Abstract
We describe a new octoploid species of African clawed frog (Xenopus) from the Lendu Plateau in the northern Albertine Rift of eastern Democratic Republic of the Congo. This species is the sister taxon of Xenopus vestitus (another octoploid), but is distinguished by a unique morphology, vocalization and molecular divergence in mitochondrial and autosomal DNA. Using a comprehensive genetic sample, we provide new information on the species ranges and intra-specific diversity of African clawed frogs from the Albertine Rift, including the details of a small range extension for the critically endangered Xenopus itombwensis and previously uncharacterized variation in Xenopus laevis. We also detail a new method for generating cytogenetic preparations in the field that can be stored at room temperature for up to 3 weeks. While extending our understanding of the extant diversity in the Albertine Rift, this new species highlights components of species diversity in ancestral African clawed frogs that are not represented by known extant descendants.
Keywords: allopolyploid evolution, Albertine Rift, whole genome duplication, advertisement calls, DNA barcode
Introduction
African clawed frogs in the genus Xenopus have a complex evolutionary history characterized by both bifurcating and reticulating modes of speciation (reviewed in Evans, 2008). Described species include eight tetraploids, six octoploids and two dodecaploids; additional undescribed species have been inferred using molecular markers (Evans et al., 2004; Evans et al., 2005; Evans, 2007). Some polyploid species, such as the octoploids Xenopus itombwensis and Xenopus wittei, are sister taxa that probably evolved by ‘regular’ bifurcating speciation from an octoploid ancestor. Other species pairs, such as the octoploids Xenopus andrei and Xenopus vestitus, share recent common ancestry of half of their genomes while the other half is more divergent. This observation is best explained by allopolyploid speciation – speciation by genome duplication that is associated with hybridization among species. This mode of speciation is further suggested by the creation of allopolyploid individuals by hybridizing and backcrossing different species in the laboratory (Kobel & Du Pasquier, 1986; Kobel, 1996).
Phylogenetic relationships among extant African clawed frogs (Xenopus and Silurana) suggest the previous existence of up to three ancestral diploids and three ancestral tetraploid species that are not represented by a known extant species with the same ploidy level (Evans, 2007, 2008). Even though these species may now be extinct, the genomes of these ancestors persist as a portion of the genomes of extant allopolyploid species. The prediction that these ‘lost’ ancestral species once existed provides motivation for field studies aimed at discovering their equivalent-ploidy-level descendants, if they exist.
In an ongoing effort to understand the evolution and diversity of clawed frogs, we performed field collections and molecular analyses in regions where allopolyploid descendants of the predicted tetraploid ancestors currently occur, including the Albertine Rift of Central Africa. This effort recently resulted in the discovery of the octoploid species X. itombwensis (Evans et al., 2008) and subsequent searches identified another new species that is described here. The inferred evolutionary relationships between this new species, which is octoploid, and other extant species further support the notion that tetraploid ‘lost’ ancestors once existed.
Methods
Field collection
The new species was first collected by E. G. and C. K. in July 2009 on the Lendu Plateau, northern Albertine Rift, Democratic Republic of the Congo (DRC), and molecular analyses prompted a subsequent expedition by B. J. E. and C.K. to this locality in December 2009, in order to further characterize this species in terms of its morphology, distribution and vocalization, and to provide additional voucher specimens. Other samples in this study were collected by E. G., C. K., B. J. E. and colleagues, or are reported previously (Evans et al., 2004).
Analysis of male vocalization and morphology
Male clawed frogs produce species-specific advertisement vocalizations (calls) by opening a pair of cartilaginous disks in the larynx to produce a ‘click’ (Yager, 1992). Calls of some species are composed of only one click, whereas others produce trains of clicks called a trill. In this study, we recorded advertisement calls from four males from the new species and compared these with a previous analysis of calls from X. itombwensis, X. vestitus and X. wittei (Evans et al., 2008) with data from two additional calls from X. wittei. Based on analyses discussed below, these species are closely related to the new species. We report vocal parameters from only two individuals from the new species because the intervals between clicks of the other two were not sufficiently distinguishable for individual clicks to be measured.
We measured call duration, the number of clicks per call, the inter-click interval (time between the start of one click and the start of the next click), the intensity modulation (the change in intensity between the softest and the loudest clicks calculated as [(max–min)/min], the call duty cycle (the proportion of vocalization in a bout of calls), and the two sound frequencies with the most energy (dominant frequencies; DFlow is the lower and DFhigh is the higher of the two frequencies) and inter-call interval (time between successive calls in a bout of calling). Most of these parameters have a single value for each call. The inter-click interval is a mean value calculated from all inter-click intervals in a call. Call duty cycle and inter-call interval are measured from bouts of advertisement calls. Advertisement calls, or bouts of advertisement calls, were randomly selected and analyzed. Evans et al. (2008) ranked the dominant frequencies by amplitude and presented the mean across individuals of the highest and second-highest frequencies. Here, we instead rank the two frequencies with the highest amplitude by frequency and report the mean of these frequencies (DFlow and DFhigh). This is more appropriate because the two frequencies vary in relative amplitude from call to call. For almost all of these parameters, multiple calls from at least two individuals were analyzed. To communicate information about inter- and intra-individual variation in the vocal parameters, we report the grand mean and standard deviation across all replicates, and the sum of squares and residuals divided by their respective degrees of freedom, which is the number of individuals minus one for comparisons among individuals or the number of calls minus one for comparisons among calls.
Advertisement calls were evoked by priming males with human chorionic gonadotropin (Sigma, Oakville, ON, Canada; 100 international units). Vocalizations were recorded ~ 6 h after the injection, at the start of the night cycle. An injected male and a sexually unreceptive female were placed in a 75.7L aquarium, two-thirds filled with unchlorinated water at 20 °C. Recordings were made with a hydrophone (HighTech Inc.; Gulfport, MS, USA sensitivity = −164.5 dB re 1VuPa−1), stored on CDs (Marantz, Mahawah, NJ, USA; model CDR300) and analyzed using signal software version 5.0 (Engineering Design, Berkeley, CA, USA).
To provide information on comparative morphology, we measured 12 external morphological characteristics following Tinsley (1973) from preserved specimens of the new species and of a sympatric population of Xenopus laevis. Additional information on specimens examined is available in the supporting information.
Cytogenetics
We previously reported a new method to obtain Xenopus cytogenetic preparations by directly harvesting and fixing liver tissue (Evans et al., 2008). Here, we adapted this method to allow the cells to be harvested and fixed in the field, and stored at room temperature for up to 3 weeks during transport back to the laboratory. Animals were killed by immersion in an ~1% MS222 (Sigma) solution. The liver was accessed through a lateral incision and an ~8 mm3 piece was placed on a pre-cleaned glass slide, wetted with three to four drops of 0.04 M KCl and minced with a new razor blade. The resulting tissue/KCl slurry was then transferred to a 15 mL centrifuge tube and 0.04 M KCl solution was added to produce a final volume of 13.5 mL. The mixture was gently inverted/agitated and incubated at room temperature for 1 h with occasional agitation and inversion. During this time, a fresh 3:1 methanol:glacial acetic acid fixative was prepared. After a final agitation, the tissue/KCl mixture was left for 2 min to allow debris to settle to the bottom of the tube. Ten millilitres was drawn from the top and transferred to a new 50 mL tube. The remaining cell suspension and tissue debris were discarded. Two mL of the fixative were added to the 50 mL tube, followed by gentle agitation. An additional 10 mL of fixative was then added, followed by gentle agitation. The tube was then topped off with fixative to a total of 50 mL, capped and gently agitated. At this point, the samples were stored at room temperature for up to 3 weeks until they were transported back to the laboratory.
In the laboratory, 5–12 mL of the field-fixed samples were transferred to a 15 mL tube, topped up to 13 mL with a fresh 3:1 fixative and washed four times by centrifugation at 500 g for 8 min. For each wash, the supernatant was removed by aspiration down to 0.25 mL and resuspended in 13 mL of a fresh fixative by vortexing. In the final resuspension, 0.5–3 mL of fixative was then added to yield a suitable concentration for slide making. Cleaned slides were wetted with a drop of 3:1 fixative and left to almost dry before a drop of cell suspension was allowed to fall on the surface from a height of ~10 cm. After ~7 s, an additional drop of 3:1 fixative was allowed to fall onto the slide before it was moved to a humid environment and allowed to dry completely. Slides were incubated at 90 °C for 1 h, cooled to room temperature, stained in 8% (v/v) Giemsa in Gurr buffer (pH 6.8) for 2–5 min, rinsed three times in milli-Q H2O and dried. Using standard bright-field microscopy, metaphases were located on slides and then captured as digital images. The remaining cell suspensions were stored at −20 °C.
Molecular data, phylogenetic analysis
We estimated the phylogenetic history of the new species using molecular data from mitochondrial DNA (mtDNA) and autosomal DNA (aDNA). The mtDNA data included 819–2924 base pairs (bp) from 12S and 16S ribosomal DNA, the intervening transfer RNAVal and cytochrome c oxidase subunit I (CO1) genes from all known species of African clawed frog, including 2920 or 2921 bp from four samples of the new species. Pipa pipa was used as an outgroup to African clawed frogs (Roelants & Bossuyt, 2005). Each species in the mtDNA dataset had at least one full-length (> 2900 bp) sequence; shorter sequences were included to characterize species’ ranges. The aDNA data included cloned paralogs from 317–3054 bp of paralogs of the RAG1 from all known species of African clawed frogs, including 1141 bp from three paralogs from one individual of the new species. MtDNA and aDNA were amplified using previously published primers and aDNA was cloned using Invitrogen cloning kits (Burlington, ON, Canada) as described previously (Evans et al., 2004, 2005; Ivanova, deWaard & Hebert, 2006; Evans, 2007). Data are deposited in Genbank (accession numbers are in supporting information Table S1 and Evans et al., 2004, 2008; Evans, 2007).
Phylogenetic relationships among mtDNA sequences were estimated using MrBayes version 3.1.2 (Huelsenbeck & Ronquist, 2001) using a partitioned analysis with a stem-loop (doublet) model for the rDNA genes and a general time-reversible model with a proportion of invariant sites and a gamma-distributed rate heterogeneity parameter (GTR + I + Γ) for the CO1 sequence, which was selected using the Akaike Information Criterion with MrModeltest (Nylander, 2004). The RAG1 phylogeny was also estimated using MrBayes with partitioning by codon position and a GTR + I + Γ model for each position, with model selection using Bayes factors described in Evans (2007). Both analyses used Dirichlet priors for the rate matrix. Two independent MCMC runs were performed for 2.6 and 1.8 million generations for mtDNA and RAG1, respectively. A burnin of 500 000 and 100 000 generations (mtDNA and RAG1 datasets, respectively) was discarded before the construction of a consensus topology based on plots of the posterior distribution of tree likelihoods and parameter estimates.
Species concept
Essentially all species concepts share a common theme of referring to evolutionarily distinct lineages that persist through time, multiple lines of evidence can be used to identify species, and the chronology by which these lines of evidence become diagnostic varies (de Queiroz, 1998). Even at a genetic level, the onset of genetic barriers to reproduction can be a gradual process that occurs at different times in different parts of the genome (Wu, 2001). For these reasons, we adopt a general lineage concept of species, where a species is defined as a separately evolving lineage of connected subpopulations (metapopulations) through time (de Queiroz, 1998, 2007). We provide information on morphological, behavioral and genetic divergence as evidence for lineage separation.
Taxonomic account
Xenopus lenduensis, new species Lendu Plateau clawed frog
(Fig. 1)
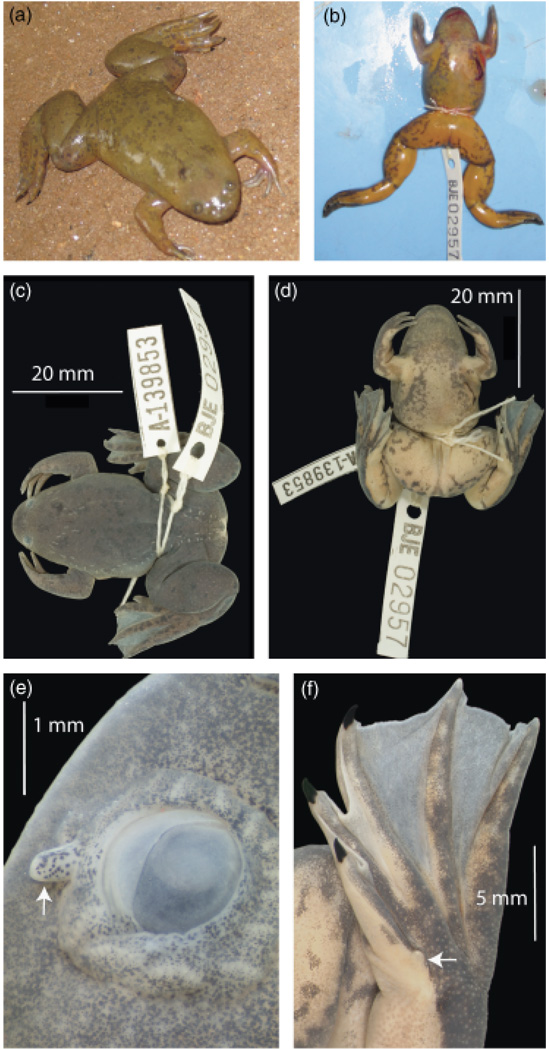
Figure 1.
Pictures of holotype MCZ A-139853, (field number BJE2957) including (a) dorsal view in life, (b) ventral view immediately after death, (c) dorsal and (d) ventral view after preservation, and close-ups after preservation indicated by arrows of the (e) subocular tentacle and (f) metatarsal tubercle. (b) Red coloration on the left side, snout and neck of the animal is blood from the incision made for genetic sampling, which appears as a maroon ‘J’ on the left side of the vent just below the attachment point of the arm.
Holotype
MCZ A-139853 (field number BJE2957), adult male: Democratic Republic of the Congo, Orientale Province, Bunia District,Djugu Territory,AboroRegion, AboroWest Village, 2.0164°N 30.8311°E, 2081ma.s.l. December 8, 2009, collected by B. J. E. and C. K.
Paratypes
MCZ A-139855 (field number BJE2945), adult male: Democratic Republic of the Congo, Orientale Province, Bunia District, Djugu Territory, Jiba Region, Goki Lake, near Jiba, 1.8688°N, 30.6959°E, 1835 m a.s.l. December 11, 2009, collected by B. J. E. and C. K.
MCZ A-139856 (field number BJE2951), other information the same as MCZ A-139855 except collection site is at 1.8604°N, 30.6880°E, 1847 m a.s.l.
MCZ A-139854 (field number BJE2958), adult female: Democratic Republic of the Congo, Orientale Province, Bunia District, Djugu Territory, Aboro Region, Ngonseneyu Town 1.9851°N, 30.8118°E, 1979 m a.s.l. December 6, 2009, collected by B. J. E. and C. K.
UTEP20186 (field number EBG2343), adult male: Democratic Republic of the Congo, Orientale Province, Bunia District, Djugu Territory, Aboro Region, 2.0087°N, 30.8352°E, 2065 m a.s.l. July 6, 2009, collected by E. G and C. K.
UTEP20192 (field number EBG2350), adult female: other information the same as UTEP20186.
Diagnosis
Evolutionary analyses discussed below indicate that X. lenduensis is a member of the vestitus-wittei subgroup of African clawed frogs (Kobel, Loumont & Tinsley, 1996), which includes X. itombwensis (Evans et al., 2008), X. wittei (Tinsley, Kobel & Fischberg, 1979) and X. vestitus (Laurent, 1972; Tinsley, 1973). While this group is not a clade because it does not include all descendants of the groups’ most recent common ancestor, it is nonetheless descriptive because it does include all descendants of one of the inferred tetraploid ancestors – ancestor ‘B.’ As detailed below, Xenopus lenduensis can be distinguished from other members of the vestitus-wittei subgroup and other clawed frogs by: (1) its range on the Lendu Plateau; the only other African clawed frog in this region is X. laevis; (2) a unique combination of morphological characters including the absence of a metatarsal claw, short toes, variable presence of dorsal and ventral patterning, a small- to medium-sized subocular tentacle and a rounded snout; (3) its male advertisement call, which is shorter than other members of the vestitus-wittei group and characterized by several unique spectral characteristics not found in other clawed frog species; (4) divergent mitochondrial and autosomal genes.
Morphology and variation
Xenopus lenduensis is a medium-sized African clawed frog with the snout–vent length (SVL) of females and males averaging 48 and 40 mm, respectively (Table 1). This species has three claws, a prominent but small metatarsal tubercle with no claw, and a subocular tentacle that sometimes extends beyond the margin of the head when viewed from above. Snout shape is variable but generally more rounded than triangular, and dorsal coloration of X. lenduensis is light green to olive (Figs 1 and 2a–p). Darker olive spots are usually present on the dorsum, variable in size from ~0.5 to 3 mm, sometimes more densely distributed caudally, or instead present as larger dark olive and irregularly shaped patches. Some individuals have dark brown or black spots on the dorsum, dark olive triangular patches sometimes posterior to the eyes and/or a dark brown patch between the nostrils and the eyes (Fig. 2d–h). Some individuals have a light band connecting the eyes. One individual was observed with a dark olive hood over the dorsal surface of the head, similar to the commonly observed condition in X. vestitus (Tinsley, 1973). Some individuals of the new species have a light olive hood similar to that variably reported in X. wittei (Tinsley et al., 1979). Nuptial pads of X. lenduensis (when present, males only) extend from the base of the arm to the tips of the fingers.
Table 1.
Morphological measurements of African-clawed frogs from the Lendu Plateau and proximal regions, Democratic Republic of the Congo, including the holotype of Xenopus lenduensis
| Xenopus lenduensis | Xenopus laevis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Females; n = 27 | Males; n = 15a | Females; n = 9 | Males; n = 14 | ||||||||||
| Measurement | Holotype | Average | StDev | Max | Average | StDev | Max | Average | StDev | Max | Average | StDev | Max |
| Body length | 37.69 | 48.10 | 5.21 | 56.43 | 40.25 | 2.76 | 46.02 | 63.19 | 4.41 | 67.75 | 49.94 | 2.67 | 57.41 |
| Body width | 15.39 | 22.01 | 4.03 | 28.26 | 16.50 | 1.39 | 18.60 | 29.58 | 4.04 | 36.28 | 19.80 | 1.22 | 22.42 |
| Head width | 8.43 | 9.50 | 0.72 | 11.02 | 8.42 | 0.69 | 10.07 | 12.34 | 1.01 | 13.36 | 10.55 | 0.41 | 11.09 |
| Snout length | 2.9 | 3.63 | 0.47 | 4.84 | 3.10 | 0.34 | 3.93 | 4.87 | 0.60 | 5.65 | 4.42 | 0.40 | 5.14 |
| Eye diameter | 1.78 | 1.88 | 0.18 | 2.24 | 1.64 | 0.22 | 2.01 | 2.76 | 0.42 | 3.27 | 2.49 | 0.19 | 2.77 |
| Interocular distance | 5.22 | 4.64 | 0.38 | 5.39 | 4.21 | 0.34 | 5.22 | 6.15 | 0.47 | 6.67 | 4.81 | 0.72 | 5.59 |
| Nostril diameter | 1.62 | 1.42 | 0.21 | 1.91 | 1.29 | 0.20 | 1.62 | 1.61 | 0.21 | 1.98 | 1.42 | 0.18 | 1.85 |
| Internarial distance | 1.98 | 1.91 | 0.21 | 2.61 | 1.67 | 0.29 | 2.09 | 2.50 | 0.33 | 3.02 | 2.31 | 0.36 | 3.30 |
| Tibia length | 16.61 | 18.33 | 1.42 | 20.48 | 16.69 | 1.10 | 18.42 | 23.84 | 2.21 | 28.05 | 20.92 | 0.91 | 22.33 |
| Fifth Toe length | 12.66 | 15.04 | 1.68 | 17.64 | 13.36 | 1.62 | 17.29 | 22.69 | 2.39 | 25.05 | 19.41 | 1.12 | 20.87 |
| Elbow-first finger | 14.62 | 15.23 | 1.00 | 16.93 | 14.57 | 0.66 | 16.30 | 21.48 | 2.24 | 24.77 | 18.54 | 0.78 | 19.70 |
| First finger length | 4.3 | 4.63 | 0.49 | 5.64 | 4.35 | 0.44 | 5.09 | 7.96 | 1.48 | 9.89 | 6.40 | 0.70 | 7.50 |
Including the holotype.
Figure 2.
Morphological variation of Xenopus lenduensis in life and after preservation. Dorsal views include preserved specimens: (a) paratype MCZ A-139856 (field number BJE2951), (b) paratype MCZ A-139854 (field number BJE2958), (c) paratype MCZ A-139855 (field number BJE2945) and live unvouchered individuals (d–h). Ventral views include preserved specimens: (i) UTEP20198 (field number EBG2356), (j) paratype MCZ A-139856, (k) paratype MCZ A-139855, and (l) paratype MCZ A-139854 and live unvouchered individuals (m–p).
The ventral surface of the legs of X. lenduensis is orange or yellow, with this color sometimes extending anteriorly over the ventral surface of the body and ventral surfaces of the arms and head, or fading to a dark creamy color over the ventral surface of the body and head, or with a fairly abrupt transition from orange to cream in the pelvic region (Fig. 2o). The legs and ventral surface sometimes have a few light spots (Fig. 2i) but more commonly with lesser pigmented patches or spots, which are sometimes pink or purple, and sometimes less or more dense on the medial portion of the legs and belly (Fig. 2j–p). Like X. vestitus (Tinsley, 1973), the digits are relatively short (Table 1).
Comparison of morphology with other species
Xenopus lenduensis can be distinguished from species in the genus Silurana, and from Xenopus amieti, X. andrei, Xenopus boumbaensis, Xenopus clivii, Xenopus fraseri, Xenopus pygmaeus and Xenopus ruwenzoriensis by the absence of a claw on the metatarsal tubercle in X. lenduensis. Xenopus lenduensis can be distinguished from Xenopus gilli and Xenopus largeni by the presence of a subocular tentacle in X. lenduensis, and by unique dorsal longitudinal blotches present in X. gilli but absent in other species of African clawed frog, including X. lenduensis. Xenopus lenduensis can be distinguished from X. laevis, Xenopus borealis, Xenopus muelleri and X. new tetraploid 1 (Evans et al., 2004) by the size of the eyes (Tinsley et al., 1979), which are much smaller in the new species. Xenopus lenduensis can be distinguished from Xenopus longipes by the comparatively short digits of X. lenduensis – for example, the fifth toe is shorter than the tibia length in X. lenduensis (Table 1), but longer than the tibia in X. longipes (Kobel et al., 1996).
Distinguishing X. lenduensis from other species in the vestitus-wittei subgroup based on morphology is best achieved with a combination of other data on provenance, vocalization and/or molecular variation. The size of X. lenduensis is similar to X. wittei and X. vestitus, which have average adult female SVL typically 46 and 47 mm, respectively, and a male SVL of 37 and 38 mm, respectively (Tinsley et al., 1979; Kobel et al., 1996). Xenopus itombwensis is slightly smaller than these species, with female SVL averaging 35 mm and male SVL around 30 mm, although this may be an underestimate of the typical size of adults (Evans et al., 2008). Dorsal coloration of X. vestitus is a marbled pattern of light silver-golden to bronze chromatophores over a brown background, sometimes with a dark transverse collar at the base of the head, a light post-orbital area or a light cephalic hood (Tinsley, 1973; Tinsley et al., 1979; Kobel et al., 1996), a pattern that is different from that generally observed in X. lenduensis. The dorsal surface of X. lenduensis sometimes has dark brown spots, such as in the holotype (Fig. 1a), whereas the dorsal surface of X. vestitus lacks dark spots because flecks of darker pigments are finely intermingled with flecks of lighter pigments, imparting a bronze to silvery bronze luster (Tinsley, 1973; Channing & Howell, 2006). Dorsal coloration of X. wittei has been reported to be a uniform gray–green, yellow–green or olive without prominent markings or spots (Tinsley et al., 1979; Channing & Howell, 2006), which would be readily distinguished from the usually mottled dorsal pattern of X. lenduensis. However, we collected and sequenced X. wittei individuals from Kahuzi-Biega National Park (eastern DRC) that had some dorsal markings that are similar to X. lenduensis. For this reason, it is difficult to distinguish Kahuzi-Biega populations of X. wittei from X. lenduensis based on dorsal coloration alone. Xenopus lenduensis does not have a dark dorsal band situated caudally with respect to the eyes that is variably observed in X. itombwensis (Evans et al., 2008). Xenopus lenduensis occasionally has ventral patterning of very dense spots (Fig. 2l) that is variably observed in X. vestitus (Tinsley, 1973) and X. wittei (Tinsley et al., 1979), although the ventrum of X. lenduensis usually has sparse spots, such as on the holotype (Fig. 1c). A key difference between X. vestitus and X. wittei is the distance between the eyes (Tinsley et al., 1979); the eyes of X. vestitus are relatively large and usually more than half of the inter-ocular distance (Tinsley, 1975), whereas the eyes of X. wittei are comparatively smaller and the inter-ocular distance is about 2.2 times the eye diameter (Tinsley et al., 1979). Surprisingly, the spacing and size of the eyes of X. lenduensis are more similar to X. wittei than to its sister taxon X. vestitus; the inter-ocular distance averages 2.5 (females) or 2.6 (males) times the eye diameter (Table 1). Another (related) difference is the shape of the snout, which tends to be more pointed in X. vestitus than in X. itombwensis and X. wittei (Tinsley et al., 1979; Evans et al., 2008). The snout of the new species tends to be rounded (Fig. 2).
Xenopus lenduensis is readily distinguished by morphology from X. laevis, a species with which it is often found (in the same ponds). Xenopus laevis is significantly larger in size than X. lenduensis (P < 0.0001 for comparisons with each sex, Student’s t-test, Table 1) and has significantly larger eyes, even after dividing by body size (P < 0.001, P < 0.0001 for comparisons between females and males, respectively, Student’s t-test). Xenopus laevis is also slimier in the hand than X. lenduensis, possibly due to more viscous and plentiful skin secretions and/or its larger size.
Size dimorphism
Females are significantly larger than males (P < 0.0001, Student’s t-test, Table 1), which is consistent with other species of African clawed frog (Kobel et al., 1996).
Vocalization
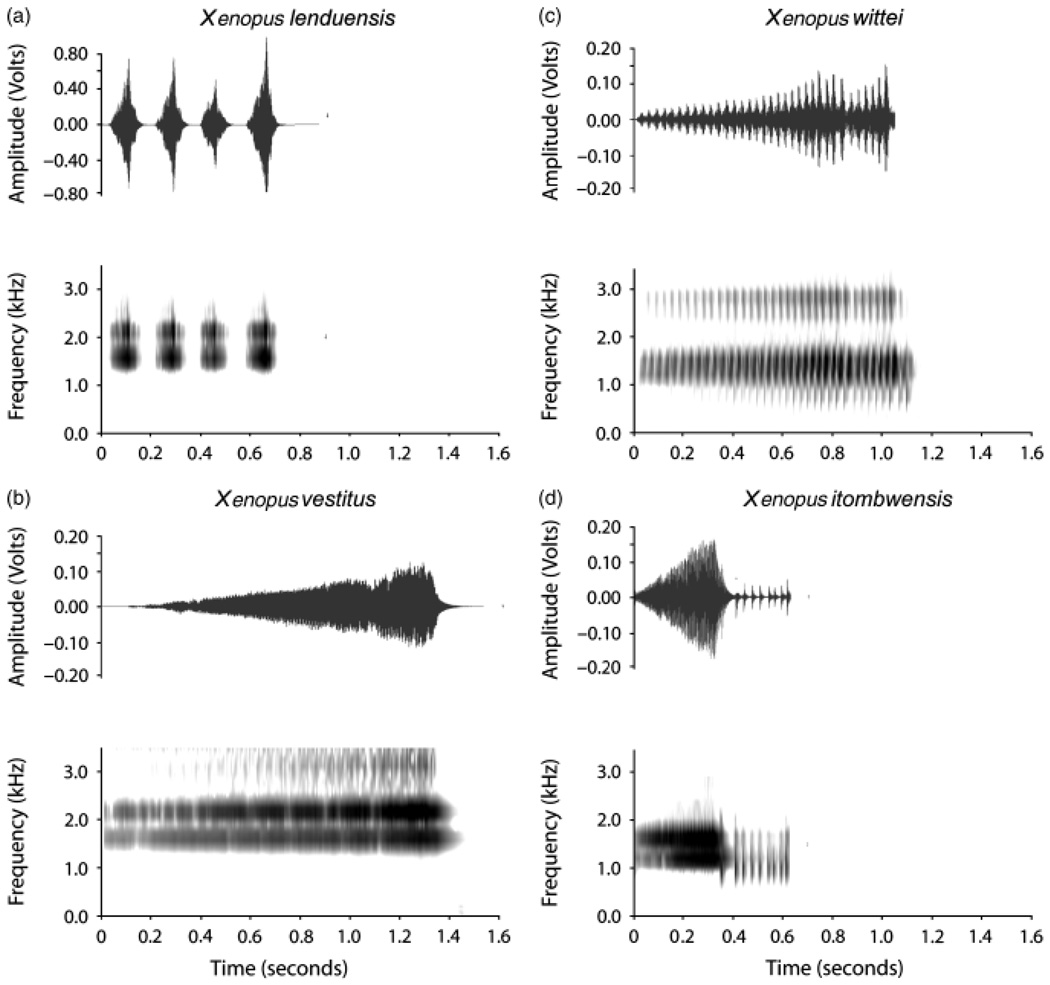
The male advertisement call of X. lenduensis is a brief, rapid, trill with short intervals between calls (Table 2, Fig. 3). The interval between clicks is so short (~12 milliseconds) that individual clicks are often difficult to distinguish. The inter-call interval is much shorter in X. lenduensis than X. vestitus and X. wittei. Another major difference between the vocalization of the new species and its closest relatives is the much shorter duration of the X. lenduensis call. The dominant frequencies of X. lenduensis are very similar to X. vestitus, but not X. itombwensis or X. wittei (Table 2). The call of X. lenduensis lacks two distinct phases that characterize the call of X. itombwensis and some other species such as X. laevis (Kobel et al., 1996). A recording of the call of X. lenduensis has been submitted to Amphibiaweb (http://www.amphibiaweb.org).
Table 2.
Advertisement call characteristics of Xenopus lenduensis, Xenopus vestitus, Xenopus wittei and Xenopus itombwensis
| X. lenduensis | CN | Duration | IM | IclickI | DF (low) | DF (high) | CDC | IcallI |
|---|---|---|---|---|---|---|---|---|
| Grand mean | 9.84 | 143.86 | 13.25 | 11.97 | 1608.62 | 2148.01 | 0.89 | 169.71 |
| stdev | 0.97 | 9.70 | 4.37 | 1.04 | 24.20 | 20.77 | 0.04 | 14.86 |
| SSbetween(1) | 2.86 | 1321.23 | 127.10 | 16.30 | 20936.60 | 15288.70 | 0.00 | 4008.80 |
| Residuals(36) | 0.89 | 60.08 | 16.13 | 0.66 | 20.40 | 18.70 | 0.00a | 115.40 |
| X. vestitus | ||||||||
| Grand mean | 89.75 | 1167.02 | 35.69 | 12.72 | 1595.64 | 2154.50 | 0.25 | 1918.52 |
| stdev | 16.92 | 220.17 | 23.00 | 0.36 | 5.80 | 9.71 | 0.04 | 658.82 |
| SSbetween(1) | 132.25 | 7425.00 | 72.84 | 0.10 | 19.13 | 5.55 | 0.00 | 436065.00 |
| Residuals(2) | 363.25 | 68997.00 | 756.81 | 0.14 | 40.87 | 138.67 | 0.00b | 433542.00b |
| X. wittei | ||||||||
| Grand mean | 45.20 | 1397.10 | 6.74 | 31.06 | 1288.78 | 1521.97 | 0.64 | 4995.63 |
| stdev | 17.60 | 486.28 | 5.24 | 4.18 | 33.55 | 32.44 | 0.24 | 4553.92 |
| SSbetween(1) | 90.13 | 8.00 | 70.62 | 69.01 | 4480.40 | 4043.80 | 0.00 | 9567602.00 |
| Residuals(3) | 382.89 | 315293.00 | 13.03 | 0.34 | 7.00 | 55.40 | 0.08 | 21536114.00c |
| X. itombwensis (fast trill) | ||||||||
| Grand mean | 18.82 | 326.20 | 4.89 | 17.57 | 1284.16 | 1536.69 | 0.88 | 681.08 |
| stdev | 3.84 | 59.08 | 2.44 | 2.47 | 174.21 | 165.20 | 0.09 | 102.02 |
| SSbetween(1) | 5.60 | 713.00 | 35.57 | 4.24 | 56202.00 | 25232.00 | 0.01 | 49067.00 |
| Residuals(9) | 15.78 | 3800.00 | 2.68 | 6.33 | 27477.00 | 27518.00 | 0.01d | 7186.00e |
| X. itombwensis (slow trill) | ||||||||
| Grand mean | 8.82 | 326.66 | 0.91 | 36.43 | 1145.73 | 1644.96a | – | – |
| stdev | 2.40 | 101.09 | 0.84 | 9.55 | 141.36 | 11.37 | – | – |
| SSbetween(1) | 3.10 | 9412.00 | 2.35 | 9.22 | 160590.00 | – | – | – |
| Residuals(9) | 6.06 | 10309.00 | 0.43f | 100.39 | 2406.00g | – | – | – |
Mean, standard deviation (stdev), sum of squares between individuals divided by the degrees of freedom (SSbetween), and residuals divided by the degrees of freedom (Residuals) are provided for call duration in milliseconds (duration), number of clicks per call (CN), intensity modulation (IM), interclick interval in milliseconds (ICI), two dominant frequencies in kiloHertz (DFlow and DFhigh), and call duty cycle (CDC). See ‘Methods’ for more information on these parameters. Measurements of the fast and slow trill portions of the X. itombwensis advertisement call are indicated separately. For X. lenduensis, results are sumarized for two individuals for three calls per individual. One hundred and twenty-three and 251 clicks were analysed, respectively, for each individual. For SSbetween and Residuals degrees of freedomare indicated in parentheses by row except where noted, or not indicated when data were obtained from only one individual, or not indicated for CDC for the slow trill of X. itombwensis because CDC is a parameter calculated from both portions of the call.
Four measurements from one individual.
4 d.f.
14 d.f.
13 d.f.
12 d.f.
6 d.f.
8 d.f.
Figure 3.
The male vocalization of (a) Xenopus lenduensis is distinct from that of closely related species (b) Xenopus vestitus, (c) Xenopus wittei and (d) Xenopus itombwensis. A bout of four advertisement calls is shown for X. lenduensis; a single advertisement call is shown for the other species.
Description of the holotype and color of the holotype in life
Holotype an adult male, small subocular tentacle present, claws present on toes I–III, metatarsal tubercle prominent but without a claw, webbing complete. Nuptial pads visible on the ventral surface of arms extending from the base of arms over the humeral area and forearm. Dorsum olive, with a dark brown spot on the dorsal surface, ~2 mm in diameter and positioned caudally with respect to the right eye and anterior to the arms (Fig. 1a). Smaller lighter brown spots variably present, becoming denser caudally and on the dorsal surface of the toes, less dense on the legs. Ventral surface bright orange with dense purplish grey stipples under the head and belly, with a clear orange band in the center and with medium-sized purple spots on the flanking region, especially in a ring around the insertion point of each arm. Inguinal region and ventral surface of legs and feet orange with larger purple spots on the margins of the dorsal surfaces of the legs and feet. Morphological measurements of the holotype are listed in Table 1.
Color of the holotype in preservative
Dorsum dark olive with brown spots over the back and the dorsal surface of the hind legs, transitioning laterally on flanks to a cream-colored venter; dorsal surface of head dark olive; dorsal surface of limbs olive with dark olive spots with a high density of spots on toes; ventral side of the head speckled with small dark olive spots; venter cream over a central band with fairly dense brown spots on lateral sides of venter; ventral surface of the hind feet cream with some brown spots near the margin with the dorsal surface on upper legs.
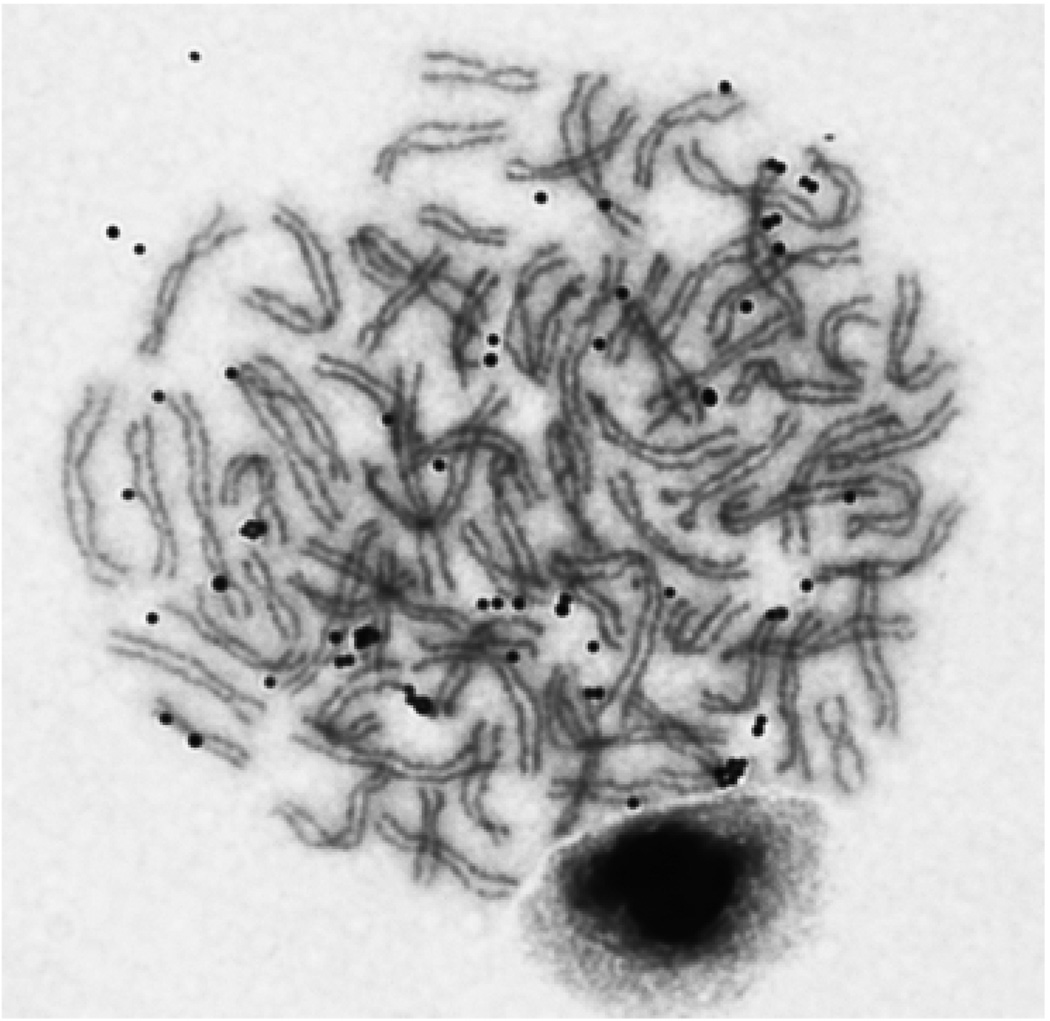
Ploidy
Xenopus lenduensis is octoploid and has 72 chromosomes (Fig. 4). The ploidy level revealed by cytogenetic analysis was consistent with ploidy inferences based on phylogenetic analysis of divergent paralogs of the recombination activating gene 1 (RAG1).
Figure 4.
Giemsa-stained metaphase stage cell from an Xenopus lenduensis illustrating octoploidy (8n = 72 chromosomes). This individual was sampled in Ngensengeyu (DRC), MCZ A-140087; fieldcode BJE2929.
Xenopus lenduensis barcode
The Genbank accession number of the holotype barcode based on the sequence from the CO1 gene (Hebert et al., 2003) is HQ225707.
Ecology and distribution
Xenopus lenduensis was collected from multiple locations on the Lendu Plateau, northern Albertine Rift, DRC (supporting information Table S1; Fig. 5). Xenopus lenduensis was present in small populations of <40 individuals in small bodies of standing water that were occasionally encountered in disturbed agricultural areas. When the habitat was highly disturbed or cleared of vegetation, for example in areas used for water supply by people, this species was not observed, although X. laevis was present. Xenopus lenduensis therefore seemed to be detrimentally impacted by habitat modification. The Lendu Plateau was probably once a savannah forest mosaic, with forests occurring in patches above 1500 m (Fishpool & Collar, 2006). In the areas where we collected the new species, the forest has been completely destroyed; the plateau is now essentially entirely grassland with a few isolated trees, many of which are introduced. We did not see any primary or secondary forest during our surveys; dense weedy vegetation was sometimes observed in small, highly disturbed valleys.
Figure 5.
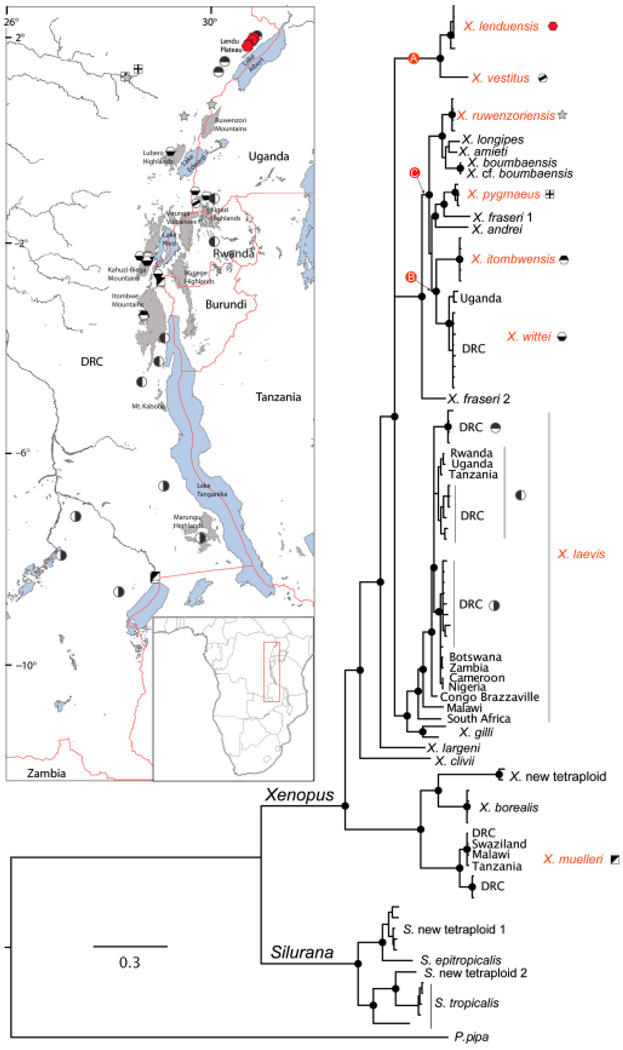
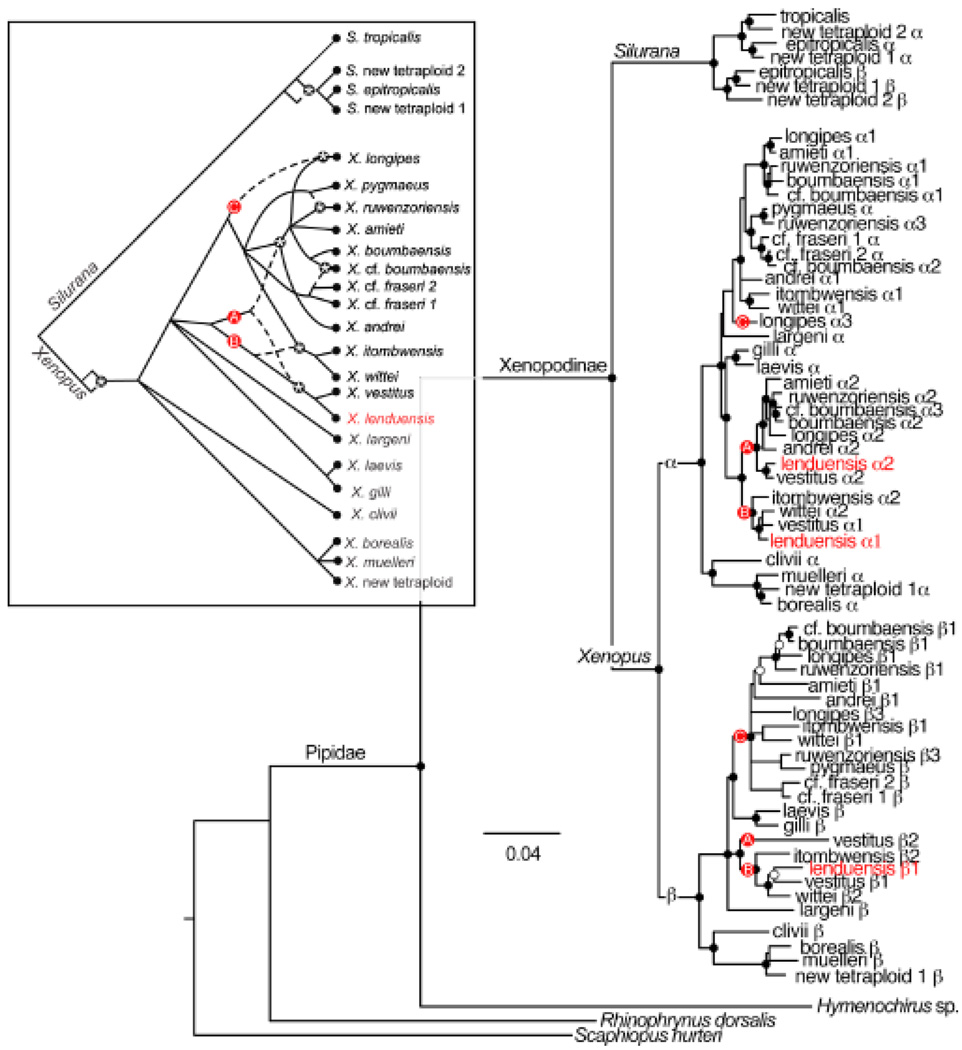
Mitochondrial DNA phylogeny and location of samples from the Albertine Rift (insert). In the phylogeny, black dots indicate nodes with ≥95% posterior probability. For clarity, some of these nodes are not indicated on terminal branches. At least three tetraploid ancestors are inferred and labeled A, B and C; different combinations of the genomes of these ancestors gave rise to extant octoploid and dodecaploid species (see text). Species with ranges in the Albertine Rift have red names on the phylogeny and symbols indicate their geographic location on the insert. On the insert, regions greater than 2000 m a.s.l are indicated in gray; lakes are blue.
Evolutionary relationships with other African clawed frogs
Independent analyses of unlinked genomic regions suggest with strong support that X. lenduensis and X. vestitus are sister species derived from the same allo-octoploid ancestor. mtDNA of these species places them together in a strongly supported clade (Fig. 5). Using the full mtDNA dataset, the uncorrected p-distance between X. lenduensis and X. vestitus is 0.04048. The uncorrected p-distance between these species including only the COI ‘barcode’ region is 0.09386. The uncorrected p-distance between these species including only the 16S fragment proposed as a barcode fragment for amphibians minus 78 bp on the 3′ end (Vences et al., 2005) is 0.03766. This mtDNA relationship supports the previous existence of a maternal (i.e. mtDNA-contributing) tetraploid ancestor of this pair of octoploid species. In Figs 5 and 6, this tetraploid ancestor is indicated by lineages labeled ‘B.’
Figure 6.
RAG1 phylogeny including Xenopus lenduensis supports a sister relationship with the octoploid species Xenopus vestitus. Nodes with posterior probabilities equal to 90–95% or >95% are indicated with open and solid dots, respectively. Lettered nodes correspond with each of the two paralogs from the inferred tetraploid ancestors labeled in Fig. 5. Lineages labeled ‘A’ correspond with paralogs from an inferred paternal tetraploid ancestor of X. lenduensis and X. vestitus and another octoploid ancestral species that gave rise to Xenopus amieti, Xenopus andrei and Xenopus boumbaensis, and the dodecaploid species Xenopus longipes, Xenopus ruwenzoriensis and Xenopus cf. boumbaensis. Lineages labeled ‘B’ correspond with paralogs from an inferred maternal tetraploid ancestor of X. lenduensis and X. vestitus, which was also the paternal tetraploid ancestor of Xenopus wittei and Xenopus itombwensis. Lineages labeled ‘C’ correspond with paralogs of an inferred maternal tetraploid ancestor of the octoploid ancestral species that gave rise to X. amieti, X. andrei, X. boumbaensis and X. long-ipes, X. ruwenzoriensis and X. cf. boumbaensis. An insert depicts a summary of bifurcating and reticulating relationships in clawed frogs with paternally inherited lineages represented by dashed lines and biparentally or maternally inherited lineages represented by solid lines. In the insert, ‘A’, ‘B’ and ‘C’ refer to ancestral tetraploid lineages instead of the ancestral paralogs. Nodes with asterisks refer to putative instances of allopolyploidization.
We cloned alleles from three of the four expected RAG1 paralogs from X. lenduensis, and each has a close relationship with an orthologous paralog of X. vestitus (Fig. 6). Data were not obtained from RAG1 paralog β2 from X. lenduensis. This is consistent with an absence of data from closely related paralogs of X. amieti, X. andrei, X. longipes, X. ruwenzoriensis and X. cf. boumbaensis (Evans, 2007), and could be a result of deletion in an ancestor. Data from RAG1 paralog β3 of X. cf. boumbaensis were not obtained, which could be due to deletion or poor amplification (Evans, 2007). Relationships based on aDNA are consistent with previous inferences that at least three tetraploid species contributed their genomes to extant allo-octoploid species such as X. lenduensis, but are not represented by a known extant representative with the same ploidy level (Evans, 2007, 2008). In Fig. 6, these inferred tetraploid ancestors are labeled ‘A’, ‘B’ and ‘C.’ By focusing on nodes with at least 95% posterior probabilities in the mtDNA and aDNA trees to infer ancient instances of allopolyploidization, at least three independent allooctoploidization events are suggested. These events include the following pairs of tetraploid ancestors (with the maternal tetraploid ancestor listed first): B+A gave rise to an ancestor of X. lenduensis and X. vestitus, C+A gave rise to an ancestor of X. amieti, X. andrei, X. boumbaensis, X. longipes, X. ruwenzoriensis and X. cf. boumbaensis and C + B gave rise to an ancestor of X. itombwensis and X. wittei (Fig. 6). This inference is consistent with those of Evans (2007), which did not include data from X. lenduensis.
Interestingly, X. lenduensis RAG1 paralog β1 has three frameshift mutations, whereas RAG1 paralog α1 and RAG1 paralog α2 appear to be functional at the coding level. This provides additional support for the finding of Evans (2007) that pseudogenization of RAG1 paralogs occurred in a phylogenetically biased fashion, such that RAG1 paralogs inherited from one diploid ancestor (the ‘β’ ancestor) became pseudogenes more frequently than paralogs inherited from the other diploid ancestor (the ‘α’ ancestor).
Etymology
The new species is named after the plateau where it occurs – the Lendu Plateau of the Albertine Rift in Orientale Province, DRC, which lies just west of Lake Albert. This region is also known as the Blue Mountains.
African clawed frog mtDNA phylogeography in the Albertine Rift
With the exception of X. laevis, no species we sampled had substantial levels of intra-specific mtDNA divergence in the Albertine Rift. Our analysis slightly extends the known range of the critically endangered X. itombwensis to proximate localities on the Itombwe Plateau. This species was formerly known only from its type locality, whose GPS coordinates were incorrectly reported in the species description, but are correctly reported here (supporting information Table S1; Evans et al., 2008). Because the range extension here is so small, this does not warrant changing the conservation status of this species. The Itombwe Plateau has the most threatened species of amphibians and the highest number of endemic amphibians in the Albertine Rift; this is also the case for birds (Plumptre et al., 2007).
Our analysis indicates that X. ruwenzoriensis also occurs in the DRC (Fig. 5; Greenbaum & Kusamba, 2010); this species was formerly only known from the type locality near Bundibugyo in the Semliki Valley, western Uganda (Fischberg & Kobel, 1978), and from the Budongo Forest, east of Lake Albert, also in western Uganda (IUCN, 2010).
We identified three diverged mtDNA lineages in X. laevis in the Albertine Rift (Fig. 5). One is closely related to mtDNA lineages sampled from X. laevis populations in East Africa (Uganda, Rwanda, Tanzania) and another was closely related to mtDNA lineages sampled in X. laevis populations in portions of west and south-central Africa (Cameroon, Nigeria, Zambia, Botswana). The third X. laevis mtDNA lineage from the Albertine Rift is only known from the Lendu Plateau, and to our knowledge, has not been reported previously.
Other inferences of species ranges are generally consistent with previous species distributions that were based on morphology (Tinsley, Loumont & Kobel, 1996). Xenopus muelleri occurs over much of east and south-east Africa, extending up Lake Tanganika to the xeric area south of Lake Kivu (near Bukavu). The distribution of X. pygmaeus over much of Central Africa reaches its easternmost limit at the region west of lakes Kivu (data not shown) and Albert. The distribution of X. wittei was confirmed to include portions of southwestern Uganda, western Rwanda and the Kahuzi-Biega and Lubero Highlands in DRC (Tinsley et al., 1979; Kobel et al., 1996). Xenopus wittei mtDNA sequences from the Kahuzi-Biega and Lubero highlands are slightly diverged from those of the type locality in south-western Uganda (Fig. 5). We do not have additional samples of X. vestitus beyond the one used in Evans et al. (2004); other studies report a broader distribution for this species, but not north of Lake Edward (Tinsley, 1973; Tinsley, 1975).
Discussion
Distinctiveness of X. lenduensis
We distinguish a new species of African clawed frog, Xenopus lenduensis, from other species based on morphology, vocalization and molecular divergence – characteristics that together identify X. lenduensis under the general lineage concept of a species. The most straightforward way to narrow down the identity of the new species is its provenance on the Lendu Plateau, DRC, which is the northern extent of the Albertine Rift. The only other African clawed frog that shares this habitat with X. lenduensis is X. laevis. Xenopus lenduensis is easily distinguished from X. laevis by the much larger size and the larger eyes of X. laevis. Within the vestitus-wittei subgroup, the new species is similar in size to its sister taxon X. vestitus and to X. wittei, but larger than X. itombwensis. Dorsal and ventral coloration and morphology distinguish X. lenduensis from its sister taxon X. vestitus, the former species having an olive dorsum with variable darker olive spots, a usually rounder snout and more widely spaced eyes than X. vestitus. Distinguishing X. lenduensis from some populations of X. wittei that have variable spots on the dorsum could be challenging, although other X. wittei populations lack dorsal spots. The fundamental characteristics of the vocalization of the new species, including call duration and inter-call interval, are distinct from other species of African clawed frog, including X. vestitus and other members of the ‘vestitus-wittei’ subgroup (Table 2; Fig. 2). On a molecular level, mtDNA and nuclear DNA divergence between X. lenduensis and X. vestitus is substantial and comparable to that seen between other closely related species of African clawed frog such as X. fraseri and X. pygmaeus or X. amieti and X. longipes (Figs 5 and 6).
Allopolyploid speciation and the search for descendants of ‘lost ancestors’
Building on previous molecular phylogenetic studies of African clawed frogs (Carr, Brothers & Wilson, 1987; Kobel, Barandun & Thiebaud, 1998; Measey & Channing, 2003; Evans et al., 2004; Evans et al., 2005; Evans, 2007), the evolutionary relationships between X. lenduensis and other species of clawed frog are evaluated here using mtDNA and sequences from cloned paralogs of the RAG1 gene. Although some paralogs of RAG1 were not sequenced, possibly because they have been deleted, together, these markers illustrate a sister relationship between X. lenduensis and X. vestitus. We speculate that X. lenduensis and X. vestitus are derived from a recent common octoploid ancestor generated from the fusion of two tetraploid genomes indicated by lineages labeled ‘B’ and ‘A’ in Figs 5 and 6. However, we cannot rule out the less parsimonious possibility that each species was formed from simultaneous, but independent allopolyploidization events involving the same combination of maternal and paternal tetraploid ancestors. In any case, X. lenduensis and X. vestitus share recent common ancestry of one half of their genome with X. itombwensis and X. wittei, and the other half with X. amieti, X. andrei, X. boumbaensis, X. longipes, X. ruwenzoriensis and X. cf. boumbaensis.
The discovery of X. lenduensis supports previous work that inferred the prior existence of ancestral tetraploid species that did not leave known extant tetraploid descendants (Evans et al., 2005, 2008; Evans, 2007). In addition to these tetraploid ancestors, one or two ancestral diploid (2N = 18) Xenopus species are predicted, depending on whether tetraploidization occurred in Xenopus via autopolyploidization or allopolyploidization. In Silurana, analysis of aDNA suggests that tetraploidization occurred by allopolyploidization; hence, another diploid (2N = 20) ancestral species is predicted to have existed in addition to the ancestor of the diploid species S. tropicalis (Evans et al., 2005; Evans, 2007).
The discovery of descendants of these predicted species would be an unusual example of unsampled species being predicted by phylogenetic inference. The discovery of diploid descendants of the predicted 2N = 18 Xenopus diploid ancestors would be particularly useful for studies of gene duplication in tetraploid species such as X. laevis. Analysis of these descendants would allow, for example, the partitioning of paralogs and singletons (duplicates where one copy became a pseudogene) into subgenomes derived from each diploid ancestor. Knowledge of this information would allow a large-scale test for evidence of phylogenetically biased pseudogenization in duplicated genetic networks after allopolyploidization. A single-locus example of biased pseudogenization was reported recently in the RAG1 proteins (Evans, 2007), and this result is further supported here by the observation that RAG1 paralog β1 is pseudogenized in X. lenduensis but RAG1 paralogs α1 and α2 seem functional at the DNA level. Another example of biased pseudogenization in Xenopus is reported by Bewick, Anderson & Evans (2010) at the DMRT1 locus.
The recent discovery of X. lenduensis and X. itombwensis is also interesting because it identifies two pairs of sister octoploid species (X. lenduensis + X. vestitus) and (X. itombwensis + X. wittei) that are distributed over much of the Albertine Rift. The distributions of these pairs of sister taxa overlap in the region between Lakes Kivu and Edward in south-western Uganda and the eastern DRC, which is where X. wittei and X. vestitus occur (Tinsley, 1973, 1975; Tinsley et al., 1979). Xenopus itombwensis, the sister taxon of X. wittei, occurs on the Itombwe Plateau to the south of X. wittei and west of Lake Tanganika. Xenopus lenduensis, the sister taxon of X. vestitus, occurs on the Lendu Plateau to the north of X. vestitus, on the west side of Lake Albert.
Phylogeography of X. laevis
Here, we use ‘X. laevis’ sensu lato following Kobel et al. (1996). This species is distributed from South Africa to southern Sudan and from Nigeria to Tanzania. Previous studies identified at least six major mtDNA lineages in X. laevis. These lineages are mostly allopatric, although populations with multiple lineages do exist in South Africa (Du Preez et al., 2009). One mtDNA lineage occurs in the Cape Region of South Africa south-west and west of the Cape Fold Mountains (Grohovaz, Harley & Fabran, 1996; Evans et al., 1997, 2004; Measey & Channing, 2003; Du Preez et al., 2009). Although subspecies are not recognized here, this lineage corresponds in geographic distribution with a portion of the ‘laevis’ subspecies (Kobel et al., 1996). A second lineage occurs in central South Africa between the Cape Fold Mountains and Lesotho (Du Preez et al., 2009); this lineage is not included in the analysis presented in Fig. 5. A third lineage occurs in Malawi and north-western South Africa (Grohovaz, Harley & Fabran, 1996; Measey & Channing, 2003; Evans et al., 2004; Du Preez et al., 2009). A fourth lineage occurs in Congo Brazzaville (Evans et al., 2004). A fifth lineage occurs in north-western and central sub-Saharan Africa in Nigeria, Cameroon, Zambia and the DRC, including the southern portion of the Albertine Rift (this study; Evans et al., 1997, 2004; Measey & Channing, 2003; Du Preez et al., 2009). This lineage corresponds in geographic distribution with the subspecies ‘sudanensis’ and ‘poweri’ (Kobel et al., 1996). A sixth lineage occurs in East Africa and the central Albertine Rift (this study; Evans et al., 1997, 2004; Du Preez et al., 2009) and corresponds in geographic distribution with the ‘victorianus’ subspecies (Kobel et al., 1996). Here, we identify a seventh mtDNA lineage within X. laevis that occurs on the Lendu Plateau and proximate areas west of Lake Albert (Fig. 5).
Multiple lines of evidence suggest that X. laevis contains substantially differentiated populations that may warrant species status (Measey & Channing, 2003). The genetic divergence between mtDNA lineages within X. laevis can be of a magnitude comparable to that observed between other species of African clawed frog. For example, in an ~870 bp region of the 16S gene, the uncorrected p-distance between the Cape Region population of X. laevis and other populations is about 3.2%, which is almost as large as the divergence over the same region of mtDNA between X. lenduensis and X. vestitus. Many X. laevis populations with divergent mtDNA also have distinct vocalizations (MLT, DBK and BJE unpublished data). It would be interesting to compare these vocalizations with that of X. laevis from the Lendu Plateau, but unfortunately, we did not obtain recordings of this population. Morphological variation is also substantial – for example, X. laevis from the West Cape Region, south-west of the Cape Fold Mountains, is significantly larger than X. laevis from other portions of South Africa (Du Preez et al., 2009). However, analysis of mitochondrial and nuclear markers in a transect of individuals running from southwest to north-east South Africa identified a zone of admixture (Du Preez et al., 2009).
The evolutionary relationships among these lineages have implications for elevation of X. laevis populations to species status. According to mtDNA, for example, the X. laevis population from the Cape Region of South Africa is equally diverged from the X. laevis populations from western Africa, from eastern Africa and from the Lendu Plateau. Additional studies are needed to sort out species from population-level relationships within X. laevis (Measey & Channing, 2003). In particular, it would be informative to use genetic data to explicitly compare a demographic hypothesis of isolation by distance within X. laevis sensu lato with a scenario involving speciation within this clade.
Biogeography of the Albertine Rift
The Great Rift Valley is an ~6000 km trench between multiple continental plates. From its northern extent in Lebanon, it runs south, dividing Jordan and Israel, down the east side of the Sinai Peninsula and under the Red Sea. It surfaces in Eritrea and Ethiopia, dividing the Ethiopian highlands, and splits into a Western and an Eastern Rift just north of the equator, around the border between Sudan and Uganda and between Ethiopia and Kenya. The Eastern Rift runs through central Kenya and Tanzania and is associated with Mts. Kenya and Kilamanjaro, the highest mountains in Africa. The Western Rift includes the Albertine Rift highlands, which run between the DRC and the western borders of Uganda, Rwanda, Burundi and western Tanzania (Plumptre et al., 2007). Further south, these rifts converge in Malawi and continue to central Mozambique.
Xenopus lenduensis and its closest relatives are endemic to the Albertine Rift. This area has a distinct flora and fauna associated with the montane rainforest highlands that grades into lowland tropical rainforests of the Congo Basin to the west and forest savannah mosaic habitat in the east and south (Burgess et al., 2004). The Albertine Rift is a high priority for biodiversity conservation and is recognized, for example, as a biodiversity hotspot (Mittermeier et al., 2004), a distinct terrestrial ecoregion (Burgess et al., 2004), and an ‘Endemic Bird Area’ (Stattersfield et al., 1998). At least one bird subspecies of the White-browed Crombec, Chapin’s Crombec (Sylvietta leucophrys chapini), occurs only on the Lendu Plateau – although this subspecies may be extinct due to habitat destruction (Fishpool & Collar, 2006). The exceptional species diversity in this region is in part a consequence of the varied ecosystems associated with the altitudinal gradient and its location between distinct habitat types. Pleistocene and Pliocene climatic oscillations probably affected patterns of speciation in Central Africa and the Albertine Rift, for example in rodents (Huhndorf, Kerbis Peterhans & Loew, 2007), gorillas (Anthony et al., 2007), birds (Bowie et al., 2006; Voelker, Outlaw & Bowie, 2010) and frogs (Evans et al., 2004; Blackburn, 2008).
The recognition of X. lenduensis further characterizes the considerable biological diversity in the Albertine Rift, contributing to our growing understanding of amphibian diversity in this region (Plumptre et al., 2007; Behangana, Kasoma & Luiselli, 2009; Greenbaum & Kusamba, 2010 Roelke et al., in press). Unfortunately, the biological diversity of the Lendu Plateau and Albertine Rift faces severe threats due to a very dense human population (Burgess et al., 2007) and the associated anthropogenic disturbance that has destroyed natural vegetation on the Lendu Plateau (B. J. Evans, E. Greenbaum, C. Kusamba pers. obs., Fishpool & Collar, 2006). Based on our locality records, we estimate the extent of the occurrence of X. lenduensis to be < 5000 km2. Given the catastrophic deforestation and severe habitat fragmentation on the Lendu Plateau and the expected continuing decline of the extent and quality of this habitat, we recommend that this species be classified as endangered under the IUCN Red List Criteria (IUCN, 2001).
Supplementary Material
Acknowledgments
We are indebted to Richard Tinsley for providing constructive comments on this manuscript, genetic samples of X. vestitus and many insightful discussions about African clawed frogs. We thank Jonathan Dushoff for statistical advice, Adam Bewick and Jonathan Woodward for assistance with photography, José Rosado and Joseph Martinez for curatorial assistance, Miguel Vences and two anonymous reviewers for constructive comments on this manuscript and Mwenebatu M. Aristote, Wandege Mastaki Moninga, John and Felix Akuku, Mululema Zigabe, Maurice Luhumyo, Jean Marie Chambu, Angalikiya Mulamba Marcel and the late Asukulu M’Mema for field assistance. We thank l’Institut Congolais pour la Conservation de la Nature for our Autorisation de Recherche in DRC, Baluku Bajope and Muhimanyi Mununu of the Centre de Recherche en Sciences Naturelles for providing project support and permits. This research was supported by the Canadian Foundation for Innovation, the National Science and Engineering Research Council and the Museum of Comparative Zoology at Harvard University. Fieldwork by EG was funded by the Percy Sladen Memorial Fund, the Department of Biology at Villanova University, a National Geographic Research and Exploration Grant (no. 8556-08), an IUCN/SSC Amphibian Specialist Group Seed Grant and the University of Texas at El Paso.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Specimen, locality, and sequence accession information for Albertine Rift samples. There is no voucher for some samples and GPS coordinates (in decimal degree format) of some localities are approximate. Information for other samples analyzed in this study is available in Evans et al. (2004).
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Anthony NM, Johnsin-Bawe M, Jeffery K, Clifford SL, Abernethy KA, Tutin CE, Lahm SA, White JT, Utley JF, Wickings EJ, Bruford MW. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc. Natl. Acad. Sci. USA. 2007;104:20432–20436. doi: 10.1073/pnas.0704816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behangana M, Kasoma PMB, Luiselli L. Ecological correlates of species richness and population abundance patterns in the amphibian communities from the Albertine Rift, East Africa. Biodivers. Conserv. 2009;18:2855–2873. [Google Scholar]
- Bewick AJ, Anderson DW, Evans BJ. Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus) Evolution. 2010 doi: 10.1111/j.1558-5646.2010.01163.x. Online. [DOI] [PubMed] [Google Scholar]
- Blackburn DC. Biogeography and evolution of body size and life history of African frogs: Phylogeny of squeakers (Arthroleptis) and long-fingered frogs (Cardioglossa) estimated from mitochondrial data. Mol. Phylogenet. Evol. 2008;49:806–826. doi: 10.1016/j.ympev.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Bowie RCK, Fjeldså J, Hackett SJ, Bates JM, Crowe TM. Coalescent models reveal the relative roles of ancestral polymorphism, vicariance, and dispersal in shaping phylogeographical structure of an African montane forest robin. Mol. Phylogenet. Evol. 2006;38:171–188. doi: 10.1016/j.ympev.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Burgess ND, Balmford A, Cordeiro NJ, Fjeldså J, Küper W, Rahbek C, Sanderson EW, Scharlemann JPW, Soomer JH, Williams PH. Correlations among species distributions, human density and human infrastructure across the high diversity tropical mountains of Africa. Biol. Conserv. 2007;134:164–177. [Google Scholar]
- Burgess ND, D’Amico Hales J, Underwood EC, Dinerstein E, Olson D, Itoua I, Schipper J, Ricketts TH, Newman K. Terrestrial ecoregions of Africa and Madagascar: a conservation assessment. Washington: Island Press; 2004. [Google Scholar]
- Carr S, Brothers A, Wilson A. Evolutionary inferences from restriction maps of mitochondrial DNA from nine taxa of Xenopus frogs. Evolution. 1987;41:176–188. doi: 10.1111/j.1558-5646.1987.tb05780.x. [DOI] [PubMed] [Google Scholar]
- Channing A, Howell KM. Amphibians of East Africa. Ithaca: Cornell University Press; 2006. [Google Scholar]
- de Queiroz K. The general lineage concept of species, species criteria, and the process of speciation. In: Howard D, Berlocher S, editors. Endless forms: species and speciation. New York: Oxford Press; 1998. pp. 57–75. [Google Scholar]
- de Queiroz K. Species concepts and species delimitation. Syst. Biol. 2007;56:879–886. doi: 10.1080/10635150701701083. [DOI] [PubMed] [Google Scholar]
- Du Preez LH, Kunene N, Hanner R, Giesy JP, Solomon KR, Hosmer A, Van Der Kraak G. Population-specific occurrence of testicular ovarian follicles in Xenopus laevis from South Africa. Aquat. Toxicol. 2009;95:10–16. doi: 10.1016/j.aquatox.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Evans BJ. Ancestry influences the fate of duplicated genes millions of years after duplication in allopolyploid clawed frogs (Xenopus) Genetics. 2007;176:1119–1130. doi: 10.1534/genetics.106.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans BJ. Genome evolution and speciation genetics of allopolyploid clawed frogs (Xenopus and Silurana) Front. Biosci. 2008;13:4687–4706. doi: 10.2741/3033. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Carter TF, Hanner R, Tobias ML, Kelley DB, Hanner R, Tinsley RC. A new species of clawed frog (genus Xenopus), from the Itombwe Plateau, Democratic Republic of the Congo: implications for DNA barcodes and biodiversity conservation. Zootaxa. 2008;1780:55–68. [Google Scholar]
- Evans BJ, Kelley DB, Melnick DJ, Cannatella DC. Evolution of RAG-1 in polyploid clawed frogs. Mol. Biol. Evol. 2005;22:1193–1207. doi: 10.1093/molbev/msi104. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of clawed frogs: phylogeography on sub-Saharan Africa and implications for polyploid evolution. Mol. Phylogenet. Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Evans BJ, Morales JC, Picker MD, Kelley DB, Melnick DJ. Comparative molecular phylogeography of two Xenopus species, X. gilli and X. laevis, in the southwestern Cape Province, South Africa. Mol. Ecol. 1997;6:333–343. doi: 10.1046/j.1365-294x.1997.00198.x. [DOI] [PubMed] [Google Scholar]
- Fischberg M, Kobel HR. Two new polyploid Xenopus species from western Uganda. Experientia. 1978;34:1012–1014. doi: 10.1007/BF01915316. [DOI] [PubMed] [Google Scholar]
- Fishpool LDC, Collar NJ. The taxonomic and conservation status of Chapin’s Crombec. Bull. Afr. Bird Club. 2006;13:130–135. [Google Scholar]
- Greenbaum E, Kusamba C. Xenopus ruwenzoriensis (Ruwenzori Clawed Frog). Geographic distribution. Herpetol. Rev. 2010;41:376–377. [Google Scholar]
- Grohovaz GS, Harley B, Fabran B. Significant mitochondrial DNA sequence divergence in natural populations of Xenopus laevis (Pipidae) from South Africa. Herpetologica. 1996;52:247–253. [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc. Roy. Soc. Lond. Ser. B. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Huhndorf MH, Kerbis Peterhans JC, Loew SS. Comparative phylogeography of three endemic rodents from the Albertine Rift, east central Africa. Mol. Ecol. 2007;16:663–674. doi: 10.1111/j.1365-294X.2007.03153.x. [DOI] [PubMed] [Google Scholar]
- IUCN. IUCN Red List Categories, Version 3.1. Prepared by the IUCN Species Survival Commission. Gland: IUCN; 2001. [Google Scholar]
- IUCN. IUCN red list of threatened species. Version 2010.1. Gland: IUCN; 2010. [Google Scholar]
- Ivanova NV, deWaard JR, Hebert PDN. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes. 2006;6:998–1002. [Google Scholar]
- Kobel H, Barandun B, Thiebaud CH. Mitochondrial rDNA phylogeny in Xenopus. Herpetol. J. 1998;8:13–17. [Google Scholar]
- Kobel HR. Allopolyploid speciation. In: Tinsley RC, Kobel HR, editors. The Biology of Xenopus. Oxford: Clarendon Press; 1996. pp. 391–401. [Google Scholar]
- Kobel HR, Du Pasquier L. Genetics of polyploid Xenopus. Trends Genet. 1986;2:310–315. [Google Scholar]
- Kobel HR, Loumont C, Tinsley RC. The extant species. In: Tinsley RC, Kobel HR, editors. The Biology of Xenopus. Oxford: Clarendon Press; 1996. pp. 9–33. [Google Scholar]
- Laurent RF. Amphibiens. Exploration du parc national de Virunga. Ser II, facs 22. Bruxelles: Recherches enterprises par l’Institut National pour la nonservation de la nature de la Réublique du Zaïe; 1972. [Google Scholar]
- Measey GJ, Channing A. Phylogeography of the genus Xenopus in southern Africa. Amphib-Reptilia. 2003;24:321–330. [Google Scholar]
- Mittermeier RA, Robles Gil P, Hoffman M, Pilgrim JD, Brooks T, Mittermeier CG, Lamoreaux J, da Fonseca GAB. Hotspots revisited: Earth’s biologically richest and most endangered terrestrial ecoregions. Mexico City: CEMEX; 2004. [Google Scholar]
- Nylander JAA. MrModeltest v2. Uppsala: Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Plumptre AJ, Davenport TRB, Behangana M, Kityo R, Eilu G, Ssegawa P, Ewango C, Meitre D, Kahindo C, Herremans M, Peterhans JK, Pilgrim JD, Wilson M, Languy M, Moyer D. The biodiversity of the Albertine Rift. Biol. Conserv. 2007;134:178–194. [Google Scholar]
- Roelants K, Bossuyt F. Archaeobatrachian paraphyly and Pangean diversification of crown-group frogs. Syst. Biol. 2005;54:111–126. doi: 10.1080/10635150590905894. [DOI] [PubMed] [Google Scholar]
- Roelke CE, Greenbaum E, Kusamba C, Aristote MM, Smith EN. Systematics and conservation of Albertine Rift Leptopelis. J. Herp. in press. [Google Scholar]
- Stattersfield AJ, Crosby MJ, Long AJ, Wege DC. Endemic Bird Areas of the World: priorities for biodiversity conservation. Cambridge: BirdLife International; 1998. [Google Scholar]
- Tinsley RC. Studies on the ecology and systematics of a new species of clawed toad, the genus Xenopus, from western Uganda. J. Zool. (Lond.) 1973;169:1–27. [Google Scholar]
- Tinsley RC. The morphology and distribution of Xenopus vestitus (Anura: Pipidae) in Central Africa. J. Zool. (Lond.) 1975;175:473–492. [Google Scholar]
- Tinsley RC, Kobel HR, Fischberg M. The biology and systematics of a new species of Xenopus (Anura: Pipidae) from the highlands of Central Africa. J. Zool. (Lond.) 1979;188:69–102. [Google Scholar]
- Tinsley RC, Loumont C, Kobel HR. Geographical distribution and ecology. In: Tinsley RC, Kobel HR, editors. The Biology of Xenopus. Oxford: Clarendon Press; 1996. pp. 35–59. [Google Scholar]
- Vences M, Thomas M, van der Meijden A, Chiari Y, Vieites DR. Comparative performance of the 16S rDNA gene in DNA barcoding of amphibians. Front. Zool. 2005;2:1–12. doi: 10.1186/1742-9994-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker G, Outlaw RK, Bowie RCK. Pliocene forest dynamics as a primary driver of African bird speciation. Glob. Ecol. Biogeogr. 2010;19:111–121. [Google Scholar]
- Wu C-I. The genic view of the process of speciation. J. Evol. Biol. 2001;14:851–865. [Google Scholar]
- Yager DD. A unique sound production mechanism in the pipid anuran Xenopus borealis. Zool. J. Linn. Soc. 1992;104:351–375. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.