Abstract
Purpose
This study was performed to discover prognostic genomic markers associated with post-operative outcome of stage I-III non-small cell lung cancer (NSCLC) that are reproducible between geographically distant and demographically distinct patient populations.
Experimental design
American patients (n=27) were stratified on the basis of recurrence and microarray profiling of their tumors was performed to derive a training set of 44 genes. A larger Korean patient validation cohort (n=138) was also stratified by recurrence and screened for these genes. Four reproducible genes were identified and used to construct genomic and clinicogenomic Cox models for both cohorts.
Results
Four genomic markers, DBN1 (drebrin 1), CACNB3 (calcium channel beta 3), FLAD1 (PP591; flavin adenine dinucleotide synthetase), and CCND2 (cyclin D2), exhibited highly significant differential expression in recurrent tumors in the training set (P<0.001). In the validation set, DBN1, FLAD1 (PP591) and CACNB3 were significant by Cox univariate analysis (P≤0.035), whereas only DBN1 was significant by multivariate analysis. Genomic and clinicogenomic models for recurrence free survival (RFS) were equally effective for risk stratification of stage I-II or I-III patients (all models P<0.0001). For stage I-II or I-III patients, 5-y RFS of the low- and high-risk patients was ∼ 70 vs. 30% for both models. The genomic model for overall survival (OS) of stage I-III patients was improved by addition of pT and pN stage (P<0.0013 vs. 0.010).
Conclusion
A 4-gene prognostic model incorporating the multivariate marker DBN1 exhibits potential clinical utility for risk stratification of stage I-III NSCLC patients.
Keywords: NSCLC recurrence, DBN1, CACNB3, FLAD1, CCND2
Introduction
The discovery of genomic markers that are prognostic of NSCLC recurrence could change clinical practice by identifying patients who would do well regardless of adjuvant chemotherapy, thus sparing those patients considerable treatment-related toxicities. Recently, patients identified as low-risk for death from lung cancer on the basis of stratification by a 15-gene signature exhibited worse outcomes when treated with adjuvant cisplatin/vinorelbine (1). Many microarray studies of NSCLC gene expression have been performed with the purpose of finding gene markers predictive of overall survival (OS) (2, 3, 4, 5, 6), but few individual genes are reproducible.
Although earlier microarray studies successfully identified gene markers associated with OS, in some cases independent of stage, some studies didn't distinguish between death from NSCLC and other causes. Lung cancer markers linked to survival alone exhibit limited clinical utility, because there are significant competing causes of mortality including postoperative mortality (7, 8), the occurrence of second primary cancers including second primary lung cancer, cardiovascular disease and chronic obstructive pulmonary disease (COPD). Among stage I NSCLC patients, 5-y disease-specific survival is 77% for pT1N0M0 patients and 62% for pT2N0M0 patients (9), while 5-y OS is 67 and 57% for these groups (10). This comparison indicates that even for stage I patients there is a significant death rate from competing causes of death. Recently, there has been an emphasis on disease-specific survival, which has been more helpful (1). Another approach is to study RFS, which has led to significant progress in identification of genomic markers that are associated with lung cancer-specific outcomes (11, 12). Although these studies have led to identification of multivariate gene groups, to the best of our knowledge, they have not led to the discovery of single gene multivariate markers for RFS, whereas pT (9) and pN1 stage (13) are multivariate.
An as yet unrealized goal is to develop stage-independent genomic models for NSCLC that are prognostic for RFS and reproducible between differing patient populations. This goal, if accomplished, would make it possible to predict the likelihood of recurrence within 5-y of surgery based solely on genomic markers, thus giving the patient and oncologist information to make adjuvant therapy decisions that are related to patient's individual tumor biology rather than stage (I-III). This type of model would be very useful clinically, because patients seen in the NSCLC clinic exhibit a wide range of stages, but may exhibit personalized risk that may differ from that predicted by stage alone. To achieve this goal, reproducible multivariate genomic markers for RFS are needed. Most studies are internally validated by dividing a relatively homogeneous patient cohort, such as one consisting of North American patients, the majority of whom are current or former smokers, into smaller test and larger validation sets. These studies suffer from demographic and geographic similarity of the two populations. Recent studies have successfully identified genomic markers associated with RFS; nonetheless, the lead genomic markers derived within an exclusively North American or Asian patient cohorts are not generally reproducible between demographically distinct and geographically distant populations (11, 12). Differences between genomic markers for RFS may be related to chance or reflect differing mechanisms of cancer recurrence. Differences in recurrence patterns between distinct populations could relate to genetics or interaction of genetics with diet, exercise, and second hand smoke exposure. Although much can be learned about cancer outcome disparities by studying genomic markers within specific racial or ethnic groups, much can also be learned about what is similar between differing groups. Genomic markers that remain the same between differing patient groups are therefore more likely to be broadly useful and independent of confounding factors. Our hypothesis is that improved reproducibility of genomic markers can be achieved by identifying markers that are reproducible between demographically distinct and geographically distant populations.
A novel approach is to identify genomic markers that are of univariate significance in a small training set and then re-test them for univariate or multivariate significance in a larger validation group that is demographically distinct. We therefore took a group of 44 lead genomic markers for RFS from a small American training group (stages I-III) and found four genes that also exhibited either univariate or multivariate significance in a larger Korean validation group (11). These 4 genomic markers were strongly associated with recurrence in both groups and resulted in a genomic model that was prognostic for recurrence independent of clinical variables, including stage and histology.
Materials and Methods
Training set patients
This study was approved by the Indiana University Purdue University at Indianapolis (IUPUI)/Clarian IRB (#0201-58), and the banking of tissue was performed under a separate protocol approved by the same IRB (#9401-17) (IU-Lilly Tissue Bank). A longitudinal database of consented patients undergoing NSCLC resection with curative intent from 12-07-1999 to 02-01-2002 was searched for banked NSCLC tumor tissue with sufficient tumor content. The tumor resections in the single institution training set were performed at Indiana University by two thoracic surgeons who used consistent procedures for lobectomy or pneumonectomy during the period of the study (8, 14). The lobectomy/pneumonectomy procedure involved ligation of the vein leading to the tumor-containing lobe or lung before arterial ligation. All patients had complete peribronchial and mediastinal lymph node dissections. Patients treated on this study were subsequently followed by the multidisciplinary Thoracic Oncology Program at Indiana University.
All patients (n=27) with stage Ia-IIIb NSCLC, who were evaluable for recurrence at 2 years of follow-up after surgery were included in the study. The median follow-up time for the recurrence-free patients was 57 months. NSCLC recurrence, if suspected, was histologically confirmed. All patient-related data were de-identified by the tissue bank staff so that the investigators could access only coded frozen tissue specimens, coded paraffin slides and relevant variables for multivariate analysis that were associated with the code number for the patient.
While none of the patients in the training set received adjuvant chemotherapy, one recurrent patient and two non-recurrent patients received carboplatin-based chemotherapy as neoadjuvant treatment. One recurrent patient (#11) received chemotherapy 8 months before resection, having participated in a phase II study of paclitaxel/carboplatin and cetuximab. The same patient was continued on cetuximab at the time of recurrence, which was 8 months following resection. One non-recurrent patient (#13) received paclitaxel/carboplatin two months before resection. Another non-recurrent patient (#21) received paclitaxel/carboplatin for two cycles before resection, which was performed 7 weeks after initiation of chemotherapy.
Pathology
NSCLC tumor histology was confirmed by hematoxylin/eosin (H+E) staining of associated tissue blocks. The frozen samples included in the study ranged from 90 to 40% tumor tissue with a mean of 59% ± 11% SD. Of the 30 specimens that were available for study, 3 were rejected because they either contained mainly stroma or necrosis, or exhibited less than 40% tumor epithelium. The percentage of malignant cells in the frozen tumor specimen could not be determined definitively from the single 5 micron frozen section, in part, because of the difficulty of completely counting the admixture of interspersed normal stromal, endothelial cells and immunocytes, which were much smaller than the tumor cells.
RNA isolation and purification
Tumors were collected by a tissue procurement service in the operating room and stored in liquid nitrogen immediately following resection and remained frozen until RNA isolation was performed. Specimens were removed from liquid nitrogen, transported on dry ice to the laboratory and homogenized using a Polytron homogenizer (Brinkmann Instruments, Westbury, NY) in ice-cold Trizol reagent (Gibco BRL, Gaithersburg, MD). RNA was extracted with chloroform and precipitated using isopropanol with glycogen as carrier. The RNA pellet was washed in 75% ethanol and resuspended in DEPC-treated MilliQ water. The RNA was diluted in a guanidinium thiocyanate-containing RLT buffer (Qiagen Sciences, Inc., Germantown, MD) and further purified using a silica-gel-based membrane in RNeasy MinElute™ spin columns. The RNA quality was confirmed by electrophoresis on a 1% non-denaturing agarose gel or by Agilent Bioanalyser, which revealed intact 18 and 28S rRNA in all samples. UV spectroscopy (A210 –A350) was performed to confirm RNA purity; the A260: A280 ratio was 1.99 to 2.00 for all samples.
RNA hybridization
cDNA was prepared from total RNA (10 μg) using the Superscript Choice™ system (Gibco-BRL, Gaithersburg, MD). A T7-(dT) oligonucleotide primer was used for first strand synthesis. Double-stranded cDNA was purified by phenol/chloroform extraction and the Phase Lock Gel® method (Eppendorf-5 Prime, Boulder, CO). Biotin-labeled cRNA was synthesized using a BioArray™ RNA Amplification and Labeling Kit (ENZO, New York, NY), cleaned and fragmented (15). The cRNA was biotin labeled and hybridized on U133A GeneChip® arrays for 17 h (45°C) in an Affymetrix GeneChip® 640 Hybridization Oven (Affymetrix, Santa Clara, CA). Washing and staining of the chips were performed using the Affymetrix GeneChip® Fluidics Station 400. Arrays were scanned in an HP GeneArray Scanner (Hewlett Packard, Palo Alto, CA), and data were analyzed with Affymetrix Microarray Suite v5.0 software (MAS5).
Microarray analysis of discriminatory genes
Probes that were not detected as “present” in at least half of the samples using the standard MAS5 parameters were removed from analysis (16, 17). The remaining probes were analyzed using Welch's t-test, assuming unequal variance, on the log2 transformed MAS5 signals. The Affymetrix chip for one patient, sample #26, was damaged; consequently, this patient was deleted from the gene analyses.
Quantitative RT-PCR
First strand cDNA synthesis was performed using SuperScript™ III reverse transcriptase (RT) (Invitrogen Corp., Carlsbad, CA) with total RNA (5 μg) and oligo dT12-18 primer. The reaction mix was diluted (1:12.5) and used for q-PCR analysis. The primers and probes used for the PCR were purchased from Applied Biosystems (Foster city, CA). The probes used for DBN1 (Hs00365623_m1, exon boundary 9-10), PP591 (Hs00611011_m1, exon boundary 5-6), CCND2 (Hs00277041_m1, exon boundary1-2) and CACNB3 (Hs00167873_m1, exon boundary 9-10) were all FAM labeled. FAM labeled GAPDH probes (433376F) was used in separate experiments to normalize the Ct value between the samples. The Applied Biosystems master mix was used for the q-PCR reaction and the manufacturer's instructions were followed. The q-PCR reactions were performed in a I-Cycler (BioRad laboratories, Hercules, CA) at 50°C (2 minutes), 95°C (10 minutes) followed by 40 cycles of consisting of 95°C (15 seconds) and 60°C (1 minute) steps. The reactions were performed in triplicate and the GAPDH Ct values were subtracted from the raw sample Ct values to get the corrected Ct, which was converted into the relative RNA amount using the formula (2-(corrected Ct)). All the 27 patient samples were used for q-PCR analysis.
Hierarchical clustering analysis of genes associated with recurrence
For hierarchical clustering, 51 probe sets that differed between the groups (P value < 0.001) were selected. The log2 transformed MAS5 signals were normalized [(signal-mean)/std dev] and then arrays were clustered using the hierarchical clustering function of Partek ® Genomics Suite, version 6.3 © 2007 (Partek, Inc., St. Louis, MO), with Euclidean distance and average linkage. Pearson's dissimilarity and average linkage were used to cluster the probe sets. Normalization was performed on each probe set to ensure that no individual probe set would have undue influence on the clustering.
Validation set patients
The validation set consisted of 138 patients (n=138) from Samsung Medical Center, Korea. Of 138 patients, 69 exhibited no recurrence following surgery (group NR) and 69 patients exhibited recurrence after surgery (group R). The details of the patients of this validation set are described in Lee et al., 2008 (11). The patients were a mix of different NSCLC stages, established by pathologic staging after surgery: 64, 17 and 19% were stages I, II and III, respectively. The TNM staging of the patients was widely distributed: 17% patients were pT1 stage, 68% pT2, 7% pT3 and 7% pT4. Most of the patients had no nodal involvement (pN0, 71%), while 20% were pN1 and 9% were pN2. Of the total, 63 patients had adenocarcinoma and 75 patients had squamous cell carcinoma. The microarray data obtained was processed using gene chip robust multi-array average (GCRMA) normalization (18) with perfect match (PM) and perfect match/mismatch (PM/MM) modeling.
Among the 69 non-recurrent patients, three received adjuvant combination chemotherapy. Specifically, one patient received a combination of fluorouracil, leucovorin, ifosfamide and dexamethasone, another patient received combination of etoposide and dexamethasone, and a third patient received a combination of cisplatin and paclitaxel. Nine of the recurrent patients received different adjuvant chemotherapy or biologically targeted regimens including: gefitinib, etoposide/dexamethasone, cisplatin/paclitaxel, cisplatin/etoposide/dexamethasone, and docetaxel/cisplatin/dexamethasone/gemcitabine.
Clinical, genomic and clinicogenomic models of RFS in the training set
The objective was to derive three models for RFS, models based solely on patient characteristics (i.e. clinical model), solely on gene expression levels (i.e. genomic model), and on both (i.e. clinicogenomic model). To model RFS outcomes in the training set, multivariate analyses were performed using a stepwise Cox proportional hazards model on the 27 patients (stage I-III). For the clinical model, patient characteristics including age, sex, race, and smoking history were considered. Smoking history was categorized as: current smoker (C, defined as smoking <1 year before surgery), former smoker (F, defined as quit >1 year before surgery), and never smoker (N). For each of the models, a subsequent Cox score was calculated and dichotomized at its median value into low- and high-risk groups. The recurrence-free survival curves were estimated by using the Kaplan-Meier method and compared by log-rank test. The variables used for the clinical model of the training set were: histology, pathologic stage (pStage), sex, race, and smoking. For the genomic and clinicogenomic models the natural logarithm of gene expression levels determined by qPCR was included, using the genes DBN1, FLAD1 (PP591; flavin adenine dinucleotide synthetase), CACNB3 and CCND2, which were identified as being differentially expressed in both the training and validation sets. Median value of the Cox score was determined as previously described (11) and was used to dichotomize the patients into low- and high-risk groups for Kaplan-Meier plots. The equations used for the modeling are given in the Fig. 2 legend. The SAS program codes used to derive the models are provided in the Supplementary Material (SAS Codes).
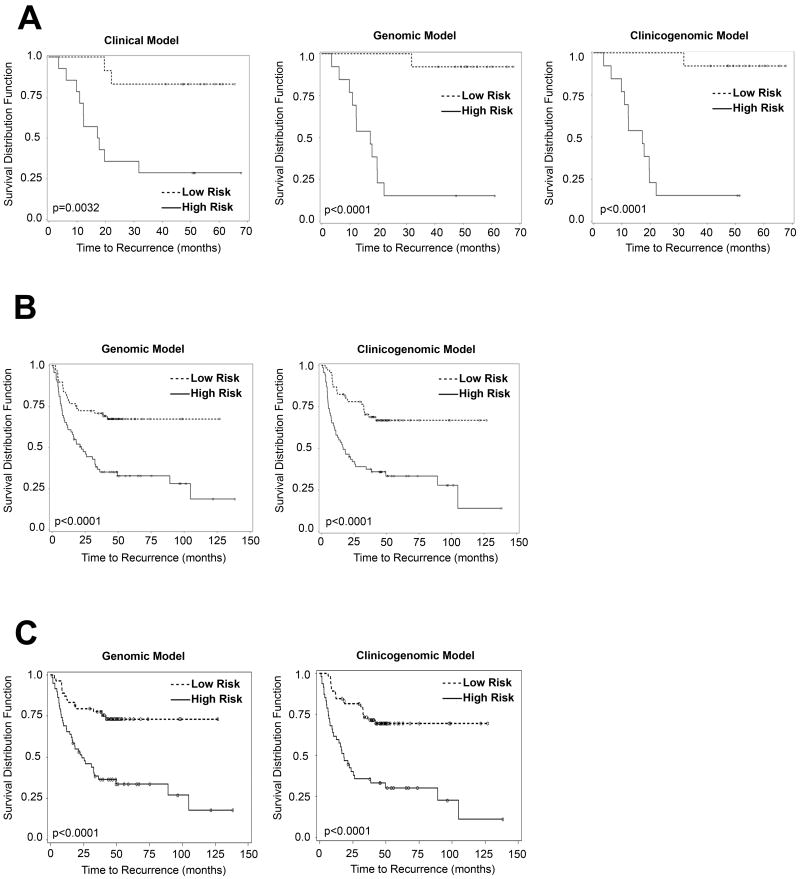
Figure 2.
A. Clinical, genomic and clinicogenomic RFS models of training cohort (stage I-III patients). The clinical model is: Cox score = Histology (-16.12716 × Squamous - 1.03510 × other) + 0.95865 × pStage + 0.22791 × Gender + Race (-0.57444 × African American - 0.95745 × Undetermined); the genomic model is: Cox score = -713.79613 × DBN1 -1090 × CCND2 + 668.28496 × CACNB3 + 383.51066 × PP591; and the clinicogenomic model is: Cox score = -1218 × DBN1 - 2072 × CCND2 + 1710 × CACNB3 + 507.39229 × PP591 + Histology (-14.22074 × Squamous - 2.78616 × other) + 0.60136 × pStage + 1.40561 × gender + Race (-3.16002 × Black - 3.44106 × Undetermined). The cutoffs (i.e. median Cox scores) for the clinical, genomic and clinicogenomic models were 1.18655, -30.8891 and -45.2830, respectively. Time to recurrence is expressed in months; number of patients: 27.
B. Genomic and clinicogenomic RFS models of validation cohort (stage I-III patients). The genomic model is: Cox score = 0.38040 × DBN1 + 0.30160 × CACNB3 - 0.93964 × CCND2 + 0.28898 × FLAD1; the clinicogenomic model is: Cox score = pT stage (0.58862 × T2+2.19489 × T3+0.31153 × T4) + pN stage (0.75430 × N1+1.24674 × N2) + 0.56392 × DBN1 + 0.43517 × CACNB3 - 0.61023 × CCND2 + 0.39543 × FLAD1. The median Cox scores for the genomic and clinicogenomic models were 3.66 and 7.43, respectively. Time to recurrence is expressed in months; number of patients: 138. The clinical model for this set of patients has been previously published (11).
C. Genomic and clinicogenomic RFS models of validation cohort (stage I-II patients). The genomic model is: Cox score = 0.37488 × DBN1 + 0.47831 × CACNB3 - 0.98259 × CCND2 + 0.33511 × FLAD1; the clinicogenomic model is: Cox score = pT stage (0.82325 × T2 + 2.78736 × T3) + 1.02786 × pN stage + 0.54254 × DBN1 + 0.46196 × CACNB3 - 0.99139 × CCND2 + 0.36875 × FLAD1. Time to recurrence is expressed in months; number of patients: 112.
Genomic and clinicogenomic RFS models of the validation set
Genomic and clinicogenomic models for the stage I-III patients of the validation set were developed similar to a prior study of the same cohort, for which the clinical model has already been published (11). The genomic markers used for the validation set model were: DBN1, FLAD1 (PP591; flavin adenine dinucleotide synthetase), CACNB3 and CCND2. For the validation set, there were several Affymetrix probes for each gene: 2 for DBN1, 2 CACNB3, 5 for CCND2 and 2 for FLAD1 (Table S2). To optimize the model, all the probe combinations were considered (2 × 2 × 5 × 2 = 40). Single Affymetrix probes were selected for each gene using a stepwise parameter method (11) that allowed selection of an optimized model of 4 unique probes, (Table S2). To develop the clinicogenomic model for the validation set, the same stepwise parameter selection was also used for filtering of clinical variables. Pathologic T stage (p-value=0.0003) and pathologic N stage (p-value=0.0038) were identified as being of utility among the considered variables, whereas age, gender, cell type, tumor size, smoking status and tumor differentiation were not.
Median value of the Cox score was determined as previously described (11)and was used to dichotomize the patients into low- and high-risk groups for Kaplan-Meier plots. The equations used for the modeling are given in the Fig. 2 legend. The SAS program codes used to derive the models are provided in the Supplementary Material (SAS Codes).
Genomic and clinicogenomic RFS modeling of Stage I and II patients in the validation set
Stage I-II patients of the validation set were selected for Cox analysis. The genomic markers used for the genomic modeling were those described above (DBN1, FLAD1, CACNB3 and CCND2). The clinicogenomic model incorporated the genomic information with additional pT and pN stage data. The Cox equations used for the modeling are given in the Fig. 2 legend. The SAS program codes used to derive the models are provided in the Supplementary Material (SAS Codes). Due to the relatively small number of patients, this type of analysis was not performed on the training set.
Genomic and clinicogenomic OS modeling of validation set
Stage I-III patients of the validation set were included in this Cox analysis. All the deaths included in this analysis are death due to lung cancer, including complications associated with lung cancer (disease-specific OS). To optimize the OS model, a Cox proportional hazards regression model with stepwise parameter selection method was developed. The Cox equations used for the modeling are given in the Fig. 3 legend. The SAS program codes used to derive the models are provided in the Supplementary Material (SAS Codes). Due to the relatively small number of patients, this analysis was not performed on the training set.
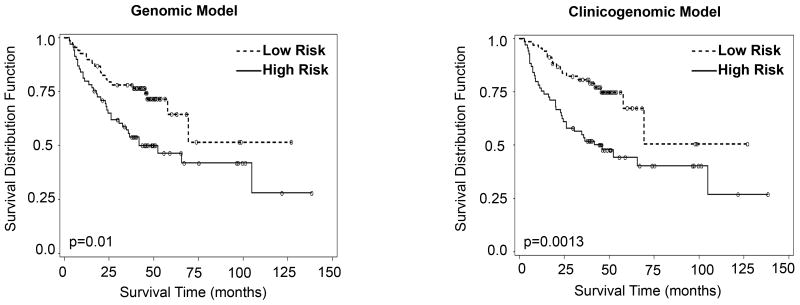
Figure 3. Genomic and clinicogenomic disease-specific OS models of validation cohort (stage I-III patients).
The genomic model is: Cox score = 0.39443 × DBN1 + 0.13635 × CACNB3 - 0.77471 × CCND2 + 0.28103 × FLAD1; the clinicogenomic model is: Cox score = pT stage (1.03717 × pT2+2.34391 × pT3+0.69257 × pT4) + pN stage (0.54998 × pN1+1.22555 × pN2) + 0.48697 × DBN1 + 0.25821 × CACNB3 - 0.69815 × CCND2 + 0.32732 × FLAD1.
Overall survival is expressed in months; number of patients: 138.
Results
American patient training set
The training set of patients accrued at the Indiana University Thoracic Oncology Program consisted of 27 patients with stage Ia-IIIb NSCLC who were evaluable for recurrence at two years of follow-up. All histologies of NSCLC were included. This patient cohort was reflective of a broad patient population seen in an American academic medical center without selection for histology or stage. This approach could be useful because genomic markers derived from this type of study could potentially be applied to clinical decision-making in a general thoracic oncology practice. There were 11 patients who experienced recurrence within two years of resection (group R) and 16 patients who did not (group NR). Patient characteristics, including stage of disease, recurrence status, histology, age at surgery, smoking history, survival after surgery, time to recurrence, gender, and adjuvant therapy are described in Table 1. The majority of the NSCLC patients exhibited adenocarcinoma histology (n=19). Mean age of the patients at operation was 61.8 (SD=12.8) years (range 34-81 years).
Table 1. Description of training set patients.
| Training set (n=27) | |||
|---|---|---|---|
| Age at operation (years) | Mean | 61.8 (SD=12.8) | |
| Range | 34-81 | ||
| Gender | Male | 15 | 55.5% |
| Female | 12 | 44.5% | |
| Smoking History | Never smoker | 5 | 18.5% |
| Former smoker | 14 | 51.9% | |
| Current Smoker | 8 | 29.6% | |
| Histology | Adenocarcinoma (ADC) | 18 | 66.7% |
| Squamous carcinoma (SQC) | 3 | 11.1% | |
| Other | 6 | 22.2% | |
| Histological differentiation | Well differentiated | 25 | 92.6% |
| Poorly differentiated | 2 | 7.4% | |
| Stage | IA | 5 | 18.5% |
| IB | 10 | 37.0% | |
| IIA | 1 | 3.7% | |
| IIB | 3 | 11.1% | |
| IIIA | 5 | 18.5% | |
| IIIB | 2 | 7.4% | |
| Not known | 1 | 3.7% | |
| T | 1 | 7 | 25.9% |
| 2 | 15 | 55.6% | |
| 3 | 1 | 3.7% | |
| 4 | 1 | 3.7% | |
| Not known | 3 | 11.1% | |
| N | 0 | 16 | 59.3% |
| 1 | 3 | 11.1% | |
| 2 | 4 | 14.8% | |
| 3 | 1 | 3.7% | |
| Not known | 3 | 11.1% | |
| M | 0 | 9 | 33.3% |
| X | 13 | 48.1% | |
| Not known | 5 | 18.5% | |
| Time to recurrence (number of patients) |
0-12 months | 6 | 22.2% |
| 13-24 months | 5 | 18.5% | |
| 25-36 months | 1 | 3.7% | |
| 37-48 months | 0 | 0% | |
| 49-60 months | 2 | 7.4% | |
| Not recurred | (up to max follow up of 67 months) | 13 | 48.1% |
| Overall survival | 0-12 months | 2 | 7.4% |
| 13-24 months | 4 | 14.8% | |
| 25-36 months | 2 | 7.4% | |
| 37-67 months (max follow up) | 19 | 70.4% | |
| Adjuvant therapy | Neo adjuvant | 3 | 11.1% |
| Adjuvant | 0 | 0% | |
The median time to recurrence (TTR) of group R was 12 months and the median overall survival (OS) was 20 months. This study had a 57-month median follow-up and the 2-year cut-off captured 81% of the recurring patients. Among the NR group, 19% of the patients recurred after 2 years, the recurrences being at 32, 50 and 53 months and these patients were still alive at the time of data collection closure. Age at operation, race, gender and smoking status were analyzed in conjunction with gene expression data in subsequent Cox multivariate analysis (see below).
Genomic markers associated with NSCLC recurrence in the training set
To identify genes that were differentially expressed in recurring patients, genomic microarray analysis of tumor gene expression was performed by Affymetrix U133A chip hybridization. Candidate genomic recurrence markers (Table 2) were identified from 12,956 evaluable probes in the microarray by statistical analysis of log2-transformed signals (Welch's T-test; P≤0.001). This analysis resulted in 51 probes corresponding to 44 genes that were differentially expressed between the R and NR groups (Table 2) (raw data available at Geo database; GSE9971) (Table S1).
Table 2. Differentially Expressed Genes in Recurrent NSCLC (Group R) Compared to Non-Recurrent NSCLC (Group NR) – Differential analysis based on P values <0.001.
| Gene Symbol | Source id | Fold Change | Welch's T-test Log(Signal) | Description |
|---|---|---|---|---|
| CYP3A5 | 205765_at | 18.66 | 1.30E-04 | Cytochrome P450, family 3, subfamily A, polypeptide 5 |
| DPM3 | 219373_at | 1.87 | 4.60E-04 | Dolichyl-phosphate mannosyltransferase polypeptide 3 |
| NES | 218678_at | 1.85 | 2.20E-04 | Nestin |
| TACSTD1 | 201839_s_at | 1.84 | 5.80E-04 | Tumor-associated calcium signal transducer 1 |
| DECR1 | 202447_at | 1.74 | 1.50E-04 | 2,4-dienoyl CoA reductase 1, mitochondrial |
| DBN1 | 202806_at | 1.71 | 6.40E-04 | Drebrin 1 |
| KIAA1598 | 221802_s_at | 1.62 | 5.30E-04 | KIAA1598 |
| CACNB3 | 34726_at | 1.58 | 1.10E-04 | Calcium channel, voltage-dependent, beta 3 subunit |
| PP591 | 212541_at | 1.56 | 8.40E-04 | FAD-synthetase |
| PSMD4 | 200882_s_at | 1.56 | 6.20E-04 | Proteasome 26S subunit, non-ATPase, 4 |
| DNAJB12 | 202866_at | 1.52 | 3.50E-04 | DnaJ (Hsp40) homolog, subfamily B, member 12 |
| MAN2A2 | 202032_s_at | 1.52 | 2.60E-04 | Mannosidase, alpha, class 2A, member 2 |
| D2LIC | 203763_at | 1.51 | 0.001 | Dynein 2 light intermediate chain |
| DKFZp566O084 | 220690_s_at | 1.51 | 7.00E-05 | DKFZP566O084 protein |
| FLJ20343 | 217964_at | 1.46 | 1.00E-05 | Hypothetical protein FLJ20343 |
| BCAP31 | 200837_at | 1.45 | 3.00E-04 | B-cell receptor-associated protein 31 |
| SMBP | 217758_s_at | 1.42 | 8.10E-04 | SM-11044 binding protein |
| ECHS1 | 201135_at | 1.39 | 8.80E-04 | Enoyl Coenzyme A hydratase, short chain, 1 |
| SCRIB | 212556_at | 1.38 | 5.00E-04 | Scribbled homolog |
| SUPV3L1 | 212894_at | 1.35 | 8.40E-04 | Suppressor of var1, 3-like 1 |
| XPNPEP1 | 209045_at | 1.35 | 9.90E-04 | X-prolyl aminopeptidase 1 |
| C10orf119 | 217905_at | 1.33 | 4.00E-04 | Chromosome 10 open reading frame 119 |
| ACTR1A | 200721_s_at | 1.32 | 3.30E-04 | ARP1 actin-related protein 1 homolog A |
| PSMB4 | 202244_at | 1.23 | 9.80E-04 | Proteasome subunit, beta type, 4 |
| TEX27 | 218020_s_at | -1.19 | 5.90E-04 | Testis expressed sequence 27 |
| MLL | 212076_at | -1.24 | 6.50E-04 | Myeloid/lymphoid or mixed-lineage leukemia |
| ENO1 | 216554_s_at | -1.28 | 4.50E-04 | Enolase 1 |
| C6orf69 | 214849_at | -1.35 | 8.30E-04 | Chromosome 6 open reading frame 69 |
| SPAG9 | 212470_at | -1.37 | 2.40E-04 | Sperm associated antigen 9 |
| STK17B | 217503_at | -1.37 | 6.50E-04 | Serine/threonine kinase 17b |
| FLJ10996 | 219774_at | -1.38 | 9.20E-04 | Hypothetical protein FLJ10996 |
| C14orf118 | 219720_s_at | -1.41 | 8.00E-05 | Chromosome 14 open reading frame 118 |
| TBC1D5 | 201815_s_at | -1.41 | 2.20E-04 | TBC1 domain family, member 5 |
| CLN5 | 204085_s_at | -1.43 | 9.10E-04 | Ceroid-lipofuscinosis, neuronal 5 |
| AHCYL1 | 207464_at | -1.45 | 4.10E-04 | S-adenosylhomocysteine hydrolase-like 1 |
| STAT2 | 205170_at | -1.48 | 1.10E-04 | Signal transducer and activator of transcription 2 |
| ATF7IP | 218987_at | -1.52 | 1.30E-04 | Activating transcription factor 7 interacting protein |
| CD44 | 210916_s_at | -1.62 | 4.20E-04 | CD44 antigen |
| HIPK3 | 210148_at | -1.72 | 1.80E-04 | Homeodomain interacting protein kinase 3 |
| HIPK3 | 207764_s_at | -1.85 | 1.40E-04 | Homeodomain interacting protein kinase 3 |
| CD44 | 204490_s_at | -1.88 | 2.50E-04 | CD44 antigen |
| CCND2 | 200952_s_at | -1.89 | 1.90E-04 | Cyclin D2 |
| CD44 | 209835_x_at | -1.99 | 5.40E-04 | CD44 antigen |
| TRA@ | 209671_x_at | -2.1 | 3.10E-04 | T cell receptor alpha locus |
| CD44 | 212014_x_at | -2.12 | 8.80E-04 | CD44 antigen |
| HLA-DOA | 206313_at | -2.2 | 1.60E-04 | Major histocompatibility complex, class II, DO alpha |
| SOD2 | 221477_s_at | -2.25 | 6.60E-04 | HepG2 3′ region MboI cDNA, clone hmd2a08m3 |
| SOD2 | 216841_s_at | -2.52 | 7.40E-04 | Superoxide dismutase 2, mitochondrial |
| SOD2 | 215223_s_at | -2.77 | 8.80E-04 | Superoxide dismutase 2, mitochondrial |
| CCND2 | 200951_s_at | -3.49 | 4.50E-04 | Cyclin D2 |
| CXCL13 | 205242_at | -5.31 | 4.00E-04 | Chemokine (C-X-C motif) ligand 13 |
Affymetrix source id is indicated because of the identification of multiple Affymetrix probes for several genes, including CD44, SOD2, HIPK3 and CCND2.
Hierarchical clustering analysis of training set genes associated with NSCLC recurrence
Hierarchical clustering using the 51 probes associated with recurrence separated the training set patients exhibiting recurrence. Based on recurrence or not at 2 y of follow-up, this clustering identified three subgroups of genes exhibiting up-regulation and one exhibiting down-regulation in recurrence (Fig.1). The most statistically significant markers up-regulated in recurrence were, in order of significance by Welch's T test, FLJ20343, DKFZp566O084, CACNB3, CYP3A5, and DBN1 (Table 2). The first three markers were associated with one cluster of genes up-regulated in patients exhibiting recurrence (Group 1), while CYP3A5 was associated with a second cluster (Group 2) and DBN1 with a third (Group 3). Genes in groups 1-3 were associated with a broad range of T test values (ranging from 0.001 to 0.00001). The most statistically significant markers that were down-regulated in recurring patients were, in order of significance, C14orf118, STAT2, ATF7IP, HIPK3 and HLA-DOA (Table 2) and this group clustered together (Group 4). Group 4 exhibited a smaller distribution of T test values (ranging from 0.0009 to 0.00008), consistent with the larger size of this group.
Figure 1. Gene expression profile and hierarchical clustering separating the recurrent and non-recurrent groups on the basis of discriminatory genes.
Twenty six evaluable tumors were clustered hierarchically on the basis of 51 probes corresponding to 44 genes, using Partek ® Genomics Suite v6.3. The dendrograms of individual patient samples and overall patterns of gene expression data are exhibited. The recurrent group is labeled R and non-recurrent NR. Individual tumor specimen numbers are indicated at the bottom of the figure and the tumor specimen dendrogram is given along the top. Gene symbols are indicated on the right side of the figure and relatedness of gene expression is indicated by the dendrogram on the left side of the figure. The clustering is of log2 transformed Affymetrix MAS5 signals. Red color indicates increased expression and blue color lower, relative to the mean level of gene expression, indicated in grey. The color scale indicates the mean log2 expression level above (red) and below (blue) the mean of all genes (grey), over a range of 2.9 log2 units above and below the mean.
Screening of a Korean patient validation set for concordance of candidate genomic markers
We hypothesized that screening of a geographically distant and demographically distinct patient population for recurrence-associated genomic markers would lead to the identification of more reproducible genes for the study of NSCLC prognosis. To find a distinct and larger validation set, the GEO database was screened for NSCLC studies with similar stage grouping and no selection based on histology, performed on a similar genomics platform. A Korean study of NSCLC recurrence-associated genomic markers, GSE8894, met the criteria for comparison with the American training set and was probed for genomic markers associated with recurrence common to both sets (11). The K-M curve for the Korean patients exhibited a median disease-specific survival time of 69.4 months and a 5-y survival percentage of 56.2%, comparable to but perhaps slightly shorter than North American patients with resected NSCLC (9) (Fig. S1).
Each of the 44 genes identified in the American training set was tested in the Korean data set for significance by univariate and multivariate Cox analysis. The four genes that were most significant by univariate Cox analysis were, in order of significance, DBN1, FLAD1 (PP591), CACNB3 and CCND2 (Table 3). The first three genes exhibited P values <0.05 and the fourth, exhibiting a P value of 0.08, was retained for model building. By multivariate analysis, only DBN1 exhibited significance (P=0.0095) (Table 3). Nonetheless, two of the genes approached multivariate significance, FLAD1 (P=0.0720) and CCND2 (P=0.0713), while CACNB3 did not (P=0.1813) (Table 3). These results indicate that DBN1 is potentially of value as a multivariate marker and the combination of the 4 genes was effective in model building (see below).
Table 3. Validation of genes using Cox-analysis.
| A- Univariate Cox analysis | ||||
|---|---|---|---|---|
| Gene Symbol | Probe | Hazard Ratio | 95% C.I. for Hazard Ratio | Cox analysis p-value |
| DBN1 | 202806_at | 1.743 | 1.238, 2.453 | 0.0014 |
| FLAD1 (PP591) | 212541_at | 1.567 | 1.064, 2.307 | 0.0229 |
| CACNB3 | 34726_at | 1.421 | 1.025, 1.968 | 0.0347 |
| CCND2 | 200952_at, 200951_s_at |
0.296 | 0.075, 1.163 | 0.0813 |
| B-Multivariate Cox analysis | ||||
|---|---|---|---|---|
| Gene Symbol | Probe | Hazard Ratio | 95% C.I. for Hazard Ratio | Cox analysis p-value |
| DBN1 | 202806_at | 1.565 | 1.116, 2.195 | 0.0095 |
| FLAD1 (PP591) | 212541_at | 1.422 | 0.969, 2.088 | 0.072 |
| CACNB3 | 34726_at | 1.267 | 0.896, 1.792 | 0.1813 |
| CCND2 | 200952_at, 200951_s_at |
0.303 | 0.083, 1.109 | 0.0713 |
Confirmation of DBN1, CACNB3, FLAD1 (PP591) and CCND2 expression in the training set by q-PCR
The utility of prognosis-associated genes identified by microarray analysis is increased if they are also assayable by q-PCR (11). Increased expression of the DBN1, CACNB3 and FLAD1 (PP591) genes in the recurrent NSCLC tumors was confirmed in the training set by q-PCR (Table 4). Decreased expression of CCND2 in the recurring patients of the training set approached, but did not reach, statistical significance (Table 4). These expression values were used to perform subsequent Cox regression analysis of the training set.
Table 4. q-PCR analysis of DBN1, CACNB3, FLAD1 (PP591) and CCND2.
| Relative % of Expression | ||||
|---|---|---|---|---|
| Gene name | Recurrent | Non-recurrent | Fold change | P-value |
| DBN1 | 50.7 | 24.2 | 2.094 | 0.012 |
| CACNB3 | 82.1 | 28.1 | 2.921 | 0.033 |
| FLAD1 (PP591) | 72 | 20.75 | 3.471 | 0.001 |
| CCND2 | 189.9 | 405.79 | 0.468 | 0.058 |
Clinical, genomic, and clinicogenomic modeling of the training and validation sets
A clinical model of the training set was performed, based on histology, pStage, sex, race, and smoking history (Fig.2A). The clinical model was effective at separating the low- and high-risk patients in the training set (P=0.0032). The median RFS for the training set was 17.2 months, while the 5-y % RFS was 83.4 and 28.6% for the low- and high-risk groups, respectively (Fig. 2A; Table 5). Nonetheless, application of a similar clinical model to the larger validation cohort was less successful (P=0.0518) (11). A genomic model developed from the training set was more effective than the clinical model at risk stratification of the training (P<0.0001) (Fig.2A) and validation sets (P<0.0001) (Fig.2B). Using the genomic model, the 5-y RFS for the low- and high-risk groups was 92.3 vs. 15.4% and 67.5 vs. 32.8% in the training and validation sets, respectively (Table 5). Using the clinicogenomic model, patients were effectively risk-stratified in the training (P<0.0001) and validation sets (P<0.0001) (Fig.2A and B). Using the clinicogenomic model, the 5-y RFS for the low- and high-risk groups was 92.3 vs. 15.4% and 67.0 vs. 33.3% for the training and validation sets, respectively (Table 5). In summary, the genomic and clinicogenomic models exhibit clinical utility because the difference in 5-y RFS is more than 2-fold indicating a substantial clinical effect.
Table 5. Cox modeling on the training and the validation set.
| Cox Model Training set (Stage I-III): | ||||||
|---|---|---|---|---|---|---|
| Clinical model | Genomic model | Clinicogenomic model | ||||
| High Risk | Low Risk | High Risk | Low Risk | High Risk | Low Risk | |
| Median RFS time (months) | 17.167 | ∞ | 17.167 | ∞ | 17.167 | ∞ |
| Survival percentage for 5-y (%) | 28.57 | 83.37 | 15.38 | 92.31 | 15.38 | 92.31 |
| Cox Model Validation set (Stage I-III): | ||||
|---|---|---|---|---|
| Genomic model | Clinicogenomic model | |||
| High Risk | Low Risk | High Risk | Low Risk | |
| Median RFS time (months) | 21.40 | ∞ | 16.63 | ∞ |
| Survival percentage for 5-y (%) | 32.80 | 67.46 | 33.32 | 66.95 |
| Cox Model Validation set (Stage I-II): | ||||
|---|---|---|---|---|
| Genomic model | Clinicogenomic model | |||
| High Risk | Low Risk | High Risk | Low Risk | |
| Median RFS time (months) | 23.77 | ∞ | 18.33 | ∞ |
| Survival percentage for 5-y (%) | 33.84 | 73.19 | 30.33 | 69.56 |
| Cox Model Validation set (OS; Stage I-III): | ||||
|---|---|---|---|---|
| Genomic model | Clinicogenomic model | |||
| High Risk | Low Risk | High Risk | Low Risk | |
| Median disease-specific survival time (months) | 22.40 | ∞ | 46.2 | ∞ |
| Survival percentage for 5-y (%) | 36.91 | 63.31 | 44.23 | 67.35 |
Clinical Model of the validation set has been described in Lee et. al. 2008.
Clinical, genomic, and clinicogenomic modeling of stage I and II patients in the validation set
Because the decision to offer chemotherapy or not is crucial for early stage patients, we reanalyzed the stage I-II patients in the validation set using the 4 genomic markers to develop genomic and clinicogenomic models for this risk group. The genomic model risk-stratified the stage I-II patients (P value < 0.0001; Fig.2C), exhibiting 5-y RFS of 73.2 vs. 33.8 % for the low- and high-risk groups, respectively (Fig. 2C and Table 5). The clinicogenomic model also risk-stratified stage I-II patients (P value < 0.0001; Fig.2C), exhibiting 5-y RFS of 69.6 vs. 30.3% for the low- and high-risk groups, respectively (Fig. 2C and Table 5). These results support the utility of the 4 genes for risk model development for stage I-II patients.
Genomic and clinicogenomic modeling of the validation set based on disease-specific OS
Risk stratification on the basis of disease-specific OS is another important test of the utility of the 4 genomic markers. Therefore, genomic and clinicogenomic models were developed. Both models were equally effective risk-stratifying the patients into low- and high-risk groups (P<0.0001) (Fig. 3). Using the genomic and clinicogenomic models, the 5-y disease-specific survival for the low- and high-risk groups was 63.3 vs. 37.0% and 67.3 vs. 44.2%, respectively (Table 5). These differences in disease-specific OS were at least 1.5-fold, indicating clinical utility.
Analysis of multivariate marker DBN1
The genomic marker DBN1 was identified as a significant in the genomic and clinicogenomic models of RFS stage I-III, RFS stages I-II and disease-specific OS (Stage I-III RFS genomic HR=1.463 CI 1.088-1.967; Stage I-III RFS clinicogenomic HR=1.758 CI 1.248-2.476; Stage I-II RFS genomic HR=1.455 CI 1.038-2.04; Stage I-II RFS clinicogenomic HR=1.72 CI 1.165-2.541; OS genomic HR=1.484 CI 1.057-2.082; OS clinicogenomic HR=1.627 CI 1.115-2.367; Table S3). The addition of pT and pN stage information improved the significance of DBN1 for all models (RFS stage I-III, P= 0.0119 to 0.0013; RFS stage I-II, P= 0.0297 to 0.0064; OS, P= 0.0226 to 0.0117). This finding indicates that DBN1 serves as a component of the genomic model that can be improved by the addition of clinical stage.
Discussion
We hypothesized that comparison of two geographically distant and demographically distinct patient cohorts, namely American and Korean, could facilitate the identification and validation of NSCLC genomic markers associated with RFS. Four genes were associated with recurrence in the American training set and validated in the larger Korean patient cohort. Genomic and clinicogenomic modeling for RFS risk-stratified both cohorts with high statistical significance, and genomic modeling alone was sufficient to risk-stratify the groups. Genomic and clinicogenomic models also risk-stratified stage I-II patients of the validation set. For stage I-II or I-III patients, the 5-y RFS of the low- and high-risk patients differed by ≥ 2-fold. The genomic and clinicogenomic models also predicted 5-y disease-specific OS, exhibiting a 1.5-1.7-fold difference between low- and high-risk groups.
The comparison of American and Korean cohorts resulted in discovery of DBN1 as a multivariate biomarker, which by itself exhibits significant prognostic utility for RFS that is improved by the addition of the clinical markers of pT and pN stage. Because addition of clinical stage information increased the HR of the DBN1 marker in all patient groups and for all outcomes, DBN1 may be a platform on which to build future models consisting of other multivariate markers. Additional multivariate markers may be discovered by similar comparisons of demographically distinct NSCLC patient populations. For example, the validation set from this study could be used as the training set for an even larger demographically distinct patient cohort, such as a larger American patient group. This iterative process would confirm that some or all of the genomic markers derived here can contribute to further model building, thus allowing model refinement.
It is notable that the genomic markers that correlate between the training and validation groups were not necessarily the ones exhibiting the lowest univariate P values for differential expression or the greatest fold-differences in expression. This indicates that if modeling were to be performed solely on the basis of P value or fold-differences, genes that could contribute to more robust modeling would be missed. Although clustering of gene expression patterns identified four clusters, only genes belonging to three of the clusters contributed to the model. Of note, one of these clusters contained two of the four genes, including DBN1, which is the only gene significant by multivariate analysis. The approach taken here suggests that successful genomic modeling can be performed even with a relatively small number of significant genomic markers identified by comparison of very different training and validation sets. Furthermore, the approach taken may be effective at identifying more reproducible and broadly applicable genomic markers, which are essential for efficient clinical trial development of a risk model. Thus, adopting this approach, a single, committed gene set can be derived that is statistically significant across a number of patient populations, independent of demographic factors, with informative Cox models as the product.
Of importance, the four genomic markers identified are all amenable to q-PCR assay, which is an important factor in determining clinical development of the 4-gene set. The utilization of q-PCR data for Cox model development confirms their robust amplification and indicates that the differences in their expression between recurrent and non-recurrent patients is sufficient to overcome background noise (11). These findings also provide a robust assay to allow rapid confirmation of the results in other NSCLC patient cohorts.
In summary, the genomic model developed in the present study identifies recurrence-associated genes that are internationally validated and are strong candidates for broader analysis in larger patient cohorts. The validated genomic model exhibits utility, because of the magnitude of the absolute differences between 5-y RFS and OS of the low- and high-risk patients. The utility of the 4-gene set is independent of stage, and is applicable to RFS of stage I-II, I-III patients as well as OS of stage I-III patients. Together these results support the further development of the 4-gene set or its multivariate DBN1 component for future NSCLC risk model development. Of particular importance, risk-stratification has recently demonstrated that low-risk NSCLC patients may exhibit decreased disease-specific survival when treated with adjuvant cisplatin/vinorelbine chemotherapy (1). With confirmation, our 4 genomic markers could be prospectively tested in a future clinical trial to determine the effectiveness of risk stratification of patients independent of stage and could also potentially be used to test the effectiveness of novel adjuvant chemotherapy regimens in low- and high-risk patients.
Supplementary Material
The K-M curve for OS indicates median survival time of 69.4 months and 56.2% OS at 5-y.
Acknowledgments
We thank Drs. Lawrence Einhorn, Nasser Hanna, David Flockhart, David Donner, Jorge Capdevila, Ming Sound Tsao, Ignacio Wistuba, David Beer, Mitch Raponi, Joan Schiller, Peter Ravdin, Faris Farassati, Janice Blum, Penni Black, Arkadiusz Dudek and Miriam Garland for helpful discussions. We are grateful to Michael Franklin for outstanding editing help. We acknowledge a grant from the Eli Lilly Co to support the tissue bank and Carol Boyd and Christina Beard for excellent technical help with tissue banking. This paper is dedicated to the memory of Dr. Stephen Williams, past Director of the IU Simon Cancer Center whose vision led to the IU/Lilly Tumor Bank.
Grant Support: DAP acknowledges grants NIH P20-GM66403, R01 CA113570, the Flight Attendant Medical Research Institute, Walther Cancer Research Prize, the Thoracic Oncology Program at Indiana University Simon Cancer Center, the Walther Oncology Center at Indiana University, the Cancer Experimental Therapeutics Initiative of the Masonic Cancer Center, and the Dr. Barbara Bowers Oncology Fund of the Fairview Foundation. The training set microarray experiments were carried out using the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by a grant to HJE from the Indiana 21st Century Research and Technology Fund, and from the Indiana Genomics Initiative (INGEN). This work was supported in part by NIH P30 CA77598 utilizing the support of the Biostatistics and Bioinformatics Core of the Masonic Cancer Center, University of Minnesota shared resource.
Footnotes
References
- 1.Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–24. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 3.Chen G, Gharib TG, Wang H, et al. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci U S A. 2003;100:13537–42. doi: 10.1073/pnas.2233850100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–72. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 5.Yanagisawa K, Tomida S, Shimada Y, Yatabe Y, Mitsudomi T, Takahashi T. A 25-signal proteomic signature and outcome for patients with resected non-small-cell lung cancer. J Natl Cancer Inst. 2007;99:858–67. doi: 10.1093/jnci/djk197. [DOI] [PubMed] [Google Scholar]
- 6.Seike M, Yanaihara N, Bowman ED, et al. Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst. 2007;99:1257–69. doi: 10.1093/jnci/djm083. [DOI] [PubMed] [Google Scholar]
- 7.Wright CD, Kesler KA. New trends in the surgical treatment of non-small cell lung cancer. Indiana Med. 1990;83:192–4. [PubMed] [Google Scholar]
- 8.Hanna N, Brooks JA, Fyffe J, Kesler K. A retrospective analysis comparing patients 70 years or older to patients younger than 70 years with non-small-cell lung cancer treated with surgery at Indiana university: 1989-1999. Clin Lung Cancer. 2002;3:200–4. doi: 10.3816/clc.2002.n.003. [DOI] [PubMed] [Google Scholar]
- 9.Ravdin PM, Davis G. Prognosis of patients with resected non-small cell lung cancer: impact of clinical and pathologic variables. Lung Cancer. 2006;52:207–12. doi: 10.1016/j.lungcan.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Mountain CF. Revisions in the International System for Staging Lung Cancer. Chest. 1997;111:1710–7. doi: 10.1378/chest.111.6.1710. [DOI] [PubMed] [Google Scholar]
- 11.Lee ES, Son DS, Kim SH, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14:7397–404. doi: 10.1158/1078-0432.CCR-07-4937. [DOI] [PubMed] [Google Scholar]
- 12.Potti A, Mukherjee S, Petersen R, et al. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N Engl J Med. 2006;355:570–80. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 13.Kang CH, Ra YJ, Kim YT, Jheon SH, Sung SW, Kim JH. The impact of multiple metastatic nodal stations on survival in patients with resectable N1 and N2 nonsmall-cell lung cancer. The Annals of thoracic surgery. 2008;86:1092–7. doi: 10.1016/j.athoracsur.2008.06.056. [DOI] [PubMed] [Google Scholar]
- 14.Kesler KA, Hammoud ZT, Rieger KM, Kruter LE, Yu M, Brown JW. Carinaplasty airway closure: a technique for right pneumonectomy. The Annals of thoracic surgery. 2008;85:1178–85. doi: 10.1016/j.athoracsur.2007.12.046. discussion 85-6. [DOI] [PubMed] [Google Scholar]
- 15.Zhou FC, Duguid JR, Edenberg HJ, McClintick J, Young P, Nelson P. DNA microarray analysis of differential gene expression of 6-year-old rat neural striatal progenitor cells during early differentiation. Restor Neurol Neurosci. 2001;18:95–104. [PubMed] [Google Scholar]
- 16.McClintick JN, Jerome RE, Nicholson CR, Crabb DW, Edenberg HJ. Reproducibility of oligonucleotide arrays using small samples. BMC Genomics. 2003;4:4. doi: 10.1186/1471-2164-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F. A Model-Based Background Adjustment for Oligonucleotide Expression Arrays. Journal of the American Statistical Association. 2004;99:909–17. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The K-M curve for OS indicates median survival time of 69.4 months and 56.2% OS at 5-y.