Abstract
Invasiveness of tumor cells is often determined by the profile of their expressed genes. To determine the gene expression differences between an invasive and a non-invasive human breast tumor cells, we selected BT-549 (invasive) and MDA-MB-468 (non-invasive) cells, and compared their transcriptomes by cDNA microarray analysis. Among the significant differences in gene expressions, notable are the up-regulation of cytokeratins 8 and 19, and down-regulation of metallothioneins 1G and IL in MDA-MB-468 cells. Since MDA-MB-468 cells do not express SLUG, a member of a small family of E2-box-binding zinc finger silencer proteins, we studied whether the cytokeratin gene overexpressions in these cells are due to the absence of SLUG. Inducible expression of SLUG in MDA-MB-468 cells inhibited the expressions of the cytokeratin 8 and 19 but not others as was revealed by microarray analysis. Similarly, siRNA knock down of SLUG in BT-549 cells increased the expressions of those cytokeratin mRNAs. SLUG levels in the cell regulated the function of cytokeratins 8 and 19 gene promoters. We conclude that the expressions of cytokeratins and metallothioneins may be associated with the differential invasive behaviors of these breast tumor cells and SLUG may have regulatory roles in this process.
Keywords: Invasiveness, Breast tumor, SLUG, Cytokeratin 8, Cytokeratin 19, Metallothionein 1G, Metallothionein 1L, siRNA, Microarray
A serious gap in our understanding of cancer concerns malignancy and metastasis. We have yet to clearly identify gene defects that specifically permit cells to invade surrounding tissues, spread through the body, and form metastases. Local invasiveness requires a break down of the mechanisms that normally hold epithelial cells together. To determine the gene expression differences between an invasive and a non-invasive human breast tumor cells, we selected BT-549 (invasive) and MDA-MB-468 (non-invasive) cells, and compared their transcriptomes by cDNA microarray analysis. One of the known genotypic differences between these two cells is that BT-549 expresses the zinc finger repressor protein SLUG but the MDA-MB-468 cells do not [1]. Hence, we tested whether any gene overexpressed in the MDA-MB-468 cells is regulated by SLUG.
We paid particular attention to SLUG because it is implicated as one of the determinants of tumor cell invasiveness [2-6]. This protein is a member of a superfamily of transcriptional regulators that are implicated in the formation of mesoderm and neural crest, as well as in the pathological progression of epithelial tumors [2,3]. Overexpression of these proteins in epithelial cells leads to loss of expression of key cell–cell adhesion molecules, such as E-cadherin, claudins, and occludin [4-8]. SLUG contains a highly conserved region at the carboxyl-terminus of the protein containing fourto six zinc fingers of the C2H2 type [2,3]. The zinc fingers mediate sequence-specific interactions with DNA. SLUG superfamily members bind specifically to consensus-binding sites that contain the E2-box sequence CACCTG [2,3]. The amino-terminus of SLUG contains the evolutionarily conserved SNAG (for Snail/Gfi) domain [2,3] that may mediate transcriptional repression function of SLUG. It is generally believed that SLUG recruits a co-repressor, possibly CtBP1 which in turn recruits HDAC that leads to gene repression [3].
Although SLUG is not essential for normal development in the mouse and homozygous null animals are viable and fertile [9], SLUG is implicated in epithelial to mesenchymal transitions during development in other vertebrates. In embryogenesis in the chick and in Xenopus sp., SLUG function is required for specification of neural crest [3,10]. Thus, SLUG function to promote cell fate changes during development, leading to the production of migratory, mesenchymal cells. In vertebrates, SLUG is aberrantly up-regulated by the E2A-HLF oncoprotein in certain leukemias, leading to increased cell survival [11]. Further analysis of SLUG during hematopoiesis has revealed a non-pathological role for SLUG in promoting cell survival in hematopoietic progenitor cells [12,13]. Finally, overexpression of SLUG in MCF7 cells has recently been shown to induce phenotypic alterations including loss of cell–cell contacts and acquisition of invasive growth, and these recombinant cells were protected from apoptosis induced by this DNA-damaging agent such as adriamycin [8].
The two cell lines, BT-549 and MDA-MB-468, compared in this study are otherwise very similar. Both of them are negative in the functional expression of Rb, p53, and ER-β [14-17]. On the other hand, BT-549 cells are vimentin positive and MDA-MB-468 cells are vimentin negative [15]. BT-549 cells induce the formation of invasive tumors but MDA-MB-468 cells are non-invasive [15]. We initially compared the transcriptomes of BT-549 and MDA-MB-468 cells by cDNA microarray analysis to understand what gene expression differences may exist between these two phenotypically distinct cells. We found differential expression of metallothionein and cytokeratins in these two cells. We then overexpressed SLUG in MDA-MB-468 cells from a doxycycline-inducible promoter and compared the gene expression profile in the presence or absence of the inducer. We identified that cytokeratins 8 and 19 genes are repressed when SLUG expression is induced. We have verified the role of SLUG in regulating these cytokeratins by siRNA-mediated knock down of SLUG in BT-549 cells. We also report here our preliminary characterization of the promoters of these cytokeratin genes as it is regulated by SLUG expression.
Materials and methods
Cells
We used commercially available lines of human breast cells MDA-MB-468 and BT-549 [18,19]. These cells were purchased from American Type Culture Collection (ATCC, Manassas, VA). Human breast carcinoma MDA-MD-468 and BT-549 cells were maintained following standard ATCC recommended media. All cells were maintained in a humidified CO2 (5%) incubator at 37 °C [18,19]. BT-549 cells were isolated from a 72-year-old Caucasian breast cancer patient (ATCC). The MDA-MB-468 cell line was isolated from a pleural effusion of a 51-year-old African-American female patient with metastatic adenocarcinoma of the breast (ATCC).
Microarray analysis
Comparative gene expression analysis between BT-549 and MDA-MB-468 cells or between normal and SLUG overexpressing MDA-MB-468 cells was done by human cDNA microarray analysis as described before [19]. Total RNA was isolated from sub-confluent cultured human breast cells using the RNeasy kit (Qiagen). The RNA quality was checked by formaldehyde–agarose gel electrophoresis. Total RNAs (40 μg) from cells were labeled in reverse transcription reactions (Superscript II kit, Invitrogen) with dCTP-Cy5 and dCTP-Cy3, respectively (Amersham Biosciences) [19]. In every second replicate experiment, the fluorescent deoxynucleotides were swapped. Purified cDNA probes labeled with Cy3 and Cy5 were mixed per pair and hybridized to human cDNA microarray chips (Human Research Genetics 11K) in the VMSR Microarray Core Facility at the Vanderbilt University. The slides were scanned with a GenePix 4000A microarray scanner (Axon Instruments, Union City, CA), and the images were analyzed using Genepix pro 3 software. Data files were entered into the Stanford Microarray Database (genome—www5.stanford.edu/MicroArray/SMD). A uniform scale factor was applied to normalized signal intensities between Cy5 and Cy3. Flagged spots and spots with an average intensity below 2.5-fold above the background were not retained for further analysis. The log2 (Cy5/Cy3) ratio of the other spots was calculated for each slide. To compare the results from the different subjects, data from each slide were normalized in log space to have a mean of 0 and a SD of 1 by using the Cluster program [20]. Genes with significant changes in mRNA levels were identified using the significant analysis of microarrays (SAM) procedure [21], a validated statistical technique for identifying differentially expressed genes across high density microarrays. This procedure provides a list of “significant” genes and an estimate of the false discovery rate, which represents the percentage of genes that could be identified by chance [21].
Expression of recombinant SLUG in MDA-MB-468 cells
Human SLUG coding sequence was amplified from RNA isolated from BT-549 cells using N-terminal primer (5′-GACGGATCCATGCCGCGCTCCTTCCTG-3′) and FLAG-tagged C-terminal primer (5′-CGTCGACTCACTTATCGTCGTCATCCTTGTAATCGTGTGCTACACAGC-3′). The sequence-verified amplified cDNA (831 bp) was digested with BamHI/SalI and was cloned at the BamHI/SalI sites at the MCS of pRevTet-TRE plasmid (BD-Bioscience). PT67 packaging cells were transfected with either pRevTet-ON plasmid (BD-Biosciences) or the pRevTet-TRE-SLUG plasmid to collect the recombinant retrovirus. PT67 cells were grown to ~50% confluency in 100-mm culture dishes and then transfected with 4 μg plasmid DNA using Lipofectamine 2000 (Invitrogen). The culture medium was replaced at 24 h after transfection, and retrovirus-containing medium was collected at 48 h after transfection and filtered through a 0.45-μm filter. For long-term storage the retrovirus-containing medium was frozen in liquid nitrogen and stored at −80 °C. MDA-MB-468 cells were grown to ~40–50% confluency in 100-mm culture dishes and then transduced first by retrovirus containing pRevTet-ON plasmid by addition of 1:1 retrovirus-containing medium and growth medium. The culture medium was replaced after 24 h with cell growth medium. Medium containing 1 mg/ml geneticin was added to select stable cell population at 48 h. This geneticin resistant cell population was further transduced by pRevTRE-SLUG plasmid containing virus as described above and stable cell line was selected further with 500 μg/ml hygromycin B. For all the experiments this double resistant cell line was maintained in 100 μg/ml geneticin and hygromycin B. Expression of SLUG was induced by doxycycline (1 μg/ml).
Knock down of SLUG gene expression in BT-549 cells
The anti-human SLUG siRNAs were designed using the Invitrogen site software (https://rnaidesigner.invitrogen.com/sirna/) and custom synthesized from Invitrogen. The nucleotide sequences of the siRNA pair (SR) and its respective control (SRC) are as follows (the number indicates the location of the sequence in the SLUG coding region): SR230, 5′-CCGGAUACUCCUCAUCUUU-3′/5′-AAAGAUGAGGAGUAUCCGG-3′; SRC230, 5′-CCGUACTCCUCAUCGAUUU-3′/5′-AAAUCGAUGAGGAGUACGG-3′; SR661, 5′-GCAUUUGCAGACAGGUCAA-3′/5′-UUGAACUGUCUGCAAAUGC-3′; SRC661, 5′-GCAA -3′/5′-UUGAACCACUGUACGUUGC-3′; SR688, 5′-GCUCAUCUGCAGACCCAUU-3′/5′-AAUGGGUCUGCAGAUGAGC-3′; SRC688, 5′-GCUUCTACGCAGCCCAAUU-3′/5′-AAUUGGGCUGCGUAGAAGC-3′. We used Trans-messenger reagent kit (Qiagen) and protocols for the transfection of the BT-549 cells with the siRNAs. RNA was prepared by TRIZOL reagent and DNase I treated before RT-PCR analysis.
RT-PCR evaluation of SLUG and other mRNA levels
Total RNA was isolated from cells and analyzed for SLUG or cytokeratins by RT-PCR analysis (Amersham, Arlington Heights, IL). β-Actin was used as a loading control. Total RNAs (5 μg) were treated with DNase I (Promega, Madison, WI) and reverse transcribed with Superscript II (Invitrogen). The resulting cDNA was used to carry out PCR amplification of SLUG (807 bp; 5′-CCTGGTCAAGAAGCATTTCAACG-3′ and 5′-CCCCAAAGATGAGGAGTATCCG-3′), cytokeratin 8 (255 bp; CK8: 5′-TTCCTGGAGCAGCAGAACAA-3′ and 5′-GAGGACAAATTCGTTCTCCAT-3′), cytokeratin 19 (238 bp; CK19: 5′-ATTCTTGG TGCCACCATTGA-3′ and 5′-TCCTCATGGTTCTTCTTCAGG-3′), and β-actin (353 bp; 5′-GCTCGTCGTCGACAACGGCTC-3′ and 5′-CAAACATGATCTGGGTCATCTTCTC-3′). The cytokeratin primers were designed from human cDNA sequences. The forward and the reverse primers were designed from exon 2 and exon 3, respectively.
Promoter constructs and luciferase assay
CK8 and CK19 gene promoters were amplified from genomic DNA isolated from BT-549 cells using specific primer sets (5′-TCAACGGATCTCGCTC-3′/5′-TTGTCTCTCTCTTCCAAGAC-3′ for CK8; 5′-AACGCATGCTTTGGGGGATG-3′/5′-TCCCCCTTTACTCGGCCCCAC-3′ for CK19). The amplified products were initially cloned into pCRII-Topo (Invitrogen) and nucleotide sequences of the inserts were verified. Recombinant plasmids with appropriate orientation of the insert were digested out with HindIII and either with EcoRV (for CK8) or with XhoI (for CK19) and sub-cloned into the HindIII/SmaI (for CK8) or HindIII/XhoI (for CK19) site at the MCS of pGL3-Basic plasmid (Promega), as described before [18]. Restriction fragments from pCRII constructs that have reverse orientations of their inserts were subcloned into pGL3-Basic plasmid to give the negative control promoter constructs. Cells were seeded at 85% confluency in a 24-well plate for 24 h in their growth media before co-transfection with one of the pGL3-Basic plasmid constructs and pRL-TK plasmid. The latter was used as a transfection normalization control. Plasmids were mixed at 0.5 μg per well and transfection was done using the Lipofectamine Plus transfection reagent (Invitrogen) using the protocol suggested by the supplier. After 20 h of incubation in complete medium at 37 °C, the cells were lysed in 100 μl passive lysis buffer (Promega) and 5–20 μl of the lysate was assayed [18] for firefly luciferase as well as Renilla luciferase activities using Dual Luciferase Assay Reagents (Promega) following suggested protocols [18]. Firefly luciferase activity was normalized with respect to Renilla luciferase activity and presented as a ratio (relative light units, RLU). Protein contents of the extract, when needed, were determined using RC-DC reagents and protocol from Bio-Rad Laboratories [18].
Statistical analysis
All experimental data were expressed as means ± SE. A one-way analysis of variance, a two-way repeated measure analysis of variance, and Student’s t test were used to determine the significance of the difference [22].
Results and discussion
Differential expressions of genes in BT-549 and MDA-MB-468 cells
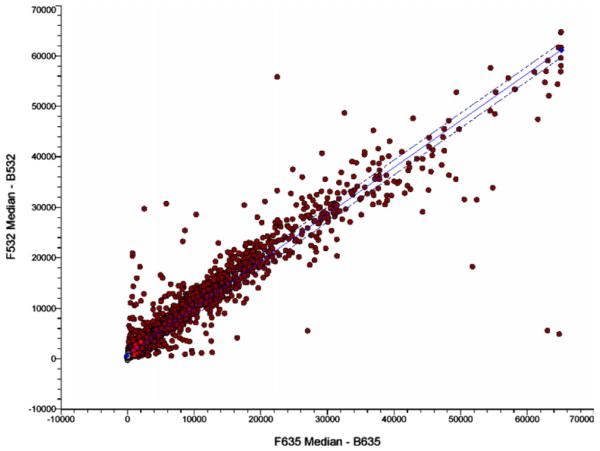
We compared the relative expressions of 11,607 human genes in the invasive BT-549 cells and non-invasive MDA-MB-468 cells using cDNA microarray. Interestingly, there were not many differences in the levels of abundantly expressed transcripts (Fig. 1). Only 12 genes showed reproducible (n =3 × 2) changes in their transcript levels in both of these cells (Table 1). Out of the 12 differentially expressed genes eight of the genes are up-regulated in MDA-MB-468 cells whereas four of them are down-regulated (Table 1). We have verified the differential expressions of all these genes by RT-PCR analysis (data not shown). Obviously, variations (if any) of low abundance transcripts are not noticeable by this technique. Thus, Table 1 only represents the differentially expressed abundant mRNAs. We only describe here our further followup studies on the remarkable up-regulation of CK8 and CK19 transcripts in MDA-MB-468 cells as these are regulated by SLUG (see below). Verifications of CK transcript levels are shown in Fig. 2.
Fig. 1.
Microarray analysis of the mRNAs from BT-549 and MDA-MB-468 cells. A representative scatter plot showing differential expressions of only a few abundant mRNAs. The cDNAs from MDA-MB-468 cells were labeled with Cy5 and those from BT-549 cells were labeled with Cy3.
Table 1.
Human genes up- or down-regulated in MDA-MB-468 cells as compared to BT-549 cells (12 out of 11,607)
| Gene name | ID# | Fold increase (+) or decrease (−) |
|---|---|---|
| Up-regulated | ||
| Cytokeratin 8 | 897781 | 6.1 (+) |
| Cytokeratin 19 | 810131 | 9.7 (+) |
| Ubiquitin like 5 | 66767 | 4.9 (+) |
| Sec61 gamma | 358456 | 11.3 (+) |
| Phosphatidylcholine transfer protein | 469924 | 13.3 (+) |
| Annexin A8 | 666879 | 4.6 (+) |
| Serine protease inhibitor, Kunitz type 2 | 814378 | 3.2 (+) |
| Protein tyrosine phosphatase, receptor type, F | 897788 | 3.3 (+) |
| Down-regulated | ||
| Metallothionein 1G | 202535 | 11.5 (−) |
| Metallothionein 1L | 297392 | 11.9 (−) |
| RNA helicase related protein | 245990 | 9.8 (−) |
| Tumor rejection antigen (gp96) 1 | 897690 | 5.3 (−) |
Fig. 2.
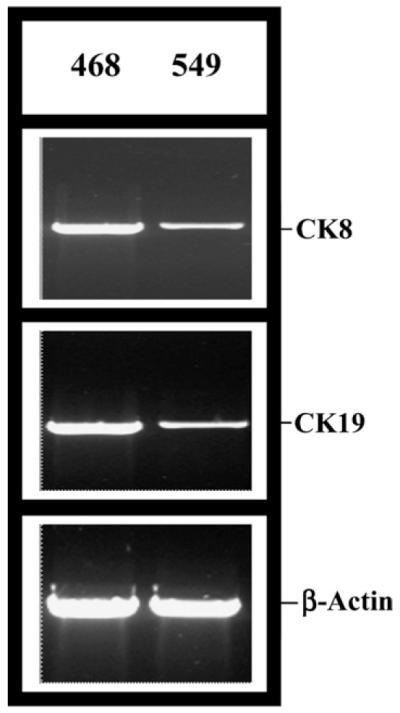
Validation of the differential expressions of cytokeratin 8 (CK8) and cytokeratin 19 (CK19) in MDA-MB-468 and BT-549 cells. RT-PCR analysis showing differential expression of CK 8 and CK 19 mRNA from RNA isolated from MDA-MB-468 (Slug −ve) cells and BT-549 (Slug +ve) cells. β-actin was used as a loading control.
Cytokeratins are the products of a large gene family of the intermediate filament genes [23]. The family is divided up into six types or sub-classes based on the sequence characteristics of the genes and their products, of which cytokeratins make up type I (CK9–CK20) and type II (CK1–CK8) groups [24]. It was recently estimated that there are at least 65 functional intermediate filament genes in the human genome, of which 54 are keratins [25]. Like other intermediate filaments, keratins are characterized by tissue-specific expression patterns. CK19 is the representative of the acidic type I cytokeratins whereas CK8 belongs to the basic type II cytokeratins [26]. On the other hand, both of these cytokeratins are of the luminal type and their expressions in the breast luminal duct may determine the grade and invasiveness of the cancer [26]. Indeed, down-regulation of a luminal cytokeratin, CK18, has recently been shown to be associated with the progression of human breast cancer [27]. Up-regulation of the luminal cytokeratins in a non-invasive breast cancer cell is thus very interesting.
Other than the cytokeratins, there are six proteins also up-regulated in the MDA-MB-468 cells (Table 1). UBL5 is a small ubiquitin homolog expressed in all of the tissues and in many organisms [28]. The biological role of UBL5 is unknown. Sec61G is a subunit of the Sec61 complex that is the central component of the protein translocation apparatus of the endoplasmic reticulum membrane [29]. Oligomers of the Sec61 complex form a transmembrane channel where proteins are translocated across and integrated into the ER membrane. This complex consists of three membrane proteins—α, β, and γ. This gamma gene is up-regulated in MDA-MB-468 cells (Table 1). Phosphatidylcholine transfer protein is a cytosolic protein that catalyzes intermembrane transfer of phosphatidylcholines [30]. Annexin A8 belongs to the family of Ca2+-dependent phospholipid-binding proteins that inhibit coagulation and phospholipase A2 activity [31]. The gene is also found to be selectively overexpressed in acute myelocytic leukemia [32]. The serine protease inhibitor, Kuniz type 2 [33], and the protein tyrosine phosphase, receptor type, F [34], take part in distinct cellular processes. The relevance of the up-regulation of these proteins is yet to be determined.
Among the genes that are up-regulated in the invasive BT-549 cells and down-regulated in the non-invasive MDA-MB-468 cells are the metallothioneins (Table 1). Metallothioneins (MTs), a group of inducible, low molecular weight, cysteine-rich proteins [35], are known to influence tumor growth by affecting both cell proliferation and death [36,37]. MT overexpression in ductal breast cancer [38] appears to be predominantly associated with more aggressive and highergrade tumors. Human MT proteins encoded by a family of genes consisting of 10 functional MT isoforms may play different roles during development or under various physiological conditions [35,38]. Specific roles played by metallothioneins 1G and 1L, if any, as is related to invasiveness are not known. Further studies on the down-regulation of these genes in BT-549 cells followed by invasiveness assays may reveal their importance in this process. The significance of the other two genes that are down-regulated in MDA-MB-468 cells, namely the RNA helicase related protein [39] and the tumor rejection antigen (gp96) 1 [40], needs to be evaluated. We have validated the cytokeratin and metallothionein gene expression patterns in the non-invasive MCF7 breast cancer cells and highly aggressive MDA-MB-231 breast cancer cells (data not shown).
Expression of SLUG gene determines the expressions of cytokeratins 8 and 19 in human breast cells
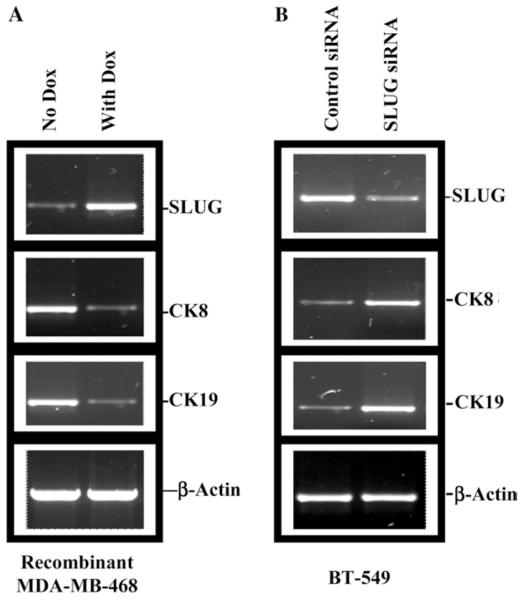
As mentioned above, one of the known genotypic differences between BT-549 and MDA-MB-468 cells is the absence of SLUG in the latter [1]. We have further verified this difference in SLUG gene expression by Northern, end point, and real-time RT-PCR analyses (data not shown). Since SLUG is a transcriptional silencer, our hypothesis was that SLUG controls at least some of the genes that are up-regulated in the MDA-MB-468 cells. To test this hypothesis we overexpressed SLUG in the MDA-MB-468 cells from a doxycycline-inducible retroviral promoter and compared the expressions of genes in the presence and the absence of doxycycline by cDNA microarray analysis. We have verified the induced expression of SLUG in these cells by RT-PCR analysis (Fig. 3A). Out of the 11,607 genes tested in the microarray analysis, only two were significantly repressed in the presence of doxycycline. They are the CK8 (8.2-fold) and CK19 (6.7-fold) genes. We recognize that the retroviral system is somewhat leaky (Fig. 3A) and degrees of repressions of the expressions of other genes in the presence and the absence of doxycycline may have been compromised by that fact. Use of more tightly regulated inducible system may reveal whether SLUG also regulates the expressions of other six genes that are up-regulated in the MDA-MB-468 cells (Table 1).
Fig. 3.
Effect of inducible expression or knock down of SLUG mRNA in MDA-MB-468 and BT-549 cells, respectively. (A) RT-PCR analysis showing doxycycline-inducible expression of SLUG mRNA from retroviral promoter in MDA-MB-468 cells and its effect on CK8 and CK19 mRNA level. β-Actin was used as loading control. +Dox: RNA isolated from cells incubated with doxycycline (1 μg/ml) for 48 h. −Dox: RNA isolated from cells incubated in parallel in absence of doxycycline for 48 h. (B) RT-PCR analysis showing the effect of SLUG mRNA knock down on CK8 and CK19 mRNA levels in BT-549 cells. β-Actin was used as loading control. Con: BT-549 cells were transfected with scramble siRNA (SRC661), RNA isolated after 48 h of treatment and used as negative control. KD: BT-549 cells were transfected with anti-SLUG siRNA (SR661), to knock down SLUG gene expression, RNA isolated after 48 h of treatment.
To further verify that SLUG regulates the expressions of the cytokeratin genes, we ablated the expression of SLUG using three different sets of anti-SLUG siRNAs (see Materials and methods) in BT-549 cells and tested the expressions of the cytokeratins by RT-PCR analysis. SLUG mRNA levels in the BT-549 cells treated with anti-SLUG siRNA decreased 2- to 3-fold as compared to that in the cells treated with corresponding control siRNA (Fig. 3B). The cells that have anti-SLUG siRNA-induced decreased levels of SLUG mRNA have increased levels of both CK8 (4- to 5-fold) and CK19 (3- to 4-fold) transcripts as compared to the control siRNA treated cells (Fig. 3B). These data further suggest that SLUG may be inhibitory to the expression of CK8 and CK19 genes in human breast cells. Data with SR661 siRNA are shown. Similar results were obtained with other two siRNAs (data not shown). Control siRNAs are made with changes in the nucleotide sequences so that they will not be able to bind to the target mRNA. We used the expression of β-actin gene as normalization control (Figs. 3A and B). Whether the SLUG ablated BT-549 cells are less invasive is under study using matrigel outgrowth assay.
The functions of the promoters of CK8 and CK19 genes are regulated by SLUG
To understand the possible mechanism of regulation of the cytokeratin genes by SLUG, we tested the promoters of these genes. We amplified ~1.7 kb genomic DNA fragments containing the putative promoters of cytokeratin 8 and cytokeratin 19 genes from total DNA isolated from BT-549 cells (Figs. 4A and B). SLUG binds to the E2 box sequence (5′-CACCTG-3′) to mediate its repressor function [2,3]. We found one potential E2 box sequence (−203 to −198) at the CK8 gene promoter and two such E2 boxes (−1016 to −1011; +483 to +488) at the CK19 promoter (Figs. 4A and B). We cloned these promoters in pGL3-Basic plasmid in proper orientations and transiently transfected the MDA-MB-468 or BT-549 cells with the constructs. Activities of both of the CK8 and CK19 promoters are repressed when the expression of SLUG was induced by doxycycline in the recombinant MDA-MB-468 cells (Table 2). On the other hand, the activities of both of these promoters were increased 2- to 4-fold in BT-549 cells that were treated with anti-SLUG siRNA as compared to that treated with the corresponding control siRNA (Table 2). Data with SR661 siRNA are shown. Other siRNAs yielded similar results (data not shown). SV40 promoter in pGL3-Control (Promega) plasmid was used as a control promoter. The activity of SV40 promoter did not alter significantly at different SLUG expression status of the cells (Table 2). It thus appears that SLUG may regulate the cytokeratin gene expressions in the breast cells tested by interacting with their promoters. Further mutational analysis of the promoter elements is underway to better understand this regulation. It will be interesting to study whether SLUG-mediated regulation of cytokeratin genes also occurs in normal breast cells. This study will add to the growing interests of scientists in understanding the role of SLUG-like proteins in the epithelial cell differentiation and cancer cell metastasis [41,42].
Fig. 4.
Nucleotide sequences of the putative promoters of human CK8 and CK19 genes indicating the locations of the E2 boxes. (A) CK8 promoter: 1703 bp sequence (−1102 to +601 consists of 1102 bp upstream sequence + 334 bp exon 1 + 267 bp from intron 1). The E2 box is located at −203 to −198. (B) CK19 promoter: 1768 bp sequence (−1174 to +594 consists of 1174 bp upstream sequence + 453 bp exon 1 + 141 bp from intron 1). There are two potential E2 boxes: E2 box 1 (−1016 to −1011) and E2 box 2 (+483 to +488). E2-box sequences (5′-CACCTG-3′) and larger size, in uppercase letters, are bold-faced and underlined. The exon 1 sequences are written in uppercase letters.
Table 2.
Activities of CK8 and CK9 gene promoters in MDA-MB-468 and BT-549 cells with different SLUG expression status
| Promoter | Luciferase activity (RLU)c |
|||
|---|---|---|---|---|
| Recombinant MDA-MB-468d |
BT-549 |
|||
| No Dox | With Dox | Control siRNAa | Anti-SLUG siRNAb | |
| CK8 | 27.6 ± 1.1 | 7.1 ± 3.3 | 47.5 ± 4.3 | 201.6 ± 3.7 |
| CK19 | 31.1 ± 2.3 | 10.5 ± 2.1 | 66.4 ± 3.1 | 169.7 ± 7.1 |
| SV40 | 42.7 ± 2.2 | 46.1 ± 3.2 | 92.1 ± 5.1 | 90.3 ± 4.1 |
‘No Dox,’ no doxycycline added; ‘with Dox,’ doxycycline (1 μg/ml) added for 48 h.
Cells treated with SRC661 control siRNA for 48 h.
Cells treated with SR661 anti-SLUG siRNA for 48 h.
Results are means (n = 12) ± SE. The differences (RLU) between ‘no Dox’ and ‘with Dox’ for MDA-MB-468 cells and ‘control siRNA’ and ‘anti-SLUG siRNA’ for BT-549 cells for the CK8 and CK19 promoters are statistically significant (p < 0.001).
MDA-MB-468 cells harboring Tet-On repressor and introduced SLUG gene under the control of doxycycline-inducible retroviral promoter.
Acknowledgments
We thank Ms. Rati M. Tripathi for technical support. This work was supported by the MMC/VICC cancer partnership Grant #1U54CA091408-010003 from NCI and the DOD Grant #DAMD17-00-1-0341 to G.C.
References
- [1].Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- [2].Hemavathy K, Ashraf SI, Ip YT. Snail/slug family of repressors: slowly going into the fast lane of development and cancer. Gene. 2000;257:1–12. doi: 10.1016/s0378-1119(00)00371-1. [DOI] [PubMed] [Google Scholar]
- [3].Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell. Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- [4].Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumor cells. Nat. Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- [5].Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- [6].Hajra KM, Fearon ER. Cadherin and catenin alterations in human cancer. Genes Chromosomes Cancer. 2002;34:255–268. doi: 10.1002/gcc.10083. [DOI] [PubMed] [Google Scholar]
- [7].Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium–mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J. Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- [8].Kajita M, McClinic KN, Wade PA. Aberrant expression of the transcription factors snail and slug alters the response to genotoxic stress. Mol. Cell. Biol. 2004;24:7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jiang R, Lan Y, Norton CR, Sundberg JP, Gridley T. The Slug gene is not essential for mesoderm or neural crest development in mice. Dev. Biol. 1998;198:277–285. [PubMed] [Google Scholar]
- [10].Ip YT, Gridley T. Cell movements during gastrulation: snail-dependent and independent pathways. Curr. Opin. Genet. Dev. 2002;12:423–429. doi: 10.1016/s0959-437x(02)00320-9. [DOI] [PubMed] [Google Scholar]
- [11].Inukai T, Inoue A, Kurosawa H, Goi K, Shinjyo T, Ozawa K, Mao M, Inaba T, Look AT. SLUG, a ces-1-related zinc finger transcription factor gene with anti-apoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Mol. Cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- [12].Inoue A, Seidel MG, Wu W, Kamizono S, Ferrando AA, Bronson RT, Iwasaki H, Akashi K, Morimoto A, Hitzler JK, Pestina TI, Jackson CW, Tanaka R, Chong MJ, McKinnon PJ, Inukai T, Grosveld GC, Look AT. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation induced apoptosis in vivo. Cancer Cell. 2002;2:279–288. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- [13].Perez-Losada J, Sanchez-Martin M, Perez-Caro M, Perez-Mancera PA, Sanchez-Garcia I. The radioresistance biological function of the SCF/kit signaling pathway is mediated by the zinc-finger transcription factor Slug. Oncogene. 2003;22:4205–4211. doi: 10.1038/sj.onc.1206467. [DOI] [PubMed] [Google Scholar]
- [14].Watts GS, Oshiro MM, Junk DJ, Wozniak RJ, Watterson S, Domann FE, Futscher BW. The acetyltransferase p300/CBP-associated factor is a p53 target gene in breast tumor cells. Neoplasia. 2004;6:187–194. doi: 10.1593/neo.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Thompson EW, Paik S, Brunner N, Sommers CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME. Association of increased basement membrane invasiveness with absence of estrogen receptor and expression of vimentin in human breast cancer cell lines. J. Cell. Physiol. 1992;150:534–544. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- [16].Carlson CA, Ethier SP. Lack of RB protein correlates with increased sensitivity to UV-radiation-induced apoptosis in human breast cancer cells. Radiat. Res. 2000;154:590–599. doi: 10.1667/0033-7587(2000)154[0590:lorpcw]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- [17].Gjetting T, Lukas J, Bartek J, Strauss M. Regulated expression of the retinoblastoma susceptibility gene in mammary carcinoma cells restores cyclin D1 expression and G1-phase control. Biol. Chem. Hoppe Seyler. 1995;376:441–446. [PubMed] [Google Scholar]
- [18].Sharan C, Hamilton N, Parl AK, Singh PK, Chaudhuri G. Identification and characterization of a transcriptional silencer upstream of human BRCA2 gene. Biochem. Biophys. Res. Commun. 1999;265:285–290. doi: 10.1006/bbrc.1999.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tripathi MK, Chaudhuri G. Down-regulation of UCRP and UBE2L6 in BRCA2 knocked-down human breast cells. Biochem. Biophys. Res. Commun. 2005;328:43–48. doi: 10.1016/j.bbrc.2004.12.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tusher GV, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Campbell MJ, Machin D. Medical Statistics: A Commonsense Approach. second ed. John Wiley; New York: 1994. [Google Scholar]
- [23].He T, Stepulak A, Holmstrom TH, Omary MB, Eriksson JE. The intermediate filament protein keratin 8 is a novel cytoplasmic substrate for c-Jun N-terminal kinase. J. Biol. Chem. 2002;277:10767–10774. doi: 10.1074/jbc.M111436200. [DOI] [PubMed] [Google Scholar]
- [24].Schutte B, Henfling M, Kolgen W, Bouman M, Meex S, Leers MP, Nap M, Bjorklund V, Bjorklund P, Bjorklund B, Lane EB, Omary MB, Jornvall H, Ramaekers FC. Keratin 8/18 breakdown and reorganization during apoptosis. Exp. Cell Res. 2004;297:11–26. doi: 10.1016/j.yexcr.2004.02.019. [DOI] [PubMed] [Google Scholar]
- [25].Uenishi T, Kubo S, Yamamoto T, Shuto T, Ogawa M, Tanaka H, Tanaka S, Kaneda K, Hirohashi K. Cytokeratin 19 expression in hepatocellular carcinoma predicts early postoperative recurrence. Cancer Sci. 2003;94:851–857. doi: 10.1111/j.1349-7006.2003.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Abd El-Rehim DM, Pinder SE, Paish CE, Bell J, Blamey RW, Robertson JF, Nicholson RI, Ellis IO. Expression of luminal and basal cytokeratins in human breast carcinoma. J. Pathol. 2004;203:661–671. doi: 10.1002/path.1559. [DOI] [PubMed] [Google Scholar]
- [27].Woelfle U, Sauter G, Santjer S, Brakenhoff R, Pantel K. Down-regulated expression of cytokeratin 18 promotes progression of human breast cancer. Clin. Cancer Res. 2004;10:2670–2674. doi: 10.1158/1078-0432.ccr-03-0114. [DOI] [PubMed] [Google Scholar]
- [28].Friedman JS, Koop BF, Raymond V, Walter MA. Isolation of a ubiquitin-like (UBL5) gene from a screen identifying highly expressed and conserved iris genes. Genomics. 2001;71:252–255. doi: 10.1006/geno.2000.6439. [DOI] [PubMed] [Google Scholar]
- [29].Hartmann E, Sommer T, Prehn S, Gorlich D, Jentsch S, Rapoport TA. Evolutionary conservation of components of the protein translocation complex. Nature. 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- [30].Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- [31].Morgan RO, Martin-Almedina S, Iglesias JM, Gonzalez-Florez MI, Fernandez MP. Evolutionary perspective on annexin calcium-binding domains. Biochim. Biophys. Acta. 2004;1742:133–140. doi: 10.1016/j.bbamcr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- [32].Chang KS, Wang G, Freireich EJ, Daly M, Naylor SL, Trujillo JM, Stass SA. Specific expression of the annexin VIII gene in acute promyelocytic leukemia. Blood. 1992;79:1802–1810. [PubMed] [Google Scholar]
- [33].Suzuki M, Kobayashi H, Fujie M, Nishida T, Takigawa M, Kanayama N, Terao T. Kunitz-type protease inhibitor bikunin disrupts phorbol ester-induced oligomerization of CD44 variant isoforms containing epitope v9 and subsequently suppresses expression of urokinase-type plasminogen activator in human chondrosarcoma cells. J. Biol. Chem. 2002;277:8022–8032. doi: 10.1074/jbc.M108545200. [DOI] [PubMed] [Google Scholar]
- [34].Blanchetot C, Tertoolen LG, Overvoorde J, den Hertog J. Intra- and intermolecular interactions between intracellular domains of receptor protein-tyrosine phosphatases. J. Biol. Chem. 2002;277:47263–47269. doi: 10.1074/jbc.M205810200. [DOI] [PubMed] [Google Scholar]
- [35].Kägi JHR, Schaffer A. Biochemistry of metallothionein. Biochemistry. 1998;27:8509–8515. doi: 10.1021/bi00423a001. [DOI] [PubMed] [Google Scholar]
- [36].Kägi JHR. Overview of metallothionein. Methods Enzymol. 1991;205:613–626. doi: 10.1016/0076-6879(91)05145-l. [DOI] [PubMed] [Google Scholar]
- [37].Schwarz MA, Lazo JS, Yalowich JC, Allen WP, Whitmore M, Bergonia HA, Tzeng E, Billiar TR, Robbins PD, Lancaster JR, Jr., Pitt BR. Metallothionein protects against the cytotoxic and DNA-damaging effects of nitric oxide. Proc. Natl. Acad. Sci. USA. 1995;92:4452–4456. doi: 10.1073/pnas.92.10.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jin R, Bay BH, Chow VT, Tan PH. Metallothionein 1F mRNA expression correlates with histological grade in breast carcinoma. Breast Cancer Res. Treat. 2001;66:265–272. doi: 10.1023/a:1010658907462. [DOI] [PubMed] [Google Scholar]
- [39].Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Maki RG, Old LJ, Srivastava PK. Human homologue of murine tumor rejection antigen gp96: 5′-regulatory and coding regions and relationship to stress-induced proteins. Proc. Natl. Acad. Sci. USA. 1990;87:5658–5662. doi: 10.1073/pnas.87.15.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kang Y, Massagué J. Epithelial–mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- [42].Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]