strand breaks in plasmid DNA by 100-1500 eV electrons. Internat. J. Radiat. Biol. 2000;76:1209–1221. doi: 10.1080/09553000050134447. [DOI] [PubMed] [Google Scholar]
- [6].Purkayastha S, Milligan JR, Bernhard WA. An Investigation into the Mechanism of DNA Strand Breakage by Direct Ionization of Variably Hydrated Plasmid DNA. Jour. Phys. Chem. B. 2006;110:26286–26291. doi: 10.1021/jp065489i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Harmon JT, Nielsen TB, Kempner ES. Molecular Weight Determinations from Radiation Inactivation. Meth. Enzymol. 1985;117:65–94. doi: 10.1016/s0076-6879(85)17008-4. [DOI] [PubMed] [Google Scholar]
- [8].Molina MC, Armstrong TK, Zhang Y, Patal MM, Lentz YK, Anchordoquy T. The stability of lyophilized lipid/DNA complexes during prolonged storage. J. Pharmaceut. Sci. 2004;93:2259–2273. doi: 10.1002/jps.20138. [DOI] [PubMed] [Google Scholar]
- [9].Purkayastha S, Milligan JR, Bernhard WA. Correlation of free radical yields with strand break yields produced in plasmid DNA by the direct effect of ionizing radiation. Jour. Phys. Chem. B. 2005;109:16967–16973. doi: 10.1021/jp0518409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Molina MC, Allison SD, Anchordoquy TJ. Maintenance of nonviral vetor particle size during the freezing step of the lyophilization process is insufficient for the preservation of activity: insight from other structural indicators. J. Pharmaceut. Sci. 2001;90:1445–1455. doi: 10.1002/jps.1096. [DOI] [PubMed] [Google Scholar]
- [11].Kempner ES. The mathematics of radiation target analysis. Bull. Math. Biol. 1995;57:883–898. doi: 10.1007/BF02458298. [DOI] [PubMed] [Google Scholar]
- [12].Pollard EC, Guild WR, Hutchinson F, Setlow RB. The Direct Action of Ionizing Radiation on Enzymes and Antigens. Prog. Biophys. 1955;5:72–108. [Google Scholar]
- [13].Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4th Ed. WH Freeman and Company; New York: 2005. [Google Scholar]
- [14].Sluis-Cremer N, Kempner ES. Radiation target analysis of DNA template/primer complexes. Biophys. J. 2006;90:L61–L63. doi: 10.1529/biophysj.106.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Varganov Y, Amosova O, Fresco JR. Third-strand mediated psoralen-induced correction of the sickle-cell mutation on a plasmid transfected into COS-7 cells. Gene Therapy. 2007;14:173–179. doi: 10.1038/sj.gt.3302850. [DOI] [PubMed] [Google Scholar]
- [16].von Sonntag C. Free-radical reactions of carbohydrates as studied by radiation techniques. Adv. Carbohydr. Chem. Biochem. 1980;37:7–77. [Google Scholar]
- [17].Kempner ES, Miller JH, McCreery MJ. Radiation target analysis of glycoproteins. Analyt. Biochem. 1986;156:140–146. doi: 10.1016/0003-2697(86)90165-x. [DOI] [PubMed] [Google Scholar]