Abstract
To understand the effects of the transient ablation of BRCA2 gene expression in dividing human breast cells we transiently knocked down BRCA2 mRNA in HMEC and other cells. Microarray analysis of mRNAs revealed the down-regulation of the mRNAs of ubiquitin cross-reacting protein (UCRP) and the E2 enzyme that help conjugating UCRP to its target proteins, namely UBE2L6 (UbcH8), in BRCA2 ablated cells. UCRP is an interferon regulated protein, involved in cell growth and cell cycle events by participating in the degradation/modulation of cell cycle regulatory proteins. Quantitative-PCR and Northern analysis confirmed down-regulation of UCRP and UBE2L6 with BRCA2 knock down, respectively. Since UCRP and UCRPylation have critical roles in the innate immunity against viral infection and during pregnancy, our observation may indicate new roles of the BRCA2 protein.
Keywords: BRCA2, UCRP, ISG15, UBE2L6, UbcH8, siRNA, shRNA, H1 RNA promoter, Antisense phosphorothioate, Microarray
BRCA2 is a tumor suppressor protein with diverse functions [1,2]. BRCA2 deficiency has been attributed as the cause for many cases of breast, ovarian, and pancreatic carcinoma [1,2]. BRCA2 deficiency may be caused by the molecular defects in the BRCA2 genes or due to sporadic reasons. BRCA2 gene is not expressed in non-dividing cells and the rate of expression of this protein is increased with the rate of cell proliferation [3-5]. This growth dependent turn-on/turn-off mechanism of this gene is not well understood. Any number of environmental cues may influence the turn-on event and may initiate a molecular domino effect leading to DNA damage and oncogenesis. While undue expression of BRCA2 protein in non-dividing cells may initiate apoptosis, failure of the cell to produce appropriate amount of BRCA2 corresponding to the growth rate of the cell may also be detrimental. Since majority of the breast cancer cases are sporadic [1-5], some of such cancers may have initiated due to the transient absence of BRCA2 during the proliferative stages of the breast cells.
The molecular consequences of transient BRCA2 deficiency in the breast cells are not known. We report here our preliminary observations on the effect of transient ablation of BRCA2 gene expression on human mammary epithelial cells (HMEC) and BT549 human breast carcinoma cells. Microarray analysis of the mRNAs isolated from these BRCA2 knocked down cells showed a limited number of genes that are down-regulated as an immediate consequence of the decrease of BRCA2 expression. Two of the proteins that are significantly down-regulated are the ubiquitin cross-reacting protein (UCRP, also known as interferon stimulated gene 15, ISG15) and UBE2L6 (UbcH8). UCRP and UBE2L6 (UbcH8) belong to the regulatory network that modulates the function and targeted degradation of specific proteins in a cell [6-10]. One significant possible consequence of the down-regulation of such a protein could be enhancement of the growth factor receptors [11,12] and hence the acceleration of the growth rate of the affected cells. We have further verified the expression of UCRP and UBE2L6 in the BRCA2 knocked down cells by different other methods.
Materials and methods
Knockdown of BRCA2 gene expression
We took three parallel approaches for the transient knock down of BRCA2 gene expression. In one, we used anti-BRCA2 antisense gapmer oligonucleotides (sense control: 5′-ATGCCTATTGGATCCAA-3′; antisense: 5′-TTGGATCCAATAGGCAT-3′). In the gapmer (custom synthesized from Tri-link Biotechnology, CA) the terminal five nucleotides have 2-methoxy ribose and the middle 7 nucleotides are phosphorothioate. These oligos were used previously [13,14] to knock down BRCA2 in human cells. In the second approach, we used BRCA2 siRNAs (25 nM) commercially available from Qiagen. We procured two sets of siRNA pairs from Qiagen (BR1: 5′-UACAGUUGGCUGAUGGUGGtt-3′ and BR2: 5′-CGAGAAGCUGCAAGUCAUGCtt-3′). BR1 has a target at 7962–7982 nt area and BR2 has a target at 1002–1022 nt in human BRCA2 mRNA. In the third approach, we have cloned an inverted repeat sequence (5′-GATCCCCGAGAAGCTGCAAGTCATGCttcaagagaGCATGACTTGCAGCTTCTCTTTTTGGAAA-3′ and the complementary strand) behind the H1 RNA promoter in the plasmid pSUPER and transfected the breast cells using lipofectamine (Invitrogen). Inside the cells, RNAP III bound the H1 RNA promoter and transcribed short hairpin RNA (shRNA) that was cleaved by dicer to produce BR2 siRNA. We constructed pSUPER in our laboratory following the published protocols [15]. The levels of knock down of BRCA2 mRNA and protein were determined following standard protocols [16,17]. We used two breast cell lines for these studies: human mammary epithelial cells (HMEC, Clonetics) and BT549 cells (ATCC).
Transfection
HMEC and BT549 cells were grown according to Clonetics and ATCC guidelines. We used Oligofectamine (Invitrogen) to transfect the cells with the oligonucleotides (25 μM). Lipofectamine 2000 (Invitrogen) was used to transfect the pSUPER plasmid constructs. We used Trans-messenger reagent kit (Qiagen) and protocols for the transfection of the cells with the siRNAs.
RT-PCR evaluation of BRCA2 and other mRNA levels
Total RNA was isolated from control scrambled siRNA-treated or BRCA2 siRNA-treated cells and analyzed by RT-PCR (Amersham, Arlington Heights, IL). β-Actin was used as a loading control. Total RNAs (5 μg), isolated from the same breast cell samples that were used for the microarray hybridization, were treated with DNase I (Promega, Madison, WI) and reverse transcribed with Superscript II (Invitrogen). The resulting cDNA was used to carry out PCR amplification of BRCA2 (5′-GTACAGGAAACAAGCTTCTGA-3′ and 5′-GACTAACAGGTGGAGGTAAAG-3′), UCRP (5′-TGACGGTGAAGATGCTGG-3′ and 5′-CAGGCTCTGTGCCGCCTC-3′), and β-actin (5′-GCTCGTCGTCGACAACGGCTC-3′ and 5′-CAAACATGATCTGGGTCATCTTCTC-3′). The cDNA was also subjected to quantitative PCR amplification for 13 other different transcripts (Table 1) that were identified as differentially expressed by microarray analysis (data not shown in this communication). For each of the 16 transcripts (14 differentially expressed, BRCA2 and β-actin), primers were designed using the Primer Express 1.5 software (PE Applied Biosystems, Foster City, CA).
Table 1.
Human genes up- or down-regulated in association with the transient knock down of the BRCA2 gene (14 out of 11,607)
| Gene name | SGD ID# | Fold increase (+) or decrease (−) |
|---|---|---|
| Down-regulated | ||
| Tryptophanyl-tRNA synthetase | 855786 | 2.7 (−) |
| PMA induced gene | 814353 | 3.0 (−) |
| INF-induced transmem protein | 755599 | 3.9 (−) |
| UCRP | 742132 | 6.4 (−) |
| UBE2L6 (UbcH8) | 725395 | 2.6 (−) |
| Cullin-5 | 562811 | 2.5 (−) |
| ESTs | 289496 | 4.0 (−) |
| Ladinin | 121551 | 2.6 (−) |
| Cig5 RNA | 120600 | 4.8 (−) |
| Up-regulated | ||
| Keratin 6A | 592111 | 3.0 (+) |
| SCF (proton coupled) | 291059 | 2.7 (+) |
| ESTs | 290597 | 3.3 (+) |
| ESTs | 212712 | 2.6 (+) |
| ESTs | 203114 | 2.5 (+) |
Western blotting
BRCA2 and actin protein levels in the total cell extract were analyzed by Western blot analysis. BRCA2 monoclonal antibody (QED Bioscience, San Diego, CA) and anti-actin antibody (Sigma, St. Louis, MO) was used for this purpose. Cells were collected, washed with cold phosphate-buffered saline twice, and then lysed at 4 °C for 1 h in EBC buffer (0.5% Nonidet P-40, 120 mM NaCl, 50 mM Tris–HCl (pH 7.4), 1 mM EDTA (pH 8.0), 1 mM β-mercaptoethanol, and Complete protease inhibitor mixture (Roche Molecular Biochemicals)). Equal aliquots of cell lysate (30–100 μg/lane) were electrophoresed through 7% SDS–polyacrylamide gels after incubating for 15 min at 65 °C in 1× loading buffer (100 mM Tris–HCl (pH 6.8), 10% glycerol, 0.01% bromphenol blue, 2% SDS, 100 mM dithiothreitol). Proteins were then transferred to nitrocellulose membranes (Schleicher & Schüll) and blocked in TBST with 5% bovine albumin (fraction V) (Sigma). Membranes were blotted with anti-BRCA2 monoclonal antibody (QED Bio science, San Diego, CA) or anti-actin antibody (Sigma, St. Louis, MO) and then incubated with horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences). Signals were visualized using the SuperSignal chemiluminescent detection system (Pierce).
Microarray analysis
Total RNA was isolated from subconfluent cultured human breast cells using the RNeasy kit (Qiagen). The RNA quality was checked by formaldehyde-agarose gel electrophoresis. Total RNAs (40 μg) from cells were labeled in reverse transcription reactions (Superscript II kit, Invitrogen) with dCTP-Cy5 and dCTP-Cy3, respectively (Amersham Biosciences). In every second replicate experiment the fluorescent deoxynucleotides were swapped. Purified cDNA probes labeled with Cy3 and Cy5 were mixed per pair and hybridized to human cDNA microarray chips (Human Research Genetics 11K) in the VMSR Microarray Core Facility at the Vanderbilt University. The slides were scanned with a GenePix 4000A microarray scanner (Axon Instruments, Union City, CA), and the images were analyzed using Genepix pro 3 software. Data files were entered into the Stanford Microarray Data base ((genome-www5.stanford.edu/MicroArray/SMD). A uniform scale factor was applied to normalized signal intensities between Cy5 and Cy3. Flagged spots and spots with an average intensity below 2.5-fold above the background were not retained for further analysis. The log2 (Cy5/Cy3) ratio of the other spots was calculated for each slide. To compare the results from the different subjects, data from each slide were normalized in log space to have a mean of 0 and a SD of 1 by using the Cluster program [18]. Genes with significant changes in mRNA levels in response to BRCA2 deficiency were identified using the significant analysis of microarrays (SAM) procedure [19], a validated statistical technique for identifying differentially expressed genes across high density microarrays. This procedure provides a list of “significant” genes and an estimate of the false discovery rate, which represents the percentage of genes that could be identified by chance [19].
Real-time RT-PCR analysis
Quantitations of BRCA2 and UCRP transcripts were performed using a TaqMan assay (Applied Biosystems). A probe for eukaryotic 18S rRNA endogenous control (product 4319413E) was VIC/minor groove binder-labeled. The primers and FAM/minor groove binder-labeled probe for ISG15 (GIP2, Hs00192713_m1, Reporter: FAM; Quencher: TAMARA (MGB)) were purchased from ABI (TaqMan Assay-on-Demand): For BRCA2 expression analysis, probes and primers were designed using Primer Express software, so that they overlap splice junctions for Exon 26-27. RBR3′F (9794–9812) CCAAGTGGTCCACCCCAAC; RBR3′R (9895–9870) CACAATTAGGAGAAGACATCAGAAGC; BRCA2 PROBE 3′ (9818–9847) 5′-6FAM-ACTGTACTTCAGGGCCGTA CACTGCTCAAA TAMARA-3′. Eukaryotic 18S rRNA was used as an endogenous control in a multiplex PCR with a primer/probe of the gene of interest. For each reaction, 2× TaqMan universal PCR master mix (Applied Biosystems), 900 nM primers, and 250 nM probes in 20 μl were added to 384-well plate. Real-time PCR and subsequent analysis were performed with the ABI Prism 7900HT sequence detection system (SDS v2.1) (Applied Biosystems) using the following conditions: 50 °C for 2 min, 95 °C for 10 min, and then 40 cycles of amplification (95 °C denaturation for 15 s, 60 °C annealing/extension for 1 min). All PCRs were performed in triplicate for each sample and were repeated three times. All experimental data except survival were expressed as the mean ± SE. A one-way analysis of variance, a two-way repeated measure analysis of variance, and Student’s t test were used to determine the significance of the difference [20].
Northern analysis
The UBE2L6 sequence (Accession No.NM_004223) was retrieved and two primers were designed (forward 5′-ACACTCGGTCCCGACATGA-3′ and reverse 5′-AGCAAGAACATGGTTTCCGT-3′) to amplify a 1149 bp cDNA fragment from total cDNA from BT549 cells. This cDNA fragment was gel-purified (Qiagen) and radiolabeled with 32P following random primer labeling protocol (Invitrogen) and used as a probe for the analysis of the levels of UBE2L6 mRNAs in normal and BRCA2 knocked down BT549 cells [16,17].
Results and discussion
Knock down of BRCA2 transcript levels in HMEC and BT549 cells
Our objective was to transiently ablate the expression of BRCA2 gene expression and determine the effect of this ablation on the transcriptome of the cells. We employed three different hybridization-assisted degradation approaches to accomplish this goal. Our test cells were one normal human breast cell population obtained as HMEC cells from Clonetics. The other is the established breast cancer cell line BT549. We also partly verified our experimental data in other human breast cancer cells such as MCF7, MDA-MB-231, and MDA-MB-468 (data not shown). BRCA2 was previously knocked down in human breast cells with success using phosphorothioate antisense oligonucleotides [13,14]. We initially used those tested oligos for our experiments. We also procured two siRNAs designed from two different loci in the BRCA2 ORF from Qiagen. We used those anti-BRCA2 siRNAs in HMEC and BT549 cells. Thirdly, we designed shRNA genes from those siRNAs and cloned them in pSUPER to express in vivo the anti-BRCA2 shRNA in transfected breast cells. We constructed the pSUPER plasmid by cloning amplified H1 RNA promoter in pBluscript KS (+), as described by Brummelkamp et al. [15]. We isolated from control and antisense nucleic acid treated cells and evaluated the levels of BRCA2 mRNA and protein by Northern analysis, RT-PCR and real time PCR methods. Fig. 1 shows a representative evaluation data as was determined with HMEC cells and BR2 siRNA. Similar results were obtained with other cells and antisense phosphorothioate oligo or shRNA constructs (data not shown). With antisense phosphorothioate ODNs we reproducibly found 55–75% reduction in BRCA2 mRNA level and 50–80% reduction in the BRCA2 protein levels (data not shown). Among the siRNAs, BR2 produced better reduction in BRCA2 mRNA and protein than BR1. With BR2 siRNA, the effect was also relatively stable (reduced level of BRCA2 remained unchanged till 72 h whereas with BR1 the ablative effect disappeared after 48 h). Similar effect was found with BR1 and BR2 shRNA constructs (data not shown). BR2 siRNA produced best effect in reducing the levels of both the mRNA (>85%) and protein (>80%) of BRCA2 under the conditions of the experiment (Fig. 1). Availability of target site in the BRCA2 mRNA and other factors may be responsible for the differential effect of BR1 and BR2 siRNAs [21]. Knock down of BRCA2 mRNA in the antisense nucleic acid (phosphorothioate oligo, siRNA or shRNA) treated cells were also verified by real-time PCR (see Figs. 2A and B).
Fig. 1.
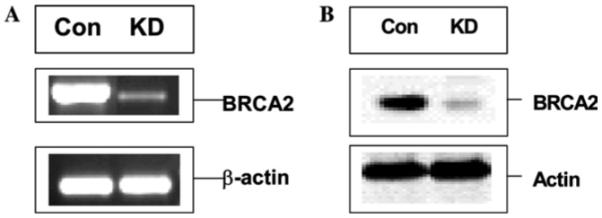
Knock down of BRCA2 mRNA (A) and protein (B) in HMEC cells by siRNA (BR2). Total RNA was isolated from control scrambled siRNA (Con)-treated or BRCA2 siRNA (BR2, KD)-treated cells and analyzed by RT-PCR analysis. β-actin was used as a loading control. Similarly, BRCA2 and actin protein levels in the total cell extract were analyzed by Western blot analysis. BRCA2 monoclonal antibody and anti-actin antibody were used for this purpose. Similar results were obtained using anti-BRCA2 phosphorothioate gapmer oligos or when cells were transiently transfected with pSUPER construct expressing BR2 shRNA from H1 RNA gene promoter in HMEC and BT549 cells.
Fig. 2.
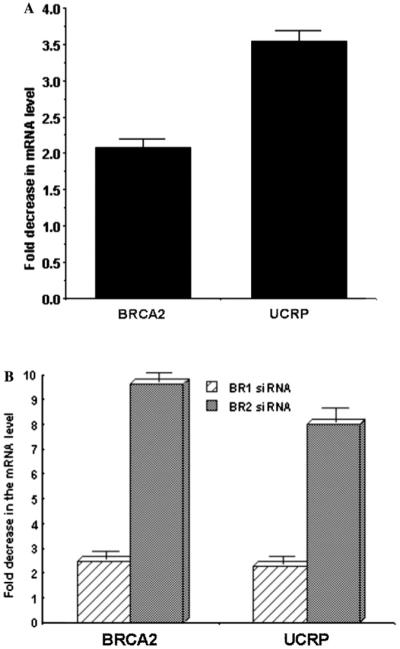
Real-time PCR analysis of BRCA2 and UCRP mRNA in (A) anti-BRCA2 phosphorothioate gapmer-treated (48 h) HMEC cells and (B) BR1 and BR2 siRNA-treated (48 h) BT549 cells. 18S rRNA was used as normalizing control. Results are means ± SE (n = 6). Sense control gapmer treated cells were used as control. Fold decrease in the UCRP mRNA level is proportional to the fold decrease in the BRCA2 mRNA level.
Down-regulation of UCRP, UBE2L6, and other transcripts in BRCA2 knocked down cells
We compared the relative expressions of 11,607 human genes in BR2 siRNA-treated or scrambled siRNA treated HMEC or BT549 cells using cDNA microarray. Interestingly, only 14 genes showed reproducible (n =3 × 2) changes in their transcript levels in both of these cells (Table 1). Out of the 14 differentially expressed genes four are ESTs and the function of these genes are not known. Nine of the genes are down-regulated in BRCA2-ablated cells whereas five of them are up-regulated (Table 1). Majority of the down-regulated genes are known to be regulated by interferons, e.g., tryptophanyl-tRNA synthetase, UCRP, UBE2L6, and Cig5 [22-26]. Interferons enhance cellular susceptibility to apoptosis in a variety of tumor cells [27]. The mechanism by which IFN promotes cell death seems to be via the regulation of the expression of different proteins involved in apoptosis [27]. Thus, negative regulation of IFN-induced genes in BRCA2-ablated breast cells may reflect another growth regulatory role of BRCA2. We have verified the differential expressions of all these genes by RT-PCR analysis (data not shown). We only describe here the down-regulation of UCRP transcripts and of the transcript of the enzyme that helps to catalyze the conjugation of UCRP to its target proteins. Verification of UCRP transcript levels are shown in Figs. 2A and B. Fold decrease in the UCRP mRNA level was proportional to the fold decrease in the BRCA2 mRNA level. Similar results were obtained using HMEC cells. Down-regulation of UBE2L6 transcripts was also verified in BR2 siRNA-treated BT549 cells by northern analysis (Fig. 3).
Fig. 3.
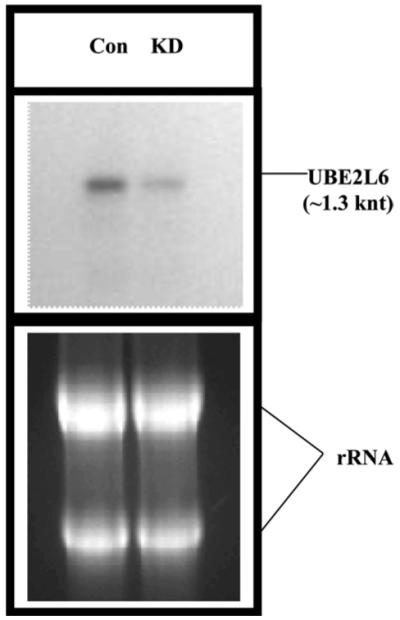
Northern analysis of UBE2L6 mRNA in scrambled siRNA (CON) and BR2 siRNA (KD) treated BT549 cells. Five microgram total RNAs were loaded per lane.
UCRP is one of the most strongly induced genes after interferon treatment and is also significantly induced by influenza B virus, lipopolysaccharide, and genotoxic stress [6,7,28-31]. The sequence of UCRP protein has significant homology to a diubiquitin sequence, accounting for its cross-reactivity with affinity purified anti-ubiquitin antibodies [32]. Several reports demonstrate that UCRP is released by various cell types and can act as a cytokine leading to proliferation of NK cells [6,7,28-31]. Most remarkably, UCRP is conjugated to intracellular proteins via an isopeptide bond in a manner similar to ubiquitin (Ub), SUMO, and Nedd8 [33,34]. Conjugation of ubiquitin-like proteins (Ubls) involves a three-step mechanism whereby specific enzymes (or enzyme complexes) activate and covalently link Ubls to their substrates [34,35]. UCRP conjugation was thought to occur via a similar but distinct pathway compared with Ub conjugation [30]. An activating enzyme for UCRP (the E1 enzyme) was shown to be UBE1L [34,35]. This enzyme is described as specific for UCRP and does not catalyze the activation of ubiquitin [10]. Recently it has been shown a common E2 enzyme UbcH8, the product of UBE2L6 gene, can catalyze the target protein modification by both UCRP and ubiquitin [9,10]. Our observation of down-regulation UBE2L6 gene product in BRCA2-ablated breast cells (Fig. 3) may thus indicate the influence of BRCA2 on UCRP metabolism but also on the general ubiquitinylation processes of the cell. Although the role of Ub, Nedd8, and SUMO conjugation has been assessed in numerous studies, the biological significance of UCRP modification remains unknown. It has been shown that serpin 2A, PLCγ1, Jak1, Stat1, and ERK1 [36,37] are among the proteins that are targeted by UCRP. Physiological roles of UCRP modification may include re-organization of the cytoskeleton after IFN stimulation and modulation of JAK-STAT signaling pathway [34,38-40]. One of the regulations of the cellular growth is the programmed modulation of growth factor receptor proteins on the cell surface [11,12]. We intend to explore whether UCRP plays any role in this process in human breast cells.
Acknowledgments
We thank Ms. Rati M. Tripathi for assistance in developing the manuscript. Supported by the MMC/VICC cancer partnership grant #1U54CA091408-010003 from NCI and the DOD Grant #DAMD17-00-1-0341 to G.C.
References
- [1].Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- [2].Keen JC, Davidson NE. The biology of breast carcinoma. Cancer. 2003;97:825–833. doi: 10.1002/cncr.11126. [DOI] [PubMed] [Google Scholar]
- [3].Chodosh LA. Expression of BRCA1 and BRCA2 in normal and neoplastic cells. J. Mammary Gland Biol. Neoplasia. 1998;3:389–402. doi: 10.1023/a:1018784031651. [DOI] [PubMed] [Google Scholar]
- [4].Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- [5].Narod SA, Foulkes WD. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer. 2004;4:665–676. doi: 10.1038/nrc1431. [DOI] [PubMed] [Google Scholar]
- [6].Haas AL, Ahrens P, Bright PM, Ankel H. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 1987;262:11315–11323. [PubMed] [Google Scholar]
- [7].Blomstrom DC, Fahey D, Kutny R, Korant BD, Knight E., Jr. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J. Biol. Chem. 1986;261:8811–8816. [PubMed] [Google Scholar]
- [8].Ardley HC, Rose SA, Tan N, Leek JP, Markham AF, Robinson PA. Genomic organization of the human ubiquitin-conjugating enzyme gene, UBE2L6 on chromosome 11q12. Cytogenet. Cell. Genet. 2000;89:137–140. doi: 10.1159/000015593. [DOI] [PubMed] [Google Scholar]
- [9].Kim KI, Giannakopoulos NV, Virgin HW, Zhang DE. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 2004;24:9592–9600. doi: 10.1128/MCB.24.21.9592-9600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. USA. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lipkowitz S. The role of the ubiquitination-proteasome pathway in breast cancer: ubiquitin mediated degradation of growth factor receptors in the pathogenesis and treatment of cancer. Breast Cancer Res. 2002;5:8–15. doi: 10.1186/bcr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qiu XB, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. USA. 2002;99:14843–14848. doi: 10.1073/pnas.232580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J. Natl. Cancer Inst. 1998;90:978–985. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- [14].Robinson-Benion C, Jensen RA, Holt JT. Analysis of cancer gene functions through gene inhibition with antisense oligonucleotides. Methods Enzymol. 2000;314:499–506. doi: 10.1016/s0076-6879(99)14125-9. [DOI] [PubMed] [Google Scholar]
- [15].Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- [16].Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. third ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- [17].Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. Wiley; New York: 2002. [Google Scholar]
- [18].Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tusher GV, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Campbell MJ, Machin D. Medical Statistics: A Commonsense Approach. second ed. Wiley; New York: 1994. [Google Scholar]
- [21].Du Q, Thonberg H, Zhang HY, Wahlestedt C, Liang Z. Validating siRNA using a reporter made from synthetic DNA oligonucleotides. Biochem. Biophys. Res. Commun. 2004;325:243–249. doi: 10.1016/j.bbrc.2004.09.222. [DOI] [PubMed] [Google Scholar]
- [22].Gromov P, Skovgaard GL, Palsdottir H, Gromova I, Ostergaard M, Celis JE. Protein profiling of the human epidermis from the elderly reveals up-regulation of a signature of interferon-{gamma}-induced polypeptides that includes manganese-superoxide dismutase and the p85{beta} subunit of phosphatidylinositol3-kinase. Mol. Cell. Proteomics. 2003;2:70–84. doi: 10.1074/mcp.M200051-MCP200. [DOI] [PubMed] [Google Scholar]
- [23].Aboagye-Mathiesen G, Ebbesen P, von der Maase H, Celis JE. Interferon gamma regulates a unique set of proteins in fresh human bladder transitional cell carcinomas. Electrophoresis. 1999;20:344–348. doi: 10.1002/(SICI)1522-2683(19990201)20:2<344::AID-ELPS344>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- [24].Fay MJ, Longo KA, Karathanasis GA, Shope DM, Mandernach CJ, Leong JR, Hicks A, Pherson K, Husain A. Analysis of CUL-5 expression in breast epithelial cells, breast cancer cell lines, normal tissues and tumor tissues. Mol. Cancer. 2003;2:40. doi: 10.1186/1476-4598-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Boudinot P, Riffault S, Salhi S, Carrat C, Sedlik C, Mahmoudi N, Charley B, Benmansour A. Vesicular stomatitis virus and pseudorabies virus induce a vig1/cig5 homologue in mouse dendritic cells via different pathways. J. Gen. Virol. 2000;81:2675–2682. doi: 10.1099/0022-1317-81-11-2675. [DOI] [PubMed] [Google Scholar]
- [27].Ruiz de Almodovar C, Lopez-Rivas A, Ruiz-Ruiz C. Interferon-gamma and TRAIL in human breast tumor cells. Vitam. Horm. 2004;67:291–318. doi: 10.1016/S0083-6729(04)67016-6. [DOI] [PubMed] [Google Scholar]
- [28].D’Cunha J, Ramanujam S, Wagner RJ, Witt PL, Knight E, Jr., Borden EC. In vitro and in vivo secretion of human ISG15, an IFN-induced immunomodulatory cytokine. J. Immunol. 1996;157:4100–4108. [PubMed] [Google Scholar]
- [29].Knight E, Jr., Fahey D, Cordova B, Hillman M, Kutny R, Reich N, Blomstrom D. A 15-kDa interferon-induced protein is derived by COOH-terminal processing of a 17-kDa precursor. J. Biol. Chem. 1988;263:4520–4522. [PubMed] [Google Scholar]
- [30].Narasimhan J, Potter JL, Haas AL. Conjugation of the15-kDa interferon-induced ubiquitin homolog is distinct from that of ubiquitin. J. Biol. Chem. 1996;271:324–330. doi: 10.1074/jbc.271.1.324. [DOI] [PubMed] [Google Scholar]
- [31].Taylor JL, D’Cunha J, Tom P, O’Brien WJ, Borden EC. Production of ISG-15, an interferon-inducible protein, in human corneal cells. J. Interferon Cytokine Res. 1996;16:937–940. doi: 10.1089/jir.1996.16.937. [DOI] [PubMed] [Google Scholar]
- [32].Raasi S, Schmidtke G, de Giuli R, Groettrup M. A ubiquitin-like protein which is synergistically inducible by interferon-gamma and tumor necrosis factor-alpha. Eur. J. Immunol. 1999;29:4030–4036. doi: 10.1002/(SICI)1521-4141(199912)29:12<4030::AID-IMMU4030>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- [33].Loeb KR, Haas AL. Conjugates of ubiquitin cross-reactive protein distribute in a cytoskeletal pattern. Mol. Cell. Biol. 1994;14:8408–8419. doi: 10.1128/mcb.14.12.8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu M, Li XL, Hassel BA. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J. Biol. Chem. 2003;278:1594–1602. doi: 10.1074/jbc.M208123200. [DOI] [PubMed] [Google Scholar]
- [35].Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hamerman JA, Hayashi F, Schroeder LA, Gygi SP, Haas A, Hampson L, Coughlin P, Aebersold R, Aderem A. Serpin 2a is induced in activated macrophages and conjugates to a ubiquitin homolog. J. Immunol. 2002;168:2415–2423. doi: 10.4049/jimmunol.168.5.2415. [DOI] [PubMed] [Google Scholar]
- [37].Malakhov MP, Kim KI, Malakhova OA, Jacobs BS, Borden EC, Zhang DE. High-throughput immunoblotting. ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- [38].Malakhova OA, Yan M, Malakhov MP, Yuan Y, Ritchie KJ, Kim KI, Peterson LF, Shuai K, Zhang DE. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 2003;17:455–460. doi: 10.1101/gad.1056303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li XL, Blackford JA, Judge CS, Liu M, Xiao W, Kalvakolanu DV, Hassel BA. RNase-L-dependent destabilization of interferon-induced mRNAs. A role for the 2-5A system in attenuation of the interferon response. J. Biol. Chem. 2000;275:8880–8888. doi: 10.1074/jbc.275.12.8880. [DOI] [PubMed] [Google Scholar]
- [40].Wong LH, Krauer KG, Hatzinisiriou I, Estcourt MJ, Hersey P, Tam ND, Edmondson S, Devenish RJ, Ralph SJ. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3gamma. J. Biol. Chem. 1997;272:28779–28785. doi: 10.1074/jbc.272.45.28779. [DOI] [PubMed] [Google Scholar]


