Summary
Proper assembly of the kinetochore, a multi-protein complex that mediates attachment of centromere DNA to spindle microtubules on each chromosome, is required for faithful chromosome segregation. Each previously characterized member of the Mis12/Mtw1 protein family is part of an essential sub-complex in the kinetochore. In this work, we identify and characterize CaMTW1, which encodes the homolog of the human Mis12 protein in the pathogenic budding yeast Candida albicans. Subcellular localization and chromatin immunoprecipitation assays confirmed CaMtw1 is a kinetochore protein. CaMtw1 is essential for viability. CaMtw1-depleted cells and cells in which CaMtw1 was inactivated with a temperature-sensitive mutation had reduced viability, accumulated at the G2/M stage of the cell cycle, and exhibited increased chromosome missegregation. CaMtw1 depletion also affected spindle length and alignment. Interestingly, in C. albicans, CaMtw1 and the centromeric histone, CaCse4, influence each other for kinetochore localization. In addition, CaMtw1 is required for efficient kinetochore recruitment of another inner kinetochore protein, the CENP-C homolog, CaMif2. Mis12/Mtw1 proteins have well-established roles in the recruitment and maintenance of outer kinetochore proteins. We propose that Mis12/Mtw1 proteins also have important co-dependent interactions with inner kinetochore proteins and that these interactions may increase the fidelity of kinetochore formation.
Introduction
The centromere is a specialized region of each chromosome on which many proteins assemble to form the kinetochore. Proper segregation of chromosomes during mitosis and meiosis is mediated by a dynamic interaction between spindle microtubules and the kinetochore. A defect in the kinetochore architecture, which disrupts proper attachment between a chromosome and the spindle microtubules, can hamper high fidelity chromosome segregation, leading to unequal distribution of chromosomes in daughter cells. In addition, centromeres/kinetochores are involved in many other associated processes that include sister chromatid cohesion, heterochromatin formation, and spindle checkpoint activity (Cleveland et al., 2003; Cheeseman & Desai, 2008).
Centromeres have been identified and characterized in many organisms ranging from the unicellular yeasts to plants and humans (Malik & Henikoff, 2009). Interestingly, centromeric DNA features, including the length, primary sequence and organization, vary widely among different yeasts. Saccharomyces cerevisiae has ‘point’ centromeres which are short (125 bp in length) and consist of conserved sequence motifs specific for centromeric protein binding (Cleveland et al., 2003). In contrast, the regional centromeres in the fission yeast Schizosaccharomyces pombe vary between 40-110 kb in length and consist of inverted repeat sequence arrays arranged around a non-homologous central core sequence on each chromosome (Clarke, 1998). Many proteins that assemble on the centromeric DNA to form the kinetochore are evolutionarily conserved in diverse systems (Westermann et al., 2003; Joglekar et al., 2008; Cheeseman & Desai, 2008). A critical analysis of kinetochore proteins from several sequenced eukaryotic genomes suggests that some of the kinetochore proteins are exclusively point centromere-specific (CBF3 complex proteins), while some proteins (Dam1 complex proteins) are specific to fungal centromeres (Meraldi et al., 2006). In spite of such species-specific protein requirements for kinetochore function, the overall tri-layer (inner, linker and outer) structure of the kinetochore in diverse organisms (Cleveland et al., 2003) appears to be similar.
The step-wise assembly of the kinetochore and its function depends on complex interactions between several kinetochore proteins. One of the universally conserved kinetochore proteins that constitute the centromeric chromatin is the centromeric histone (CENP-A/CenH3 family). CENP-A proteins replace canonical histone H3 at the centromere to form a specialized centromere-specific chromatin (Meluh et al., 1998; Buchwitz et al., 1999; Blower & Karpen, 2001; Blower et al., 2002). Since CENP-A is present in all the functional centromeres characterized to date, it is considered the fundamental marker of centromere identity (Henikoff et al., 2001). Although the mechanism of how CENP-A proteins are restricted to functional centromeres (native or neocentromeres) remains elusive, the targeting of CENP-A at the centromeres and its propagation in subsequent generations is epigenetically regulated (Ekwall, 2007; Allshire & Karpen, 2008). Besides CENP-A, four multi-protein complexes composed of evolutionarily conserved kinetochore proteins constitute the core linker layer: Mis12/Mtw1 complex, NDC80/Hec1 complex, KNL1/SPC105 complex, and Sim4/COMA complex (Meraldi et al., 2006). Moreover, the Mis12/Mtw1 complex, NDC80 complex and the KNL1 complex form the core (KMN network) of the spindle microtubule attachment site (Cheeseman et al., 2006; Joglekar et al., 2009; Wan et al., 2009). Based on biochemical studies of the recruitment of kinetochore proteins in S. cerevisiae and humans, the Mis12/Mtw1 complex was recently suggested to form a scaffold for the assembly of other kinetochore protein complexes both in the KMN network, as well as outer kinetochore proteins (Maskell et al., 2010; Petrovic et al., 2010).
The Mis12/Mtw1 protein, which is in the linker layer, was first characterized as an essential kinetochore protein in S. pombe (Goshima et al., 1999). Subsequently its functional homologs were identified and characterized in many other organisms including S. cerevisiae (ScMtw1), Drosophila melanogaster (dmMis12), Caenorhabditis elegans (CeMis12), Arabidopsis thaliana (AtMis12), Nicotiana tabaccum (NtMis12) and humans (HsMis12) (Goshima & Yanagida, 2000; Goshima et al., 2003; Sato et al., 2005; Przewloka et al., 2007; Nagaki et al., 2009). In spite of limited conservation at the primary amino acid sequence level, the Mis12/Mtw1 family of proteins is functionally conserved. Mis12/Mtw1 functions as part of a complex together with Dsn1, Nnf1, and Nsl1 (Nekrasov et al, 2003; Kline et al., 2006) to maintain proper centromere/kinetochore architecture; thus, it is essential for chromosome segregation (Goshima et al., 1999; Goshima & Yanagida, 2000; Goshima et al., 2003; Przewloka et al., 2007). Studies in S. cerevisiae and S. pombe suggest that the Mis12/Mtw1 protein family is also required for proper spindle morphogenesis (Goshima et al., 1999; Goshima & Yanagida, 2000; Goshima et al., 2003; Przewloka et al., 2007). Localization patterns of Mis12 and CENP-A (Cnp1) in fission yeast suggest that presence of either one of these proteins at the centromere is independent of the other (Takahashi et. al, 2000). However, in most other organisms the amount of Mis12/Mtw1 localized to kinetochores is influenced by CENP-A (Westermann et al., 2003; Collins et al., 2005; Liu et al., 2006; Przewloka et al., 2007).
The medically important pathogenic yeast Candida albicans is the most frequently isolated fungal pathogen from immunocompromised patients (Navarro-Garcia et al., 2001) and has regional, yet small centromeres. Each of its eight centromere regions contains unique and different 3-5 kb DNA sequences (Sanyal et al., 2004). Furthermore, de novo centromeres are not recruited to naked DNA; rather, centromere formation is epigenetically regulated (Baum et al., 2006). More recently, neocentromere formation at non-centromeric DNA has been reported in this yeast (Ketel et al., 2009) and early replication origins were shown to fire in association with each centromere (Koren et al., 2010). Thus, it is of special interest to study how the same set of kinetochore proteins can assemble on diverse centromeric DNA sequences in C. albicans. Previously, the CENP-A homolog in C. albicans, CaCse4, was characterized as an essential kinetochore protein (Sanyal & Carbon, 2002) and the CaCse4-containing centromere chromatin has been shown to have a non-canonical chromatin structure as inferred from micrococcal nuclease digestion studies (Baum et al., 2006). CaMif2, a homologue of Mif2/CENP-C, is an inner kinetochore protein that binds to the same centromeric regions and colocalized with CaCse4 (Sanyal et al., 2004).
Previous studies indicated that the numbers of microtubules that bind to a kinetochore are variable in different organisms with different types of centromeres. Only one microtubule attaches to the single Cse4/CENP-A nucleosome at each S. cerevisiae kinetochore, while about 2-3 microtubules attach to a S. pombe kinetochore, each of which contains 2-3 Cnp1/CENP-A nucleosomes (Ding et al., 1993; Winey et al., 1995). Interestingly, in C. albicans only one microtubule binds to the regional 3-5 kb centromere which associates with an average of 4 Cse4/CENP-A nucleosomes assembled per kinetochore (Joglekar et al., 2008). However, the stepwise assembly of kinetochore formation on these small, regional centromeres is not well understood.
In this work, we asked how kinetochore identity is determined at the small, regional C. albicans centromeres. We identified CaMTW1, the putative Mis12/Mtw1 homolog and characterized its role in microtubule-kinetochore mediated process of chromosome segregation in C. albicans. Interestingly, we found interdependence of CaMtw1 and CaCse4 in formation of a functional kinetochore. CaMtw1 also plays a role in the recruitment of the inner kinetochore protein CENP-C homolog in C. albicans, CaMif2 at the kinetochore. Thus, CaMtw1 is important for assembly of the inner kinetochore.
Results
CaMtw1 is a kinetochore protein in C. albicans
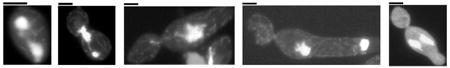
CaMtw1 is a member of the Mis12/Mtw1 family of proteins, and as such is expected to localize to the kinetochore. We verified the presence of CaMtw1 at the kinetochore by colocalizing it with CaCse4, the CENP-A homolog in C. albicans. Strain YJB10704 (Joglekar et al., 2008), where both alleles of CaMtw1 are GFP-tagged, was coimmunostained for CaMtw1 (using anti-GFP antibodies) and CaCse4 (with anti-CaCse4 antibodies) (Fig. 1A). The colocalization pattern strongly suggests that it is a kinetochore protein. Live-cell imaging of the same strain showed that CaMtw1-GFP localized in clustered centromeres in both hypha and yeast cells (Fig. 1B, 1C and 1D, see Supporting Video S1 and Video S2). In hyphal cells, two CaMtw1-GFP foci can be observed following separation of the nuclei (Fig. 1B). In all hyphal cells where the foci could be tracked, one dot (white arrowhead) moved from the septum into the daughter cell, while the other dot (red arrowhead) appeared to move back towards the mother cell (Fig. 1C) consistent with previously characterized nuclear dynamics in C. albicans hyphae (Finley and Berman, 2005). In yeast cells, division of the CaMtw1-GFP focus into two foci was also observed with one dot remaining in the mother cell and the other moving into the daughter cell (Fig. 1D).
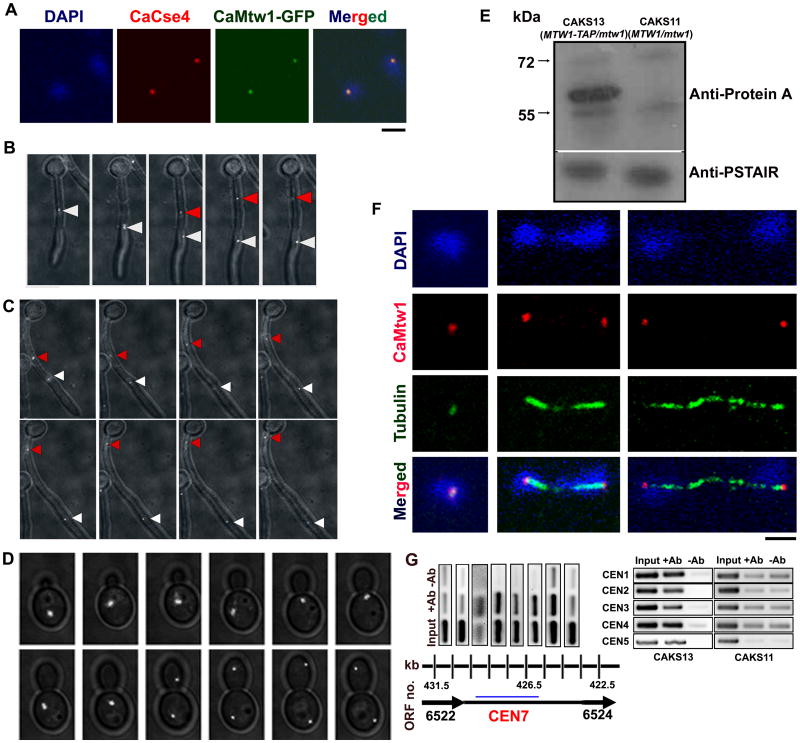
Figure 1.
CaMtw1 is a kinetochore protein in C. albicans. (A) CaMtw1 co-localizes with the centromeric histone, CaCse4. Fixed cells of YJB10704 were stained with DAPI and immuno-stained with anti-CaCse4 antibody and anti-GFP (CaMtw1) antibodies (Bar, 2μm). (B and C) Live-cell time lapse imaging of CaMtw1-GFP in hyphal cells. Cells were imaged at 4 min intervals. Images shown are overlays of DIC and GFP images. The red and white arrowheads point to two CaMtw1-GFP foci, which after segregation were moving away from each other. One focus (red) was migrating back to the mother cell, and the other (white) was moving in the opposite direction (hyphal germ-tube). (D) Live-cell time-lapse imaging of CaMtw1-GFP in yeast-form cells. Cells were imaged at 4 min intervals. Images shown are overlays of DIC and GFP images. (E) Western blot analysis of whole cell extracts prepared from CAKS11 (MTW1/mtw1) and CAKS13 (MTW1-TAP/mtw1) cells with anti-protein A antibodies shows that CAKS13 expresses TAP-tagged CaMtw1 (predicted molecular weight of 57 kDa) protein whereas CAKS11 does not. (F) CaMtw1 localizes like a typical kinetochore protein. Fixed CAKS13 cells were stained by DAPI (DNA) and coimmunostained with anti-protein A (CaMtw1) and anti-tubulin (spindle microtubules) antibodies. Dot-like CaMtw1 signals were observed in the cells at different stages of cell cycle (Bar, 2μm). (G) PCR of ChIP assays reveal that the CaMtw1 is enriched within the CEN7 region of chromosome 7. Sheared chromatin fragments of CAKS13 strain (CaMtw1-TAP) were immunoprecipitated with anti-protein A antibodies. PCR reactions with primer pairs that amplify 178- to 292-bp regions spaced approximately every 1 kb between Orf19.6522 and Orf19.6524 of CEN7 region were used to amplify total DNA (SM) and immunoprecipitated DNA with anti-protein A antibodies and (+Ab) or beads only without (-Ab) antibody ChIP DNA fractions.
We also used a fragment-mediated gene replacement strategy to construct strain CAKS13 (MTW1-TAP/mtw1), in which one allele of CaMtw1 was tagged with an tandem affinity purification (TAP) epitope after the other allele had been deleted with the CaHIS1 marker gene [CAKS11, (MTW1/mtw1)] (Fig. S1, see Experimental procedures). CAKS13, therefore, has a single full-length copy of CaMTW1 that is TAP-tagged. Western blotting of the cell extracts from this strain with anti-protein A antibodies detected a single 57 kDa band of the size expected for the fusion protein, whereas no signal was detected from the control untagged CAKS11 cell lysate (Fig. 1E). CaMtw1 was immunolocalized in this strain using anti-protein A antibodies, and confocal microscopy revealed intense dot-like signals that remained closely associated with the spindle pole bodies (visualized by tubulin staining using anti-tubulin antibodies) and always colocalized with nuclei (visualized by DAPI staining) (Fig. 1F). Taken together, these results strongly indicate that CaMtw1 is a kinetochore protein in C. albicans.
We next asked whether CaMtw1 exhibits binding to centromere DNA. For this, chromatin immunoprecipitation (ChIP) assays were performed using strain CAKS13 and anti-protein A (CaMtw1) antibodies. Coprecipitated DNA fragments were extracted and used as templates for PCR, using a set of 12 pairs of primers to examine CaMtw1 enrichment over a 61.5 kb region of chromosome 7 (nucleotides 395500- 455000, Table S2), which overlaps with the CaCse4-binding region identified previously (Sanyal et al., 2004). CaMtw1 specifically bound to the centromeric region (nucleotides 424500- 429500) on chromosome 7 (Fig. 1G). Adjacent non-ORF and ORF sequences exhibited background levels of enrichment. PCR with primer pairs from the centromeric regions of chromosomes 1-5 also confirmed the binding of CaMtw1 at the kinetochores. ChIP DNA obtained from an untagged control strain (CAKS11) showed no enrichment of binding at these centromeres (Fig. 1G, right panel). Thus, binding of CaMtw1 and CaCse4 to the same DNA regions confirms that CaMtw1 is present at the kinetochore.
CaMtw1 is essential for cell viability in C. albicans
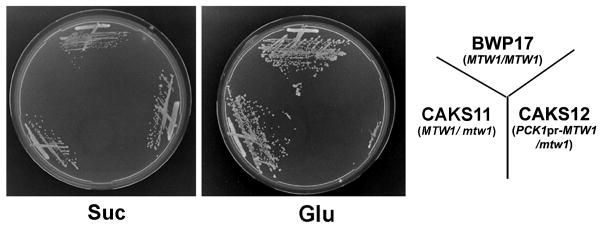
Since previous reports suggested that Mis12/Mtw1 proteins are essential for cell viability in S. pombe and S. cerevisiae (Goshima et al., 1999; Goshima et al., 2003), we examined whether the putative homolog of this gene, CaMTW1, is also essential in C. albicans. Strain CAKS12 carries the full length CaMTW1 allele under control of the regulatable promoter of CaPCK1 (Fig. S1, see Experimental procedures), which is repressed in the presence of glucose and induced in succinate media (Leuker et al., 1997). CAKS12 (PCK1pr-MTW1/mtw1) cells were grown on succinate media and shifted to glucose media to study the effect of depletion of CaMtw1 on cell cycle progression. Strains BWP17 (MTW1/MTW1) and CAKS11 (MTW1/mtw1) grew normally on both media but CAKS12 cells were unable to grow on glucose plates (Fig. 2), consistent with the conclusion that CaMTW1 is essential for C. albicans viability.
Figure 2.
CaMtw1 is essential for C. albicans growth. Shutdown of CaMTW1 expression prevents CAKS12 (PCK1pr-MTW1/mtw1) strain growth on glucose media (YPD), whereas induction of CaMTW1 expression on succinate media allows CAKS12 cells to grow. Control strains BWP17 (wildtype) and CAKS11 (MTW1/mtw1) grow on both media. Plates were incubated at 30°C for 3 days.
CaMtw1 is required for progression through G2/M stage of cell cycle
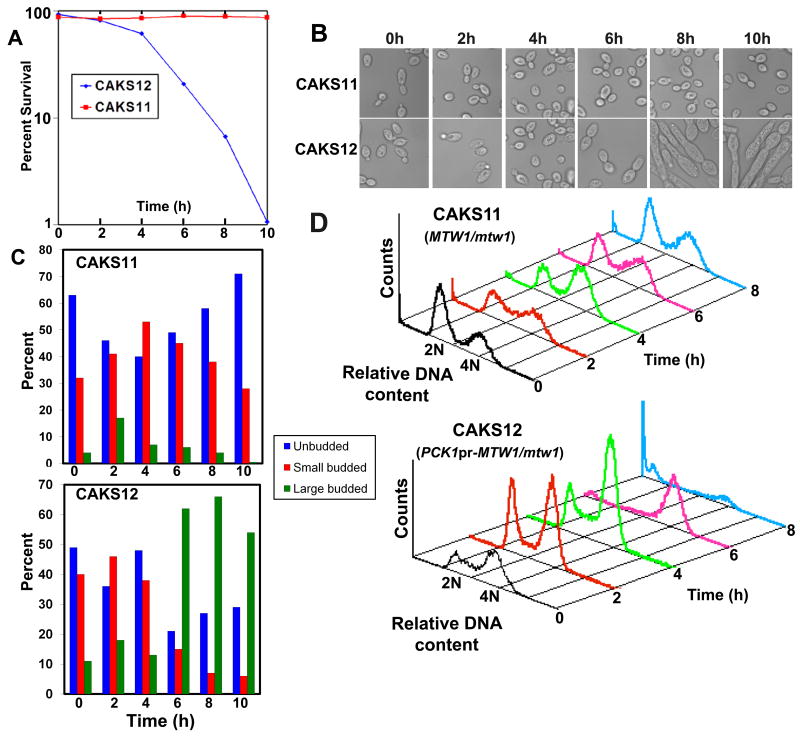
Mis12/ Mtw1 proteins are required for cell cycle progression, and proper microtubule-kinetochore mediated proper chromosome segregation (Goshima et al., 1999; Goshima et al. 2000; Goshima et al., 2003; Przewloka et al., 2007). To study the effect of CaMtw1 depletion on growth and cell cycle progression in C. albicans, CAKS11 (MTW1/mtw1) and CAKS12 (PCK1pr-MTW1/mtw1) cells were grown overnight in succinate, transferred to glucose media, and samples were collected at specific intervals (Fig. 3). Approximately 6 h after the shift to glucose media, CAKS12 cells exhibited a dramatic loss in cell viability (Fig. 3A). Morphological studies (Fig. 3B and 3C) revealed frequent accumulation of large budded cells in CAKS12 relative to parental CAKS11 cells grown under similar conditions, suggesting that the loss in viability due to CaMtw1 depletion is due to arrest of the CAKS12 cells in G2/M. After further incubation in glucose media, the large-budded CAKS12 cells showed an elongated pseudohyphal-like phenotype (Fig. 3B). Similar observations were reported previously for C. albicans cells arrested in G2/M phase in C. albicans (Bachewich et al., 2005; Berman, 2006). Flow cytometry of glucose-repressed (CaMtw1-depleted) CAKS12 cells revealed a correlation between the accumulation of large budded cells and the proportion of cells with 4N DNA content (Fig. 3D, lower), supporting the idea that cells were arrested in G2/M. As a control, CAKS11 cells grown in similar conditions did not accumulate cells with 4N DNA content.
Figure 3.
CaMtw1 is needed for high fidelity chromosome segregation in C. albicans. (A) CAKS12 (PCK1pr-MTW1/mtw1) cell viability drops dramatically between 4 h and 6 h after repression of CaMTW1 expression on glucose. CAKS12 cells were grown overnight on succinate media, washed and shifted to glucose media. Cells were then harvested at 2 h intervals. Cell viability was calculated by plating diluted aliquots of culture at indicated time points on succinate plates and counting the number of colonies that appeared after incubating the plates for 3 days at 30°C. (B) Images of CAKS11 (MTW1/mtw1) and CAKS12 cells harvested at the indicated time points. (C) Distribution of unbudded (G1), small budded (S) and large budded (G2/M) cells of CAKS11 and CAKS12 after media shift from succinate to glucose. (D) FACS profiles of CAKS11 and CAKS12 cells incubated in glucose media for indicated time points and stained with propidium iodide. The X axis uses propidium iodide staining intensity as a measure of DNA content and the Y axis reports the relative number of cells.
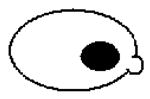
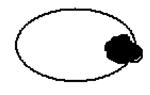
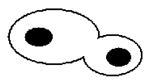
Since the majority of the CaMtw1-depleted cells show an arrest phenotype at the large bud stage, we next examined the role of CaMtw1 in nuclear divisions that leads to proper chromosome segregation. DAPI-staining of CAKS12 cells incubated for 6 h in glucose media showed that 50% of the cells (comprising almost all of the large-budded cells) had unsegregated nuclei either at or near the mother-bud neck; 13% of the cells had improperly positioned nuclei (Panel 3 on the top, Table 1). A smaller population of cells (9%) exhibited other abnormal phenotypes such as both segregated nuclei remaining within an unbudded cell or appearing only in the bud of a large budded cell (Table 1). Together, these results suggest that CaMtw1 is required for the proper chromosome segregation and the completion of mitosis.
Table 1. Percentage of cells with indicated morphology.
| Time (h) | 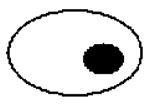 |
 |
 |
 |
 |
Total no. of cells counted | |
|---|---|---|---|---|---|---|---|
| 0 | CAKS11 (MTW1/mtw1) | 43 | 40 | <1 | 10 | 7 | 205 |
| CAKS12 (PCK1pr-MTW1/mtw1) | 63 | 23 | 1 | 12 | 1 | 191 | |
| 6 | CAKS11 (MTW1/mtw1) | 60 | 25 | 2 | 12 | 1 | 227 |
| * CAKS12 (PCK1pr-MTW1/mtw1) | 13 | 14 | 13 | 1 | 50 | 291 | |
Remaining 9% of population consisted of these types of morphologies (Bar, 5μm)
CaMtw1 has an evolutionarily conserved residue that is important for its activity
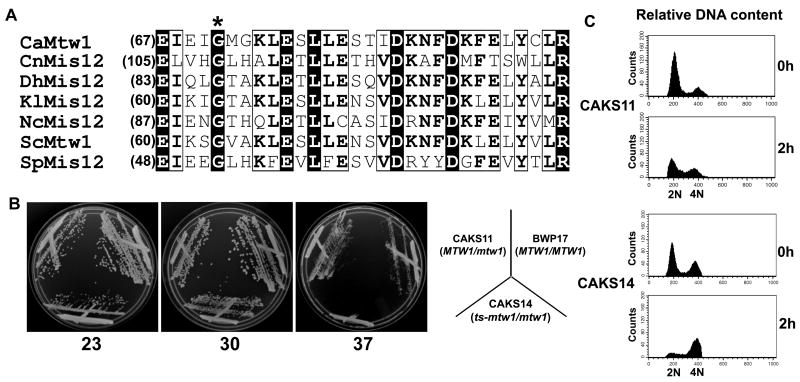
Amino acid sequence comparison of CaMtw1 with the Mis12/Mtw1 class of proteins from other fungi by BLAST analysis revealed several amino acids, including the glycine (Fig. 4A, asterisk) at amino acid 71 in CaMtw1, that are evolutionarily conserved among C. albicans, Cryptococcus neoformans, Debaryomyces hansenii, Kluyveromyces lactis, Neurospora crassa, S. cerevisiae, S. pombe. Mutations that replaced the conserved glycine with glutamate in SpMIS12 or ScMTW1 resulted in a temperature-sensitive phenotype (Goshima et al., 1999; Goshima & Yanagida, 2000). Using site-directed mutagenesis, we replaced the glycine 71 codon with a glutamate codon in C. albicans strain CAKS11 (MTW1/mtw1) to obtain strain CAKS14 (ts-mtw1/mtw1) (Fig. S1). CAKS14 grew at 23°C or 30°C but was unable to grow at 37°C (Fig. 4B). Cytological analyses of these cells grown at the non-permissive temperature showed a decrease in cell viability, cell cycle arrest at the large-budded stage, and chromosome segregation defects (Fig. S2) similar to phenotypes observed when CaMtw1 was depleted. In addition, flow cytometry of CAKS14 cells incubated at 37°C for 2 h, revealed an accumulation of 4N cells, very similar to that observed in CAKS12 (PCK1pr-MTW1/mtw1) cells grown for 6 h in glucose (Fig. 4C). Since CaMtw1 was repressed at the transcriptional level in CAKS12, the observable effect on cell viability following the switch to the non-permissive condition took longer than in CAKS14 where CaMtw1 is regulated at the level of protein function (Fig. 3, Fig. S2). These results suggest that, similar to other members of the Mis12/Mtw1 family, CaMtw1 carries an evolutionarily conserved block of amino acids including a functionally conserved glycine 71 residue that is important for its function in cell cycle progression.
Figure 4.
Mtw1/Mis12 proteins have functional domains that are evolutionarily conserved in C. albicans. (A) Amino acid sequence alignment of two blocks of conserved amino acids present at the N terminal regions of the Mis12/Mtw1 proteins in different organisms: C. albicans CaMtw1, S. cerevisiae ScMtw1, S. pombe SpMis12, N. crassa NcMis12, C. neoformans CnMis12, D. hansenii DhMis12, and K. lactis KlMis12. The conserved glycine residue (asterisk) was mutated to glutamate, resulting in a temperature- sensitive mutation in strain CAKS14 (ts-mtw1/ mtw1). Multiple sequence alignment of the Mis12 sequences was performed by T-Coffee (http://www.ebi.ac.uk/Tools/t-coffee) and representation was done by ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). (B) CAKS14 cells did not grow at 37°C, yet exhibited robust growth at 23°C and 30°C on YPDU plates for 2 days. Control parent strains BWP17 (MTW1/MTW1) and CAKS11 (MTW1/mtw1) cells grew normally at all three temperatures. (C) Flow cytometry profiles of CAKS11 and CAKS14 cells incubated at 37°C for indicated time points and stained with propidium iodide. The X axis uses propidium iodide staining intensity as a measure of DNA content and the Y axis reports the number of cells.
CaMtw1 is essential for proper spindle morphogenesis during chromosome segregation
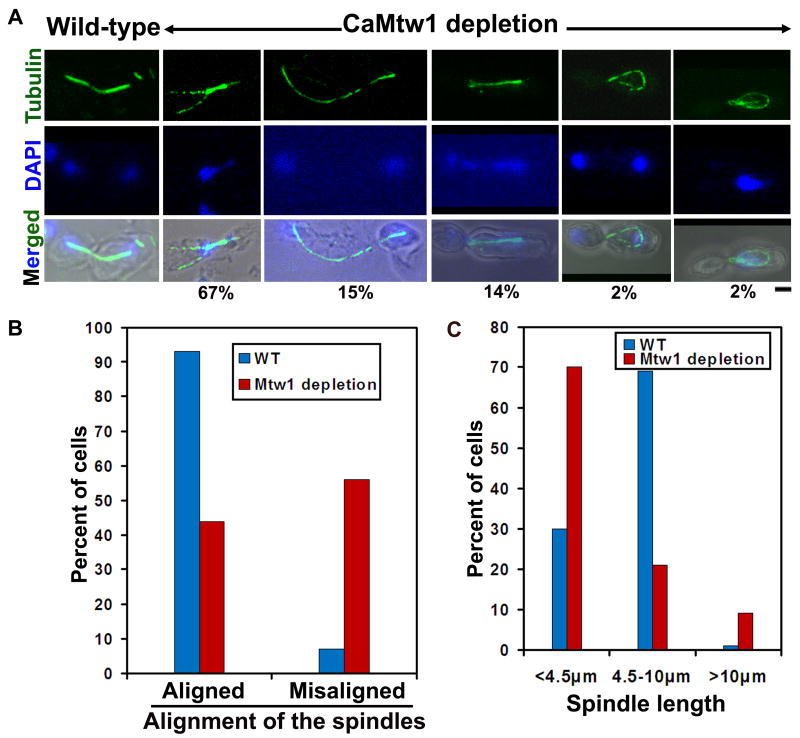
The metaphase spindle morphogenesis of Mis12/Mtw1 mutants is defective in fission yeast (mis12-537) and budding yeast (mtw1-1) (Goshima et al., 1999; Goshima & Yanagida, 2000). To determine the spindle morphologies in CaMtw1 depleted cells, BWP17 (MTW1/MTW1) and CAKS12 (PCK1pr-MTW1/mtw1) were grown overnight in succinate media, and then incubated in glucose medium for approximately seven hours. Subcellular immunolocalization with anti-tubulin antibodies was used to examine spindle morphologies. Cells were categorized based on position/alignment (with respect to mother-bud neck) and length of the mitotic spindle. Fifty-six percent of the CAKS12 cells showed defects in either spindle position and/or alignment; only 7% of BWP17 cells exhibited such defects in spindle alignment (Fig. 5A and Fig. 5B). Large budded cells were divided into three groups based on spindle length: short (less than 4.5μm), moderately long (4.5-10μm), and extra long (>10μm). Under CaMtw1 depleted conditions, 70% of cells had short mitotic spindles, and 9% cells had extra long spindles. The majority (69%) of BWP17 cells grown under similar condition had moderate spindle length (Fig. 5A and Fig. 5C). The fraction of cells with defective spindle alignment was similar for Mtw1-depleted cells and for the ts-mtw1 mutant, (CAKS14) grown at 37°C for 3h (not shown). For example, a similar proportion of CaMtw1-depleted cells and of ts-mtw1 mutant cells at 37°C had short mitotic spindles that migrated to the daughter bud prematurely, in some cases leaving unattached DNA in the mother bud (Fig. 5A, panel 3 in CaMtw1-depleted cells, Fig. S3A, B, indicated by white arrows). In some cases, CaMtw1 depleted and ts-mtw1 cells grown at 37°C contained long cytoplasmic microtubules that occasionally stretched around the cells (Fig. 5, panel 1 and Fig. S3A, panel 2). Together, these results indicate that CaMtw1 is required to maintain proper spindle positioning/alignment and length during chromosome segregation.
Figure 5.
CaMtw1 depletion causes defects in spindle alignment, position, and length. (A) BWP17 (MTW1/MTW1) and CAKS12 (PCK1pr-MTW1/mtw1) cells (grown in glucose media for 7 h at 30°C) were fixed and stained with anti-tubulin (spindle) antibodies and DAPI (DNA). (Bar, 2μm). The percent of cells showing different spindle morphologies are mentioned below every image. (B) Percentage of properly and improperly aligned spindles were counted in BWP17 (71 cells) and CAKS12 (117 cells). (C) Mitotic spindle lengths were measured by Zeiss LSM software in BWP17 (70 cells) and CAKS12 (131 cells). Percentage of cells carrying spindles of indicated length is represented.
Altered expression of CaCse4 affects kinetochore localization of CaMtw1
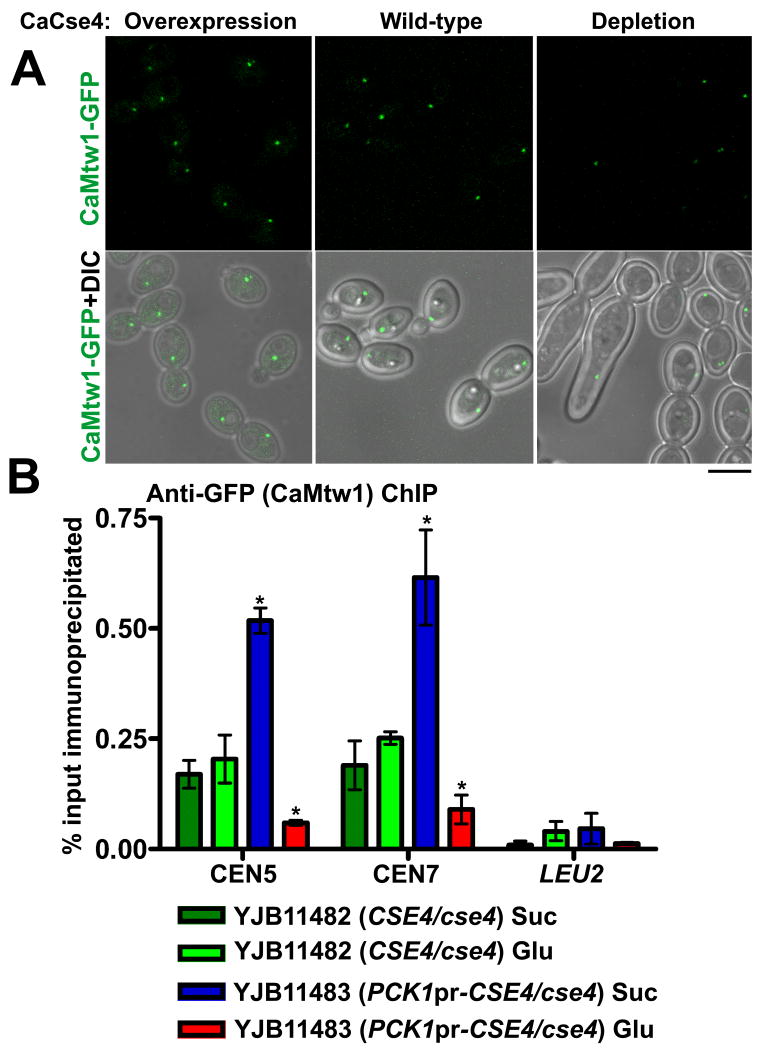
In most organisms, including S. cerevisiae, localization of Mis12/Mtw1 is dependent on CENP-A (Westermann et al., 2003; Collins et al., 2005; Liu et al., 2006; Przewloka et al., 2007), although a study in S. pombe suggested that CENP-A and Mis12 localization at the centromere are independent (Takahashi et al., 2000). To investigate the requirement of CaCse4 for CaMtw1 localization, we first looked at the localization of GFP-tagged CaMtw1 upon repression or overexpression of CaCse4. For this, one copy of CaMTW1 was GFP-tagged at the C-terminus and expressed in strains where one copy of CaCSE4 was deleted, and the other copy was either under its native promoter (YJB11482) or under control of the PCK1 conditional promoter (YJB11483). Overexpression and repression of CaCSE4 in succinate and glucose media were confirmed by quantitative real time PCR done with the total RNA isolated from YJB11482 and YJB11483 grown in succinate and glucose media (data not shown). Depletion of CaCse4 was confirmed by Western blot analysis with anti-CaCse4 antibodies of cell lysates of YJB11483 (PCK1pr-CSE4/cse4) after CaCse4 depletion, and YJB10695 (CSE4/CSE4) grown in the same conditions (Fig. S4). When YJB11483 was grown in glucose to repress CaCse4 expression, 20% of the cells did not exhibit any visible GFP signal, while the control strain had visible CaMtw1-GFP in every cell (Fig. 6A). Additionally, there was a 24% decrease in overall CaMtw1-GFP fluorescence when CaCse4 was repressed (p<0.01, indicated by *) (Fig. 6A and S5A). The moderate decrease in CaMtw1-GFP upon CaCse4 depletion is likely the result of incomplete depletion of CaCse4 protein. The residual amount of CaCse4, may have been sufficient to recruit some CaMtw1 to the kinetochore, but not adequate for proper chromosome segregation. Similar kinds of issues with incomplete depletion of kinetochore proteins were reported previously (Goshima et al., 2003). The binding of CaMtw1-GFP to centromere DNA, as measured by ChIP using anti-GFP antibodies, also was reduced approximately 3-fold when CaCse4 was repressed (p<0.01) (Fig. 6B and Fig. S6B). Conversely, when YJB11483 was grown in succinate, a condition that drives overexpression of CaCse4 and results in increased binding specifically to centromeric DNA (Fig. S6A), an increase of approximate 50% in CaMtw1-GFP localization was seen by microscopy (p<0.01, indicated by *) (see Fig. 6A and S5A) and ∼3-fold increased binding of CaMtw1-GFP to centromere DNA was observed by ChIP (p<0.01, indicated by *) (Fig. 6B and Fig. S6B). These results indicate that CaCse4 is important for the recruitment of CaMtw1 to the centromere.
Figure 6.
CaCse4 influences localization of CaMtw1 at the centromere. (A) GFP and merged GFP-DIC microscopy images showing CaMtw1-GFP signals in YJB11482 (CSE4/cse4) (wild-type) and YJB11483 (PCK1pr-CSE4/cse4) under conditions that repress (glucose) or induce (succinate) expression of the PCK1 promoter. (Bar, 5μm). (B) ChIP assays were performed on strains YJB11482 (CSE4/cse4) and YJB11483 (PCK1pr-CSE4/cse4) grown in glucose (repressing condition for PCK1pr) or succinate (inducing condition for PCK1pr) for 6h, using anti-GFP antibodies and primer pairs that amplify the central regions of CEN5 (JB3924/JB3925) and CEN7 (JB3993/JB3994). Amplification from the LEU2 ORF (JB4165/ JB4166) was also performed to detect background DNA elution in the ChIP assays. qPCR of total DNA and with (+) or without (-) antibody ChIP DNA fractions were performed. Enrichment of CaMtw1-GFP was calculated as a percentage of the total chromatin input. Shown are the averages of two representative biological replicates ± SEM. t-tests were used to compare CaMtw1-GFP recruitment at the central CEN5 and CEN7 regions in YJB11482 and YJB11483 in glucose (p<0.01) and succinate (p<0.01) (Indicated by *).
Depletion of CaMtw1 decreases CaCse4 localization to kinetochores
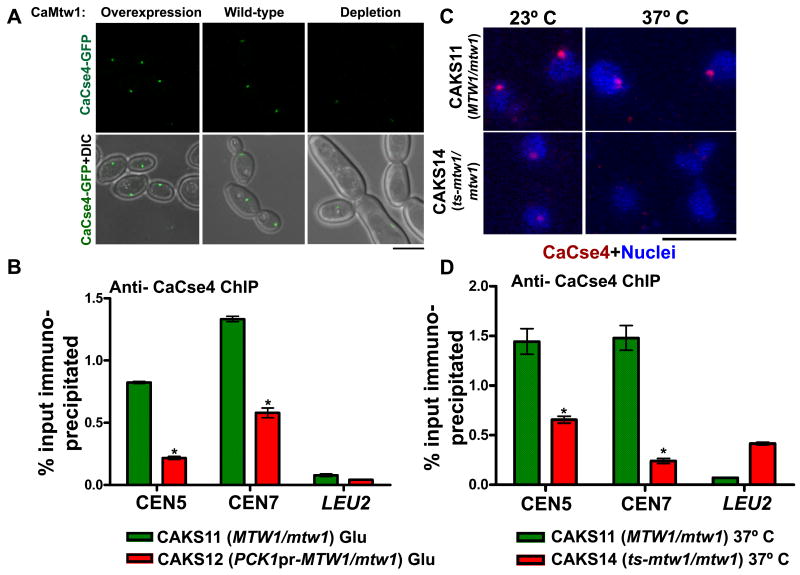
Previous work with human Dsn1, another member of the Mis12 complex, suggested that the Mis12 complex may be important for recruiting or maintaining CENP-A, as decreased levels of CENP-A were observed in cells where hDsn1 was depleted (Kline et al., 2006). To address the role of the Mis12/Mtw1 complex in CENP-A kinetochore localization in C. albicans, we used GFP to internally tag one copy of CaCSE4, expressed from its native promoter, in strains where one copy of CaMTW1 was deleted and other copy was either expressed from its native promoter (YJB11553) or expressed from the PCK1 promoter (YJB11554). Interestingly, depletion of CaMtw1 (YJB11554 grown in glucose) resulted in an approximately 4-fold decrease in CaCse4-GFP localization at the centromere (p<0.01, indicated by *) (Fig. 7A and Fig. S5B).
Figure 7.
CaCse4 localization at the centromere is affected by CaMtw1: (A) GFP and merged GFP-DIC microscopy images showing CaCse4-GFP signals in YJB11553 (MTW1/mtw1) and YJB11554 (PCK1pr-MTW1/mtw1) in repressing (glucose) and inducing (succinate) conditions for the PCK1 promoter. (Bar, 5μm) (B) ChIP assays were performed on strains CAKS12 (grown in glucose for 6h, where PCK1pr-CaMTW1 is repressed) and CAKS11 (with wild-type CaMTW1 expressed) using anti-CaCse4 antibodies and primer pairs that amplify the central regions of CEN5 (CACH5F1/CACH5R1) and CEN7 (nCEN7-3/nCEN7-4). qPCR of total DNA and with (+) or without (-) antibody ChIP DNA fractions were performed. Enrichment of CaCse4 at the centromere was calculated as a percentage of the total chromatin input. Amplification from LEU2 ORF (nLeu2- 1/nLeu2-2) was also performed to detect the background DNA elution in the ChIP assays. t-tests were used to compare CaCse4 recruitment at the central CEN5 and CEN7 region in YJB11553 and YJB11554 in glucose (p<0.01) (indicated by *). (C) CAK11 (MTW1/mtw1) and CAKS14 (ts-mtw1/mtw1) cells (grown at 23°C and 37°C) were fixed and stained with anti-CaCse4 antibodies and DAPI (nucleus). (Bar, 5μm). (D) Standard ChIP assays were performed on strains CAKS14 and CAKS11 (grown at 37°C) using anti-CaCse4 antibodies and primers that amplify the central regions of CEN5 (CACH5F1/CACH5R1) and CEN7 (nCEN7-3/nCEN7-4). PCR of total DNA and with (+) or without (-) antibody ChIP DNA fractions were performed. QPCR amplification from LEU2 ORF (nLeu2-1/nLeu2-2) was also done to detect the background DNA elution in the ChIP assays. Enrichment of CaCse4 at the centromere was calculated as a percentage of the total chromatin input. t-tests were used to compare CaCse4 recruitment at the central CEN5 and CEN7 region in CAKS11 and CAKS14 at 37° C (p<0.005) (indicated by *).
We next determined the amount of CaCse4 bound to centromere DNA using ChIP assays under conditions where CaMtw1 expression is wild-type (CAKS11, MTW1/mtw1) or depleted (CAKS12, PCK1pr-MTW1/mtw1, in glucose). Cross-linked, sheared chromatin isolated from glucose-grown CAKS11 or CAKS12 cells (see Experimental Procedures) was precipitated with anti-CaCse4 antibodies and immunoprecipitated DNA samples were analyzed by qPCR to test binding at the CEN5 and the CEN7 regions of C. albicans. CaCse4 binding at centromeres was significantly reduced by 56% (p<0.0001, indicated by *) when CaMtw1 was depleted (Fig. 7B), that CaMtw1 is required for complete localization of CaCse4 to the kinetochore (Fig. 7A, 7B and S5B). The effect of CaMtw1 overexpression on localization of CaCse4 to the kinetochore was much smaller (26% for YJB11554PCK1pr-MTW1/mtw1, CSE4/CSE4-GFP-CSE4)), yet was statistically significant (p<0.01, indicated by *) when compared to YJB11553 (MTW1/mtw1, CSE4/CSE4-GFP-CSE4) grown under similar conditions (Fig. 7A and S5B).
Since the above experiments suggest that CaMtw1 may influence CaCse4 localization at the centromeres, we further examined this dependency in strain CAKS14 (ts-mtw1/mtw1), in which CaMtw1 is no longer functional at 37° C. The control parent strain CAKS11 (MTW1/mtw1) showed normal kinetochore localization of CaCse4 both at 23° C and 37° C by indirect immunolocalization. In CAKS14 (ts-mtw1/mtw1) at 23° C, CaCse4 localized to a single dot assumed to be the centromere; at 37° C, no CaCse4 signal was detected by indirect immunofluorescence (Fig. 7C). Consistent with this result, ChIP assays indicated a dramatic reduction (p<0.005, indicated by *) in CaCse4 recruitment to the centromeres in CAKS14 cells grown at 37° C for 2 h, as compared to the wild-type CAKS11 cells grown under the same conditions (Fig. 7D). Taken together, these observations indicate that CaMtw1 is required for complete kinetochore localization of CaCse4.
CaMtw1 influences localization of CaMif2 to kinetochores
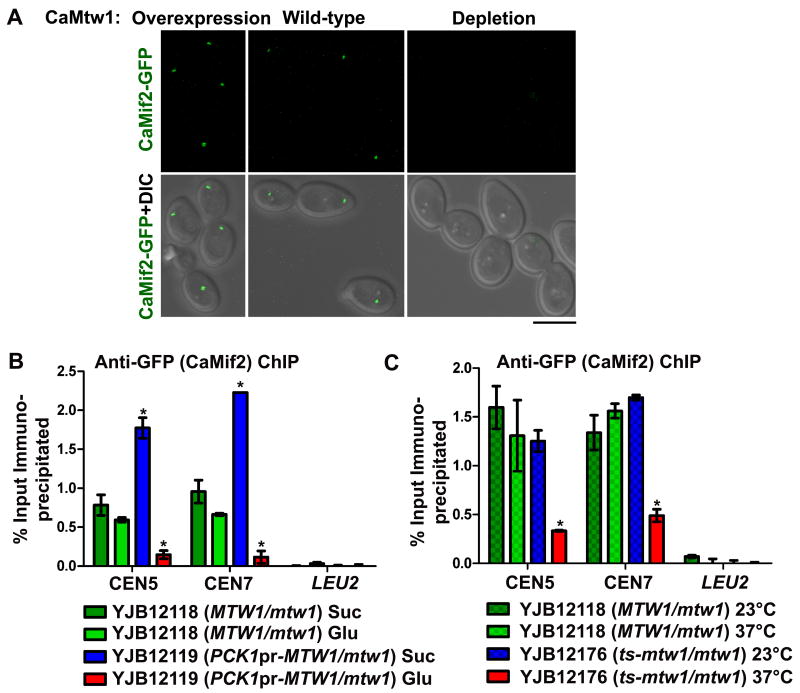
Several models of centromere structure suggest that CENP-C/Mif2 homologs are found in the inner kinetochore and may act as a linker between CENP-A/Cse4 nucleosomes and other kinetochore proteins (Milks et al., 2009; Petrovic et al., 2010; Carroll et al., 2010), including the Mis12/Mtw1 complex. To address the role of the Mis12/Mtw1 complex in CENP-C/Mif2 kinetochore localization in C. albicans, we tagged one copy of CaMIF2 expressed from its native promoter with GFP at the C-terminus, in strains where one copy of CaMTW1 was deleted and other copy was either under the control of its native promoter (YJB12118) or the PCK1 promoter (YJB12119). Interestingly, depletion or overexpression of CaMtw1 (YJB12119) resulted in a significant decrease or increase, respectively in CaMif2-GFP localization at the centromere (p<0.0001, indicated by *) (Fig. 8A and S5C).
Figure 8.
CaMif2 localization at the centromere is affected by CaMtw1: (A) GFP and merged GFP-DIC microscopy images showing CaMif2-GFP signals in YJB12118 (MTW1/mtw1) and YJB12119 (PCK1pr-MTW1/mtw1) in repressing (glucose) and inducing (succinate) conditions for the PCK1 promoter (Bar, 5μm). (B) ChIP assays were performed on strains YJB12119 [grown in succinate (CaMtw1 is overexpressed) and glucose for 6 h (CaMtw1 is repressed)] and YJB12118 (with wild-type CaMTW1 expressed) using anti-GFP antibodies and primer pairs that amplify the central regions of CEN5 (JB3924/JB3925) and CEN7 (JB3993/JB3994). qPCR of total DNA and with (+) or without (-) antibody ChIP DNA fractions were performed. Enrichment of CaMif2 at the centromere was calculated as a percentage of the total chromatin input. Amplification from LEU2 ORF (JB4165/ JB4166) was also performed to detect the background DNA elution in the ChIP assays. t-tests were used to compare CaMif2 recruitment at the central CEN5 and CEN7 region in YJB12118 and YJB12119 in glucose (p<0.001) and in succinate (p<0.05) (indicated by *). (C) ChIP assays were performed on strains YJB12118 and YJB12176 (grown both at 23°C and 37°C for 2 h) using anti-GFP antibodies and primers that amplify the central regions of CEN5 (JB3924/JB3925) and CEN7 (JB3993/JB3994). PCR of total DNA and with (+) or without (-) antibody ChIP DNA fractions were performed. QPCR amplification from LEU2 ORF (JB4165/ JB4166) was also done to detect the background DNA elution in the ChIP assays. Enrichment of CaMif2 at the centromere was calculated as a percentage of the total chromatin input. t-tests were used to compare CaMif2 recruitment at the central CEN5 and CEN7 region in YJB12118 and YJB12176 at 37° C (p< 0.001) (indicated by *).
We next determined the amount of CaMif2 bound to centromere DNA using ChIP assays under conditions where CaMtw1 expression is wild-type (YJB12118, MTW1/mtw1), overexpressed (YJB12119, in succinate), or depleted (YJB12119, in glucose). CaMif2 binding at centromeres was significantly reduced (p<0.001, indicated by *) relative to controls when CaMtw1 was depleted (Fig. 8B). CaMif2 binding at centromeres was significantly increased (p<0.05) when CaMtw1 was overexpressed. Thus, CaMtw1 is required for complete localization of the CaMif2 inner kinetochore protein.
We next examined this dependency in strain YJB12176 (ts-mtw1/mtw1, MIF2-GFP-NAT1/MIF2), which does not grow at 37°C. Localization of CaMif2-GFP by microscopy in YJB12176 was decreased upon growth at 37°C, similar to what was observed upon CaMtw1 depletion (data not shown). Binding of CaMif2-GFP to centromere DNA was also analyzed by ChIP. At 23°C, CaMif2 recruitment at the centromere was comparable to the wild-type at 23°C, but was significantly reduced (p<0.001, indicated by *) when these cells were grown for 2 h at 37°C (Fig. 8C). The control parent strain YJB12118 showed normal kinetochore recruitment of CaMif2 assayed by ChIP both at 23°C and 37°C (Fig 8C). Taken together, these observations indicate that CaMtw1 is required for complete kinetochore localization of CaMif2.
Discussion
Function of Mis12/Mtw1 homolog is conserved in C. albicans
In the present study, we identified and characterized the Mis12/Mtw1 homolog in C. albicans, a pathogenic yeast that has small, yet regional centromeres. While the overall amino acid similarity of CaMtw1 relative to other proteins of this family is restricted to two conserved blocks, our results demonstrate that the function of CaMtw1 is conserved. Immunofluorescence microscopy and ChIP assays revealed that, similar to other yeasts (Goshima et al., 1999; Goshima et al., 2000), CaMtw1 colocalizes with CaCse4. CaMtw1 localized close to the spindle poles through most of the cell cycle, indicating that, as in S. cerevisiae, kinetochores remain closely associated with spindle pole bodies in C. albicans. Similar to other members of the Mis12/Mtw1 family, CaMtw1 is an essential protein required for G2/M progression and proper chromosome segregation during mitosis. Gly71 in the CaMtw1 ORF is evolutionarily conserved across fungal species and is essential for the activity of CaMtw1 at high temperatures. Moreover, CaMtw1 affects mitotic spindle length and alignment. Interestingly, the amount of CaMtw1 in a cell influences the degree of kinetochore localization of two essential inner kinetochore proteins, CaCse4 and CaMif2, suggesting that CaMtw1 is crucial for efficient inner kinetochore assembly.
Prolonged incubation at non-permissive conditions that reduce the levels of functional CaMtw1 protein in the cell, leads to a morphological switch to polarized growth
When CaMTW1 gene expression was repressed or the mtw1-ts mutant was grown at 37°C, cells arrested in G2/M in a time-dependent manner. Prolonged incubation of these cells arrested at G2/M phase grew with pseudohyphal-like morphologies, consistent with previous reports that prolonged G2/M arrest of C. albicans leads to a pseudohyphal-like elongated bud phenotype (Bachewich et al., 2005; Berman, 2006). Similar elongated morphologies were observed in the CaCse4-depleted cells as well (Sanyal and Carbon, 2002; this study) and the population of cells with sub-G1 DNA content increased as determined by flow cytometry, suggesting that dead cells accumulate at this stage.
Role of CaMtw1 in spindle morphogenesis is conserved in yeasts
CaMtw1 depletion or incubation of the ts-mtw1 mutant at 37°C resulted in defects in nuclear positioning and in segregation of nuclear DNA in daughter cells (Table 1, Fig. S2D). Tubulin staining revealed a large proportion of CaMtw1-depleted cells and ts-mtw1 mutant cells had altered spindle length and improper spindle orientation similar to spindle morphogenesis defects previously reported in mis12-537 mutants in S. pombe and mtw1-1 mutants in S. cerevisiae (Goshima et al., 1999; Goshima & Yanagida, 2000). The mtw1-1 cells incubated at 37° C for 140 min formed short mitotic spindles with unsegregated DNA in mother-bud neck (Goshima et al., 2000; Pinsky et al., 2003). After prolonged incubation at 37° C, mtw1-1 cells exhibited abnormal expansion of the spindle length (Goshima et al., 2000). In both CaMtw1-depleted cells and the temperature-sensitive mutants, we observed the occasional presence of an unattached DNA mass in the mother bud of these cells, suggesting that defects in biorientation of chromosomes occur when CaMtw1 is not present or not functional. It was suggested (Goshima & Yanagida, 2000; Pinsky et al., 2003) that Mtw1 contributes to bi-orientation of the chromosomes, which is monitored by spindle assembly checkpoint protein Ipl1. The presence of unsegregated nuclei and short mitotic spindle in the large-budded stage of C. albicans cells indicates that the spindle assembly checkpoint was activated. The G2/M arrest of the mtw1-1 mutant in S. cerevisiae was mediated by Mad2, a component of the spindle assembly checkpoint. The similarities of mtw1 mutants of S. cerevisiae and C. albicans, suggests that spindle assembly checkpoint is probably responsible for the arrest phenotype of mtw1 mutants in C. albicans as well. It will be interesting to investigate the role of Ipl1 in CaMtw1 function.
In a small proportion of CaMtw1 depleted cells, a network of long cytoplasmic microtubules was observed. A similar phenotype was found in mtw1-1 mutants of S. cerevisiae (Pinksy et al., 2003). In addition to spindle length and alignment defects, we observed unusual bending of the spindle microtubules, similar to the spindle defects observed in other kinetochore mutants (such as dam1 or duo1 mutants) in S. cerevisiae (Hofmann et al., 1998; Cheeseman et al., 2001), as well as in Mis12 mutants in human and fly cells (Goshima et al., 2003; Przewloka et al., 2007). Overall, these results suggest that the function of Mtw1 in maintaining spindle length and orientation is conserved across species.
What is the role of Mtw1 in chromosome segregation and how does Mtw1 regulate spindle morphogenesis?
In C. elegans, CeMis12 does not bind directly to microtubules; rather, in vitro studies (Cheeseman et al., 2006) indicate that it synergistically enhances the binding of the NDC80 and KNL1 complexes to the microtubules. It is not known whether CaMtw1 directly interacts with spindle microtubules. In S. cerevisiae, improper dynamics of the spindle in mtw1-1 mutants led to the proposal that ScMtw1 may contribute to interactions between motor proteins, and the astral microtubules (Shaw et al., 1997), which in turn result in the proper alignment and migration of the mitotic spindle across the mother-bud neck (Palmer et al., 1992; Yeh et al., 2000; Pinsky et al., 2003). Repression of Mis12/Mtw1 complex components impedes the recruitment of other outer kinetochore proteins including the NDC80 complex and the Dam1-DASH complex, kinetochore Microtubule Associated Proteins (kMAP) and motor proteins (Scharfenberger et al., 2003, Kline et al., 2006, Pagliuca et al., 2009), which interact directly with the microtubules (Cheeseman et al., 2006, Westermann et al., 2005). Therefore in the absence of functional Mis12/Mtw1, the process of dynamic kinetochore-microtubule interaction is affected and the proper positioning of the spindle may also be impaired.
CaMtw1 influences recruitment of inner kinetochore proteins CaCse4 and CaMif2 at the kinetochore
Localization dependency studies between CENP-A and Mis12/Mtw1 proteins in several model systems are summarized in Table 2 (Takahashi et al., 2000; Westermann et al., 2003; Goshima et al., 2003; Cheeseman et al., 2004; Liu et al., 2006; Przewloka et al., 2007). To understand the role of CaMtw1 in kinetochore formation, we determined the dependency of CaCse4 and CaMtw1 on each other for localization to the kinetochore. Similarly, we studied the dependency of CaMif2 on CaMtw1 for recruitment at the kinetochore. Interestingly, we found that CaCse4 and CaMtw1 are interdependent. Depletion of CaCse4 affects CaMtw1 occupancy at the kinetochores, similar to the influence of CENP-A on Mis12/Mtw1 proteins reported in most organisms, including S. cerevisiae and humans (Westermann et al., 2003; Collins et al., 2005; Liu et al., 2006; Przewloka et al., 2007) and is consistent with the known role of CENP-A as an important determinant of kinetochore formation. The one exception is in an S. pombe temperature sensitive CENP-A mutant, where Mis12 reportedly remains at centromeres, as determined by semiquantitative ChIP (Takahashi et al., 2000). We suggest that this discrepancy regarding the dependency of CENP-A and Mis12 in S. pombe may be due to the particular CENP-A mutant used and to qualitative rather than quantitative interpretation of the data where Mis12 likely exhibited normal localization patterns, but reduced amounts of Mis12 protein at the centromere.
Table 2. Localization dependency of CENP-A/CenH3 and Mis12/Mtw1.
| Organism | CENP-A/CenH3 influences Mis12/Mtw1 | Mis12/Mtw1 influences CENP-A/CenH3 | Other Mis12/Mtw1 complex members influence CENP-A/CenH3 |
|---|---|---|---|
| S. cerevisiae | Yes (Westermann et al., 2003, Collins et al., 2005) | Not studied | Not studied |
| S. pombe | No - ts CENP-A mutant, data not shown (Takahashi et al., 2000) | No – ts mis12-537 mutant showed qualitative localization of CENP-A by microscopy and no effect on CENP-A binding to the central core DNA by semi-quantitative PCR ChIP (Takahashi et al., 2000) | Not studied |
| C. elegans | Yes (Cheeseman et al. 2004, data not shown) | No (Cheeseman et al., 2004, data not shown) | Not studied |
| D. melanogaster | Yes - microscopy following RNAi knockdown of CENP-A (Przewloka et al., 2007) | No (Przewloka et al., 2007, data not shown) | Not studied |
| Humans | Yes - careful quantification where reduction in CENP-A correlated with loss of Mis12 in a proportional manner (Liu et al., 2006) | No - RNAi of Mis12 by qualitative microscopy (Goshima et al., 2003); (Liu et al., 2006, data not shown) | Yes – RNAi of hDsn1 reduces CENP-A localization by ∼50% (Kline et al. 2006) |
| C. albicans | Yes (this study) | Yes (this study) | Not studied |
Interestingly, a higher level of CaCse4 expression lead to an increase in CaCse4 binding to centromere DNA and a significant increase in CaMtw1 accumulation, detected both by fluorescence microscopy and by ChIP qPCR assays (Fig. 6, Fig. S5 and S6). Fluorescence ratio imaging has shown that there are approximately 8 CaCse4 and 4 CaMtw1 molecules per centromere (Joglekar et al., 2008). Accordingly, the C. albicans centromeres, defined as the ∼ 3-5 kb CaCse4-rich regions, contain a mixture of H3- and CENP-A-containing nucleosomes (data not shown). Thus, overexpression of CaCse4 may replace the remaining centromere-associated canonical histone H3 molecules with CaCse4 molecules, which may, in turn, recruit more CaMtw1 in the kinetochore.
Interestingly, results from this study revealed that two inner kinetochore proteins, CaCse4 and CaMif2, both exhibited some dependence on CaMtw1 for their recruitment to the kinetochore. Both immunofluorescence and ChIP assays demonstrated that CaCse4 and CaMif2 levels at the centromere are significantly lower when the level of CaMtw1 was reduced either by transcriptional repression or with a temperature-sensitive CaMtw1 mutation. The dependency of CENP-C homologue ScMif2 on ScMtw1 was previously reported in S. cerevisiae (Westermann et al., 2003). Interestingly, recent results in humans found that depletion of CENP-C decreases centromeric localization of CENP-A (Carroll et al., 2010). Human Dsn1, a component of the Mis12 complex, influences the amount of CENP-A localized to human kinetochores (Kline et al. 2006). These results are consistent with the idea that interactions between components of the inner kinetochore stabilize the localization of these proteins to centromere.
The Mis12/Mtw1 complex itself exhibits interdependency among the four subunits on the level of protein and complex stability (Nekrasov et al, 2003; Kline et al., 2006). In addition, human Mis12 complex requires Hsp90 and its co-chaperone Sgt1 for stability and efficient promotion of kinetochore formation, yet excessive stability of Mis12 can lead to increased numbers of incorrect kinetochore-microtubule attachments (Davies and Kaplan, 2010). It has been suggested that Mis12 proteins may have a role in licensing the formation of the kinetochore microtubule attachment site (Cheeseman and Desai, 2008). We speculate that the co-dependency of CENP-A and Mis12/Mtw1 complex proteins in directing assembly of the rest of the kinetochore, may specify the number of microtubule attachment sites within regional centromeres and may help ensure that kinetochore proteins assemble productively onto specific subsets of CENP-A nucleosomes.
Several other studies reported no effect of Mis12/Mtw1 on CENP-A localization (Takahashi et al., 2000; Cheeseman et al. 2004; Liu et al., 2006; Goshima et al., 2003; Przewloka et al., 2007). Also, in a study examining localization dependencies in the fruit fly, CENP-C was shown to be independent of Mis12 for kinetochore localization (Przewloka et al., 2007). However, most of these studies were generally qualitative, rather than quantitative (Table 2). Our results, taken together with the more recent studies of the human Mis12 complex members (Kline et al., 2006), suggest that there is a quantitative dependency between CaMtw1 and CaCse4 complexes, perhaps mediated by CaMif2. Thus, we propose that Mis12/Mtw1 contributes to the stability of CENP-A/Cse4 at regional centromeres, such that reducing the levels of Mis12/Mtw1 affects the stability, and thus the quantity, but not the localization pattern of the remaining CENP-A nucleosomes. The altered localization efficiency of CaMif2 resulting from differential levels of CaMtw1 indicates that the level of CaMtw1 is important for inner kinetochore assembly. We suggest that the small, non-repetitive nature of C. albicans centromeres facilitates quantitative analyses of centromere structure and kinetochore protein levels, which have the potential to reveal relationships between kinetochore proteins and complexes that, are likely to be conserved in larger regional centromeres.
Experimental procedures
Strains, media, primers and transformation procedures
The C. albicans strains used in this study are listed in Table S1 in the Supporting material and strain construction strategies are detailed below. The media used for growing these strains are YPDU (1% yeast extract/2% peptone /2% dextrose/0.010% uridine), YPA-Glucose (1% yeast extract, 2% peptone, 2% glucose, 0.04mg/ml adenine, 0.08 mg/ml uridine), YPSU (1% yeast extract/2% peptone/2% succinate), YPA-Succinate (1% yeast extract, 2% peptone, 2% sodium succinate, 0.04mg/ml adenine, 0.08 mg/ml uridine) or supplemented synthetic/dextrose (SD) minimal media as described previously (Sanyal and Carbon, 2002). NatR transformants were selected on YPAD-Glucose with 400 μg/ml nourseothricin. C. albicans cells were transformed by standard techniques (Sanyal and Carbon, 2002). For all strains with PCK1 promoter constructs, cells were grown in glucose (repressing) or succinate (inducing) media for at least 6 h. We estimate 4 generations of growth during this time resulting in dilution of the protein present in the cell prior to repression by approximately 16-fold in addition to protein lost to protein degradation during that time. This is consistent with previous work analyzing the protein levels of CaCse4 under the control of the PCK1pr construct (Sanyal and Carbon, 2002). Primers of length 21-93 nucleotides used in this study are listed in Table S2 in Supporting material.
Identification of CaMtw1 in C. albicans
The C. albicans CaMtw1 sequence was identified by a BLAST search with S. cerevisiae ScMtw1 as a query sequence against Candida albicans genome database (http://www.candidagenome.org/cgi-bin/compute/blast-sgd.pl). This sequence analysis suggests that the Orf19.1367 has significant homology to ScMtw1 (BLAST score 267). This putative ORF is 945 bp long and encodes a 36 kDa protein. A pairwise sequence comparison of ScMtw1 and CaMtw1 revealed that they share overall 29% identity. The regions of high homology are restricted to two blocks localized at the N-terminus of the protein sequence, the 9-48th amino acid (1st block) and 67-102nd amino acids (2nd block) of CaMtw1 ORF (Fig. S7 in Supporting material). Pairwise alignment was performed by T-Coffee program (http://www.ebi.ac.uk/Tools/t-coffee) and representation was done by ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Construction of strains
Strains were constructed as follows:
Construction of CAKS11
An 805 bp promoter sequence and a 431 bp 3′ untranslated sequence of CaMTW1 were amplified with primers MTW1-5/MTW1-6 and MTW1-3/MTW1-4-1, respectively. These fragments were cloned into the respective sites of the E. coli plasmid pBluescript KSII (-) vector to produce pCL1. The 1229 bp CaHIS1 gene (taken from GFP-HIS1 plasmid (Gerami-Nejad et al., 2001) was cloned as a BamHI - EcoRI fragment, with the CaMTW1 upstream and downstream fragments flanking it, into the respective sites in pCL1 to yield pCL2. The pCL2 plasmid was digested with SacI and HindIII to release a 2465 bp fragment carrying the deletion cassette. This fragment was used subsequently to transform BWP17. His+ transformants were selected and the desired transformants were identified by checking the integration of the cassette at the desired locus by PCR using primers MTW1-15 and MTW1-7 (see Fig. S8A in Supporting material). The resulting strain is called CAKS11 (MTW1/mtw1).
Construction of CAKS12
A 433 bp N-terminal region of MTW1 coding sequence was amplified using primers Cal6 and Cal9 from C. albicans genomic DNA and cloned into PstI and SpeI sites in the PCK1-CSE4 plasmid (Sanyal & Carbon, 2002) that replaced CaCSE4 region with the N-terminal MTW1 sequence to get PCK1-MTW1 plasmid. This plasmid was linearized with BglII and the linearized DNA was used to transform CAKS11. The integration of the plasmid in the genome resulted in truncation of the second allele of CaMTW1 and the only full length copy of CaMTW1 gene was placed under control of the PCK1 promoter. The Ura+ transformants were obtained and confirmation of the integration of the PCK1-MTW1 cassette at the correct locus in the genome of CAKS11 was determined by PCR using primers CA26 and MTW1-8 (see Fig. S8B in Supporting material). The resulting strain was named CAKS12 (PCK1pr-MTW1/mtw1).
Construction of CAKS13
First a 1.4 kb CaURA3 sequence was cloned into pBluescript KS II (-) as an AflIII fragment, to obtain pBS-URA3. Subsequently a C-terminal CaMTW1 (443bp) fragment was amplified with primers MTW1-1/MTW1-2, digested with SacI and BamHI and subsequently cloned into the corresponding sites of pBS-URA3. A 579 bp TAP fragment carrying protein A and calmodulin binding protein (CBP) from pPK335 [(Corvey et al., 2005), a gift from Peter Koetter] was cloned as a BamHI-PstI fragment downstream of and in-frame with CaMTW1 to obtain pMT. The 420 bp 3′ UTR fragment of CaMTW1 ORF was amplified with primers MTW1-3 and MTW1-4-1, digested with EcoRI and HindIII, and cloned into the respective sites of pBluescript KSII (-). This fragment was again released by PstI and HindIII and cloned into plasmid pMT to obtain plasmid pMTU1, which had an additional EcoRV site just upstream of PstI and downstream of the HindIII in the TAP fragment, in addition to the unique EcoRV restriction site in the MTW1 sequence. Thus, a 354 bp 3′UTR fragment of CaMTW1 was amplified using primers Cal16-2 and Cal17 to replace the same fragment downstream of the TAP sequence after digesting the plasmid with HindIII and XhoI (which removes the EcoRV site of the TAP sequence) to get pMTU2. The pMTU2 was linearized with EcoRV and the purified fragment was used to transform CAKS11. The resulting Ura+ strain expressing C-terminal TAP tagged CaMTW1 under its native promoter, was named CAKS13 (MTW1-TAP/mtw1). The integration of the cassette was confirmed by Southern analysis (Fig. S8C in Supporting material).
Construction of CAKS14
Using MTW1-7 and MTW1-8 primers, a 1584 bp fragment containing 1 kb promoter region along with 584 bp of CaMTW1 ORF was PCR amplified, digested with BamHI and HindIII and cloned into the pBS –URA3 plasmid. With the help of the QuickChange kit (Stratagene), and using primers MTW1-9 and MTW1-10, the glycine at the 71st position of the cloned CaMTW1 ORF fragment was changed to glutamate. Then the plasmid was partially digested by BglII and transformed into CAKS11. The transformants, which were able to grow at 23°C and 30°C but unable to grow at 37°C, were the desired temperature sensitive mutants. The integration of this temperature sensitive mutation cassette into the CAKS11 genome was confirmed by PCR amplification using primers CAMA2con and Cal2 (see Fig. S8D in Supporting material). The resulting strain was named CAKS14 (ts-mtw1/mtw1).
Construction of YJB11482 and YJB11483
The strains with CaMtw1-GFP were constructed by transforming CAKS2b and CAKS3b (already reported in Padmanabhan et al., 2008), respectively, with the PCR products of plasmid pMG2120 (Ketel et al., 2009) and primers JB2715 and JB2759 and selecting NatR transformants. Correct inserts were checked by PCR with primers JB658 and JB2717.
Construction of YJB11553 and YJB11554
YJB11553 was constructed by transforming YJB8675 with pCL2 digested with SacI and HindIII to replace one copy of MTW1 with HIS1. Correct transformants were confirmed by PCR using primers MTW1-15 and MTW1-7. YJB11554 was constructed by transforming YJB11553 with PCK1pr-MTW1 plasmid digested with BglII to replace the promoter of MTW1 with PCK1pr marked with URA3. Correct transformants were identified by PCR using primers CA26 and MTW1-8.
Construction of YJB12118 and YJB12119
The strains with CaMif2-GFP were constructed by transforming CAKS11 and CAKS12, respectively, with the PCR products of plasmid pMG2120 (Ketel et al., 2009) and primers YJB4674 and YJB4675 and selecting NatR transformants. Correct inserts were checked by PCR with primers JB658 and JB3551.
Construction of YJB12176
Strain YJB12176 was constructed by transforming CAKS14 with the PCR products of plasmid pMG2120 (Ketel et al., 2009) and primers YJB4674 and YJB4675 and selecting NatR transformants. All manipulations of CAKS14 except the heat shock step of the transformation were performed at room temperature. Correct inserts were checked by PCR with primers JB658 and JB3551.
Cell viability, flow cytometry, and cytological analysis
These assays were performed as described previously (Sanyal & Carbon, 2002). CAKS11 (MTW1/mtw1) and CAKS12 (PCK1pr-MTW1/mtw1) cells were grown in YPSU overnight. Cells were pelleted, washed, and inoculated in YPDU at an A600 of 0.05. Samples were collected at 2 h intervals over a period of 10 h following transfer to glucose media. Cell viability, DNA content, and cell morphological analyses were done. Similarly, CAKS11 and CAKS14 (ts-mtw1/mtw1)cells also were grown in YPD overnight at 23°C and then shifted to 37°C at an A600 of 0.1 and cell samples were collected after every 30 min of growth at 37°C.
Viability tests were performed by measuring A600 and plating serial dilutions of a known number of cells on succinate plates at 30°C for 2-3 days in the case of CAKS12. The viability was determined by dividing the number of colonies grown in the plates by the theoretical number of cells plated. FACS analysis was done as described previously (Baum & Clarke, 2000). Cell morphologies were analyzed by counting different types of cells (unbudded, small-, and large-budded) and representative DIC images were captured using a confocal microscope (LSM 510 META, Carl Zeiss) after collecting the cell sample at indicated time points. The localization of nuclei of cells at different stages of growth, was determined by staining with 4′, 6-diamino-2-phenylindole (DAPI; Roche Diagnostics) as described previously (Kaiser, 1994).
Indirect Immunofluorescence
C. albicans cells of strain BWP17, CAKS12, CAKS13, CAKS14, and YJB10704 were grown asynchronously and were fixed by 37% formaldehyde at room temperature. Antibodies were diluted as described: 1:30 for rat anti-tubulin (YOL1/34) (Abcam), 1:250 for mouse anti-GFP antibody (Bangalore Genei) and for Alexa Fluor 568 goat anti-mouse IgG (Invitrogen), 1:500 for affinity purified rabbit anti-CaCse4, generated against an N terminal peptide (1-18th amino acids) of CaCse4 (Sanyal and Carbon, 2002), 1:500 for Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen) and for Alexa Fluor 488 anti-rat IgG (Invitrogen), 1:1000 for rabbit anti-protein A (Sigma). The positions of the nuclei were determined by DAPI staining. Cells were examined with a 100× magnification objective on a confocal laser scanning microscope (LSM 510 META, Carl Zeiss). Using LSM5 Image Examiner, digital images were captured, and lengths of the spindles were measured. Images were processed by Adobe Photoshop software.
Time-lapse imaging and fluorescence microscopy
Time-lapse imaging of yeast and hyphal cells were conducted as previously described (Finley & Berman, 2005; Finley et al., 2008). In each time-lapse experiment, images were collected using DIC and with an Endow GFP filter (Chroma Technology Corp, Rockingham, VT) at 4 min intervals with a Nikon E600 epifluorescence microscope using a 100× Plan-Apo (NA 1.4) objective. Images were captured with a CoolSnap HQ cooled charge-coupled-device camera (Photometrics, Tucson, AZ). Metamorph imaging software (Molecular Devices, Downingtown, PA) was used for time-lapse automation, image processing, and data collection.
Measurement of fluorescence intensities of CaCse4-GFP, CaMtw1-GFP and CaMif2 –GFP signals
ImageJ software was used to select the brightest GFP signal in each cell (the kinetochore). Equal region dimensions were used for each cell. The average pixel intensity in this region was determined and corrected for background by subtracting the lowest pixel intensity value in the cell from the average. Measurements were taken from at least 100 cells per condition. One-way ANOVA and Bonferroni post-tests were used to determine statistical significance.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) followed by PCR analysis was done as described previously (Padmanabhan et al., 2008; Sanyal et al., 2004). Rabbit anti-CaCse4 antibody or anti-protein A antibody (Sigma) was used for ChIP at a final concentration of 4 μg/ml or 20 μg/ml respectively. Asynchronous cultures of CAKS11 (MTW1/mtw1), CAKS12 (PCK1pr-MTW1/mtw1) and CAKS13 (MTW1-TAP/mtw1) grown until A600=1 were crosslinked with 37% formaldehyde for 15 min (for CaCse4) or 30 min (for CaMtw1). Subsequently, sonication was performed to get sheared chromatin fragments of an average size of 300-700 bp. The fragments were immunoprecipitated with anti-protein A or anti-CaCse4 antibody and checked by conventional PCR or real-time quantitative PCR (qPCR). ChIP DNA obtained from anti-protein A ChIP assays in CAKS13, were analyzed by conventional PCR with the primer pairs described previously [(Sanyal et al., 2004) and supporting Table S2]. ChIP DNA obtained from anti-CaCse4 ChIP assays in CAKS11, CAKS12, CAKS14 (ts-mtw1/mtw1) were analyzed by qPCR and primer pairs that amplify central regions of CEN5 (CACH5F1/CACH5R1) and CEN7 (nCEN7-3/nCEN7-4) (see supporting Table S2). Amplification from CaLEU2 ORF was also performed to detect the background immunoprecipitated DNA. For anti-CaCse4 ChIP analysis, qPCR was performed on a Rotor Gene 6000 real time PCR machine with IQ Sybr Green Supermix (Bio-Rad). Centromere primer sets are described above and in supporting Table S2. PCR of ChIP DNA was quantified by comparing (+) Ab, (-) Ab, and total input samples (diluted 10 folds). Cycling parameters were as follows: 94°C/30 sec, 55°C/30 sec, 72°C/45 sec repeated 40×. Melt curve analysis was performed from 55°C-94°C. Parameters were as follows: Premelt conditioning at 55°C/ 90 sec, rising by 1°C each step and hold for 5 sec. Each pair of primers exhibited a single melt peak. Error bars were calculated as standard error of the mean for 3 replicates.
ChIP of CaMtw1-GFP was done as previously described (Ketel et al., 2009) with anti-GFP antibody (Roche) at 4 μg/ml. Anti-Cse4 ChIP was done simultaneously with anti-Cse4 antibody produced in-house at 4 μg/ml (Ketel et al., 2009). Immunoprecipitated DNA was analyzed by qPCR using primer sets in Table S2 and the appropriate probe from the Universal Probe Library (Roche). qPCR for the CaMtw1-GFP ChIP was done on the Roche LightCycler480 according to the manufacturer's directions. The results shown are the average ± standard error of the mean of two independent biological replicates analyzed in duplicate.
ChIP of CaMif2-GFP was done essentially as described for CaMtw1-GFP. The conditional strain and the control strain were grown in inducing (succinate) or repressing (glucose) media for 6 h prior to fixation. Results shown are the average ± standard error of mean of a representative experiment from two biological replicates analyzed in duplicate. The temperature-sensitive strain and the control strain were grown at 23°C or 37°C for 2 h prior to fixation. Results shown are the average ± standard error of mean of a representative experiment from three biological replicates analyzed in duplicate.
qPCR enrichment calculation
The CaCse4 enrichment was determined by the percent input method. In brief, the Ct values for input were corrected for the dilution factor and then the percent of the input chromatin immunoprecipitated by the antibody was calculated as 100×2 (Adjusted Input Ct – IP Ct). Two-tailed t-tests were used to determine statistical significance.
Cell lysate preparation and Western Blot analysis
C. albicans cells were grown until A600=1. Cells were harvested, washed and resuspended in lysis buffer (0.2 M Tris, 1 mM EDTA, 0.39 M ammonium sulphate, 4.9 mM magnesium sulphate, 20% glycerol, 0.95% acetic acid, pH 7.8). Cells were lysed by vigorous vortexing in the presence of glass beads (Sigma, 425-600 μm) and protease inhibitors (Sigma) at 4°C. The lysis of the cells was confirmed by microscopic examination. Subsequently, the lysate was centrifuged at 4°C to pellet the cell debris, and the cleared supernatant was collected, boiled with the same volume of 2× SDS sample buffer (100 mM Tris-Cl pH 6.8, 4% (w/v) SDS, 0.2% (w/v) bromophenol blue, 20% (v/v) glycerol, 200 mM DTT) before analyzing by running on a 12% SDS-polyacrylamide gel. For Western blot analysis, the separated proteins were transferred to a nitrocellulose membrane using the transfer buffer (0.025 M Tris-Cl, 0.192 M glycine, 20% methanol) by semidry method. The membrane was then blocked using blocking solution (5% non-fat skim milk in 0.05% PBS-Tween solution). After blocking, the immunoblot was stained with rabbit anti-protein-A antibodies (Sigma), followed by goat anti-rabbit IgG-HRP antibodies (Bangalore Genei). This blot was stripped and reprobed with mouse anti-PSTAIR antibodies (Abcam), followed by goat anti-mouse IgG-HRP antibodies (Bangalore Genei).
Supplementary Material
Acknowledgments
We thank P. Koetter for the reagents, H. Hutton and A. Christensen-Quick for technical assistance, B. Suma for confocal imaging, I. Bose for help with Western blot analysis and critical comments, O. Joy for FACS analysis, Y. Thattikota for help in making line diagrams and R. Gadi for help in performing ChIP analysis. We also thank J. Thakur for constructive criticism. This work was supported by research grants from the Council of Scientific and Industrial Research and Department of Biotechnology, Government of India (K.S.). L.S.B. is supported by a Ruth L. Kirschstein NRSA Individual Fellowship from the NIH. Work in the Berman lab on this project was supported by an NIH/NIAID R01 AI075096 to J.B. We also gratefully acknowledge the financial assistance provided by Jawaharlal Nehru Centre for Advanced Scientific Research.
References
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks. Nat Rev Genet. 2008;9:923–37. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C, Nantel A, Whiteway M. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol Microbiol. 2005;57:942–959. doi: 10.1111/j.1365-2958.2005.04727.x. [DOI] [PubMed] [Google Scholar]
- Baum M, Clarke L. Fission yeast homologs of human CENP-B have redundant functions affecting cell growth and chromosome segregation. Mol Cell Biol. 2000;20:2852–2864. doi: 10.1128/mcb.20.8.2852-2864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum M, Sanyal K, Mishra PK, Thaler N, Carbon J. Formation of functional centromeric chromatin is specified epigenetically in Candida albicans. Proc Natl Acad Sci U S A. 2006;103:14877–14882. doi: 10.1073/pnas.0606958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz BJ, Ahmad K, Moore LL, Roth MB, Henikoff S. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Enquist-Newman M, Müller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Clarke L. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr Opin Genet Dev. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Collins KA, Castillo AR, Tatsutani SY, Biggins S. De novo kinetochore assembly requires the centromeric histone H3 variant. Mol Biol Cell. 2005;16:5649–60. doi: 10.1091/mbc.E05-08-0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corvey C, Koetter P, Beckhaus T, Hack J, Hofmann S, Hampel M, et al. Carbon Source-dependent assembly of the Snf1p kinase complex in Candida albicans. J Biol Chem. 2005;280:25323–25330. doi: 10.1074/jbc.M503719200. [DOI] [PubMed] [Google Scholar]
- Davies AE, Kaplan KB. Hsp90-Sgt1 and Skp1 target human Mis12 complexes to ensure efficient formation of kinetochore-microtubule binding sites. J Cell Biol. 2010;189:261–74. doi: 10.1083/jcb.200910036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993;120:141–51. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K. Epigenetic control of centromere behavior. Annu Rev Genet. 2007;41:63–81. doi: 10.1146/annurev.genet.41.110306.130127. [DOI] [PubMed] [Google Scholar]
- Finley KR, Berman J. Microtubules in Candida albicans hyphae drive nuclear dynamics and connect cell cycle progression to morphogenesis. Eukaryot Cell. 2005;4:1697–1711. doi: 10.1128/EC.4.10.1697-1711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley KR, Bouchonville KJ, Quick A, Berman J. Dynein-dependent nuclear dynamics affect morphogenesis in Candida albicans by means of the Bub2p spindle checkpoint. J Cell Sci. 2008;121:466–476. doi: 10.1242/jcs.015172. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad M, Berman J, Gale CA. Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast. 2001;18:859–864. doi: 10.1002/yea.738. [DOI] [PubMed] [Google Scholar]
- Goshima G, Kiyomitsu T, Yoda K, Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, McDonald KL, Barnes G, Drubin DG. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol. 1998;143:1029–40. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar A, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. 1994. pp. 207–217. [Google Scholar]
- Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, et al. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 2009;5:e1000400. doi: 10.1371/journal.pgen.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Tsai HJ, Tirosh I, Burrack LS, Barkai N, Berman J. Epigenetically-inherited centromere and neocentromere DNA replicates earliest in S-phase. PLoS Genet. 2010;6:e1001068. doi: 10.1371/journal.pgen.1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuker CE, Sonneborn A, Delbruck S, Ernst JF. Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans. Gene. 1997;192:235–240. doi: 10.1016/s0378-1119(97)00069-3. [DOI] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- Maskell DP, Hu XW, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol. 2010;190:823–834. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Kashihara K, Murata M. Characterization of the two centromeric proteins CENP-C and MIS12 in Nicotiana species. Chromosome Res. 2009;17:719–726. doi: 10.1007/s10577-009-9064-8. [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F, Sanchez M, Nombela C, Pla J. Virulence genes in the pathogenic yeast Candida albicans. FEMS Microbiol Rev. 2001;25:245–268. doi: 10.1111/j.1574-6976.2001.tb00577.x. [DOI] [PubMed] [Google Scholar]
- Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4931–46. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Thakur J, Siddharthan R, Sanyal K. Rapid evolution of Cse4p-rich centromeric DNA sequences in closely related pathogenic yeasts, Candida albicans and Candida dubliniensis. Proc Natl Acad Sci U S A. 2008;105:19797–19802. doi: 10.1073/pnas.0809770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca C, Draviam VM, Marco E, Sorger PK, De Wulf P. Roles for the conserved Spc105p/Kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS One. 2009;4:e7640. doi: 10.1371/journal.pone.0007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–52. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Tatsutani SY, Collins KA, Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Zhang W, Costa P, Archambault V, D'Avino PP, Lilley KS, et al. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS One. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Baum M, Carbon J. Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc Natl Acad Sci U S A. 2004;101:11374–11379. doi: 10.1073/pnas.0404318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal K, Carbon J. The CENP-A homolog CaCse4p in the pathogenic yeast Candida albicans is a centromere protein essential for chromosome transmission. Proc Natl Acad Sci U S A. 2002;99:12969–12974. doi: 10.1073/pnas.162488299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Shibata F, Murata M. Characterization of a Mis12 homologue in Arabidopsis thaliana. Chromosome Res. 2005;13:827–834. doi: 10.1007/s10577-005-1016-3. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–94. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharfenberger M, Ortiz J, Grau N, Janke C, Schiebel E, Lechner J. Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J. 2003;22:6584–97. doi: 10.1093/emboj/cdg636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Chen ES, Yanagida M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science. 2000;288:2215–9. doi: 10.1126/science.288.5474.2215. [DOI] [PubMed] [Google Scholar]
- Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–90. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–15. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.