Abstract
The escalating amount of genome-scale data demands a pragmatic stance from the research community. How can we utilize this deluge of information to better understand biology, cure diseases, or engage cells in bioremediation or biomaterial production for various purposes? A research pipeline moving new sequence, expression and binding data towards practical end goals seems to be necessary. While most individual researchers are not motivated by such well-articulated pragmatic end goals, the scientific community has already self-organized itself to successfully convert genomic data into fundamentally new biological knowledge and practical applications. Here we review two important steps in this workflow: network inference and network response identification, applied to transcriptional regulatory networks. Among network inference methods, we concentrate on relevance networks due to their conceptual simplicity. We classify and discuss network response identification approaches as either data-centric or network-centric. Finally, we conclude with an outlook on what is still missing from these approaches and what may be ahead on the road to biological discovery.
Introduction: the systems biology research workflow
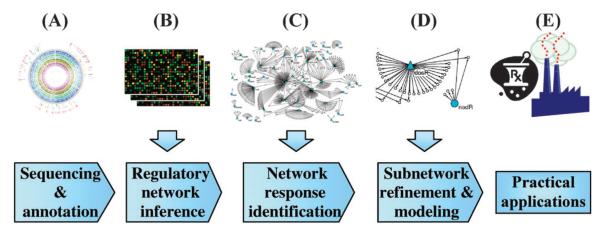
The number of completely sequenced genomes has been increasing exponentially, and is surpassing the 1K mark.1 However, the raw genomic sequence itself is of limited use for biological discovery. Once a new genome has been sequenced, its functional and regulatory regions need to be identified and annotated, which is nowadays performed simultaneously with sequencing (Fig. 1A). Nevertheless, knowing all genes of an organism is still just equivalent to a parts list of a very complex system, and tells us little about how these parts fit and operate together to initiate and control the diverse cellular responses and programs characteristic to even the simplest organisms. For true functional discovery, the annotated genomic sequence needs to pass through a systems biology research pipeline that combines it with other types of genomic data in several tiers of knowledge generation (Fig. 1), each introducing new challenges of data handling and visualization. First, the “wiring diagram” connecting thousands of biomolecular species needs to be learned, followed by charting the network regions active in specific conditions of interest. In this review, we will first concentrate on the inference of transcriptional regulatory networks (TRNs) from genomic data and then discuss the context-dependent response of these networks. Subnetworks identified this way can then serve as initial coarse-grained blueprints for experimental refinement of TRN structure and detailed quantitative modeling followed by practical applications. A prominent example of such a workflow has lifted the fragmented understanding of gene regulation in the archaeon Halobacterium salinarum NRC-1 to a level comparable to model organisms studied for many decades.2
Fig. 1.
The systems biology research workflow. Turning genomic data into practical applications demands a multistep “systems biology research workflow”, including network reconstruction and network response identification to reveal the network-level underpinnings of the organism's behavior.
Moving from the annotated genome tier to the TRN tier representation of the organism can be accomplished experimentally by genome-wide location analysis3 or computationally, by network inference (NI) methods (Fig. 1B). NI algorithms learn interactions of the type “gene product X regulates the synthesis of gene product Y” from high-throughput data. In the case of newly sequenced organisms, NI can be applied to generate a first draft of the genome-wide regulatory network; then, parts of this network can be validated by experimental approaches, followed by iterative cycles of network learning. However, at this stage it is still unclear how the network can be used for biomedical applications due to the vast number of interactions integrated into a highly complex structure.
On the bright side, the condition-dependent utilization of various TRN regions could provide the simplification necessary for quantitative tractability. This simplification is based on the assumption that, upon receiving an extracellular or intra-cellular signal, the cellular response involves not the entire network, but just specific modules responsible for processing the signal.4 Thus, identifying these network modules (conditionally active subnetworks) suffices for understanding the bases of cellular response. Charting condition-dependent TRN utilization is also important for understanding the molecular underpinnings of specific cellular phenotypes under different conditions (such as disease vs. normal). Moving from the global network tier to the context-dependent network tier is achieved by network response identification approaches (Fig. 1C). Based on the full TRN and large-scale data collected in a given condition, network response identification methods infer network regions utilized in a condition-specific manner, providing the list of active subnetworks in a particular condition.
Once the steps outlined above have been completed, the final tier of knowledge generation consists of “zooming in” on condition-dependent subnetworks responsible for cellular phenotypes of interest. For example, TRN regions identified as responsive to drug treatment can be studied in detail to combat drug resistance. Molecular biologists and computational modelers can now join forces to refine the condition-dependent network, accounting for hidden molecular components and interactions (metabolites, feedback loops, synergistic cross-talk between regulators), and chemical kinetic parameters such as binding affinities, production and degradation rates. Ultimately, this fine-grained description can be encoded into a formal mathematical or computational representation and becomes a quantitative predictive model. At this stage, predictive models can be applied to study the network behavior in unseen scenarios, by testing hypotheses in silico. For instance, it becomes feasible to ask “what if” questions, such as “What if the concentration of metabolite X increases”, or “what if we remove the inhibition over X”. This ability to predictively model and then manipulate network behavior can finally lead to reliable, new therapeutic interventions and biotechnological applications (Fig. 1E).
In the next two sections we will briefly review some of the available computational tools used in two crucial steps of the systems biology research workflow: network reconstruction (Fig. 1B) and network response identification (Fig. 1C), trying to follow progressively method development within each category of methods described.
Regulatory network inference
Biological cells are built from myriads of interacting molecules. Studying biomolecular interactions is crucial for understanding the normal and pathologic forms of the living state.5 While co-localized molecules in the cell can interact with each other regardless of their type, currently known biomolecular interactions have been assembled separately into transcriptional, signaling, metabolic, and protein-protein interaction networks.6 Of these, transcriptional regulatory and signaling networks are most closely associated with cellular information processing carried out through altered levels or activities of specific molecules. Here, we chose to focus on TRNs, due to the preponderance of data relevant for inferring these networks. Importantly, intercalating metabolites or protein interactions will not alter the regulatory nature of TRNs. Therefore, in a general sense TRNs are networks that contain a substantial fraction of directed transcriptional and post-transcriptional regulatory interactions.
Experimental network reconstruction
Early on, gene regulatory or signaling interactions were experimentally mapped one by one. More recently, the combination of chromatin immunoprecipitation and microarray technology (ChIP on chip) has revolutionized the identification of transcription factor (TF) binding sites, and has been used extensively to directly determine genome-scale TRNs in yeast and human cells.3,7 New assays built upon next-generation sequencing techniques (ChIP-Sequencing),8 or reporter cloning (TB 1-Hybrid)9 are quickly generating experimental snapshots of genome-scale TRNs. Likewise, protein arrays have recently been proposed for mapping global signaling networks.10 With the advent of systems biology, signaling and transcriptional regulatory interactions inferred by various experimental approaches were assembled into massive bibliomic databases and genome-wide TRNs.11-13 Still, the experimental scale and the costs involved in direct TRN mapping, or the effort necessary for literature-based TRN assembly, remain non-trivial. For this reason, the number of (partially) known TRNs is orders of magnitude below the number of annotated genomes, and represents an important bottleneck in the research workflow shown in Fig. 1, although this could soon be surpassed through computational literature mining14 and innovative network synthesis approaches.15 Moreover, it is usually unclear how the quality of various bibliomic and experimentally inferred genome-scale TRNs compare to each other, with some exceptions,16 and network validation by alternative approaches remains critical. The most complete TRNs are currently known for the model organisms Saccharomyces cerevisiae17 and Escherichia coli.11 However, TRN assembly has also been initiated for Bacillus subtilis,18Corynebacterium glutamicum19,20 and Mycobacterium tuberculosis,21 and many more are likely to follow. The performance of computational NI methods described below is usually assessed by overlaying the links they infer on experimentally constructed networks, further emphasizing the urgent need of advancing reliable experimental TRN mapping.
Computational network inference
Considering these difficulties, could we speed up TRN inference by seeking complementary approaches to the experimental mapping of direct regulatory interactions? “Omics” experiments are providing floods of functional data characterizing different aspects of transcriptional regulation and signal transduction. A quick search in NCBI GEO and ArrayExpress (two main repositories of expression data) results in tens of thousands of gene expression arrays for human tissues, model organisms, and clinically relevant pathogens. It is expected that deep sequencing technologies will soon produce in one year as much data as array-based approaches produced in the last 20 years. How can we exploit such omics datasets to learn molecular interactions indirectly from such data? This challenge has motivated systems biologists involved in reverse-engineering, or “learning” TRNs indirectly from gene expression and genomic sequence data. Their goal is to assemble the immense TRN puzzle from pieces such as transcription factors, sigma factors, kinases and non-coding RNAs, given genome-scale data reflecting the effect of these molecules. For example, to learn the TRN of a given organism, the task comes down to identifying TFs and their target genes.
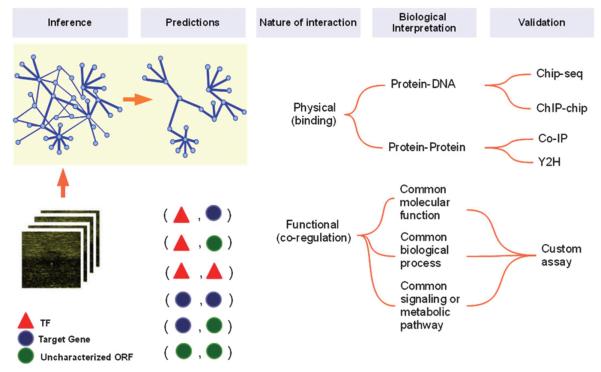
Computational NI from large-scale data has been an active area of research for the past decade, and there are excellent reviews discussing a plethora of methods.22-24 Since gene products can perform several roles in biomolecular networks, NI methods can recover many types of regulatory interactions broadly classifiable into physical and functional relationships (Fig. 2). Nevertheless, it is always critical to confirm computationally predicted TRNs by experimental approaches and independent datasets in iterative cycles of NI. In this section, without attempting completeness, we discuss a number of approaches that learn direct or indirect gene regulatory interactions from gene expression data. We will focus primarily on relevance networks,25 due to their relatively simple and intuitive construction scheme, but we will also mention briefly other NI approaches based on more elaborate algorithms.
Fig. 2.
Various interactions predicted by NI methods and their biological meaning. Consider that components in the dataset under study assume only three roles, namely TF, characterized gene/protein (known molecular function other than TF) and uncharacterized ORF or protein (unknown molecular function). In principle, NI methods are able to find either physical or functional interactions between all six pairs that arise when combining those three components. Physical and functional interactions can be direct or indirect. The biological interpretation of these interaction guides follow-up experiments for validation of in silico predictions. The highlighted region in the top left corner illustrates the inference of relevance networks by pruning links weaker than a certain threshold.
Relevance networks
The construction of relevance networks consists of two steps.25 First, a pairwise comparison of all gene expression profiles is performed using a similarity measure (metric). This results in a fully connected network among all genes, with the weight of each link equal to the metric. Second, the complete set of comparisons (links) is filtered by their strength, using threshold values. Links that “survive” thresholding constitute the relevance network and can be represented as a graph (see the highlighted region in Fig. 2).
A high value of the similarity measure between two expression profiles usually indicates coexpression at least in some conditions (although it can also appear by chance, even in completely random data). Possible biological causes of gene coexpression are direct or indirect regulation of one gene by the other, or coregulation of both by a third gene. Therefore, gene coexpression can indicate a direct or indirect regulatory relationship, and relevance networks can be inferred by hard25 or soft26 thresholding applied to various pairwise similarity metrics of gene expression profiles.
For example, relevance networks have been built using cross-correlation (Box 1) from several yeast and nematode micro-array datasets.27 The resulting networks had special properties (fat-tailed degree distribution and strong correlations) that were robust to the dataset used for NI, but disappeared when networks were regenerated using randomized data. Similar properties are shared by many other biological networks,6,28 indicating that relevance networks are biologically meaningful. Moreover, hubs (genes with many links) in these correlation-based networks were enriched in essential genes, as shown earlier for proteins with many interacting partners.29 The correlation between node degree and essentiality in biological networks is captured by the concept of “hub gene significance”, also observed in networks constructed by soft thresholding.26
Box 1. Similarity measures used for generating relevance networks.
Similarity measures quantify the dependence between gene expression profiles gi = gi(cn) and gj = gj(cn), n∈{1, 2,…, N} across a set of conditions, {c1, c2, c3,…,cN}.
The cross-correlation coefficient ρ(gi,gj) is defined as , where the brackets denote averaging over n. In terms of linear regression, cross-correlation is the part of the variance in gi(cn) explained by gj(cn) and vice versa. It takes values in the interval [−1,1].
The partial correlation ρ(gi,gj|gk) between gene expression profiles gi(cn), gj(cn), conditioned on the expression profile of a third gene, gk(cn), represents the correlation of genes i and j taking into account the effect of gene k, and is defined as . It can also be found by: (i) computing the residuals εi by linearly regressing gi(cn) as a function of gk(cn); (ii) computing the residuals εj by linearly regressing gj(cn) as a function of gk(cn); and (iii) computing the correlation between residuals εi and εj, ρ(gi,gj|gk) = ρ(εi,εj). When ρ(gi,gj|gk) ≈ 0, we say that the correlation between genes i and j is due to the effect of gene k, or alternatively, that gene k explains the correlation ρ(gi,gj).
The time-lagged correlation ρ[gi(cn),gj(cn–t)] is defined as the cross-correlation between expression profiles shifted by a time lag, τ.
The mutual information (MI) is defined as , where p is the probability density function, and the double brackets indicate averaging over both gene expression variables. This formula relies on the estimation of probability densities, which is most appropriately performed by fuzzy binning (using, for example, B-splines), especially for short gene expression profiles.106
The synergy is the opposite of the three-way mutual information between gene expression profiles gi(cn), gj(cn), and gk(cn), defined as Syn(gi,gj;gk) = −I(gi;gj;gk) = I(gi;gj|gk) − I(gi;gj). The actual quantity that augmented the MI between gi(cn) and gj(cn) in sa-CLR was the maximum of all Syn(gi,gj;gk) values calculated for every possible k different from i and j.
Partial correlation is another similarity measure that has been the basis of several methods aimed to learn both physical and functional interactions from data. The partial correlation (PCor) between gene i and gene j conditioned on gene k represents the correlation of genes i and j taking into account the effect of gene k (Box 1). A vanishing partial correlation indicates that gene k explains the correlation between genes i and j. Therefore, testing if the partial correlation vanishes can be used to filter out indirect links from correlation-based relevance networks. de la Fuente et al.30 proposed to build gene networks applying second-order PCor, based on the fact that most indirect interactions were eliminated when conditioning the correlation to every other pair of genes in the dataset. In another study, PCor was used to construct an isoprenoid gene network in Arabidopsis thaliana.31 An initial network of 40 genes was built by drawing genes from relevant pathways, followed by the subsequent recruitment of genes to the network using the criterion of high first-order PCor. Also, PCor was applied to infer regulatory interactions in E. coli.32 A genome-wide screen, testing second-order dependence between 261 TFs (putative and validated) and target genes, resulted in 75 new testable TF–operon interactions.
The correlation-based relevance networks described above are non-directional (a high correlation coefficient between two expression profiles is compatible with either gene regulating the other). Time-lagged correlation (Box 1) is a generalization of pairwise cross-correlation that compares time-shifted expression profiles to identify causal regulatory relationships from time-course data. The method was capable of identifying causal relationships in a metabolic chemical reaction system in vitro,33 and could be highly useful for reverse-engineering directed gene regulatory networks. Unfortunately, the requirement to deliver precisely controlled complex perturbations into the cell, followed by measurement of long time courses at the genome scale, has prevented the wide application of this technique, with a couple of exceptions.34,35
Correlation is geared to discover linear relationships between gene expression profiles, which do not always reflect the biological situation. For example, genes coexpressed only in specific conditions will not have a linear dependence between their expression levels. Multiple binding sites and saturation effects can make the dependence highly non-linear. Therefore, alternate similarity metrics are necessary that capture more information contained in the data, improving the reliability of NI methods.
Mutual information (MI, Box 1) is a generalized relatedness measure that overcomes some of the above limitations by quantifying any type of dependency (linear or non-linear) between two continuous variables.36 Relevance networks based on MI thresholding proved to be efficient in detecting functional relationships among yeast genes.37 Subsequent methods were aimed to discover physical interactions using MI.
ARACNe raised the challenge and tackled the problem of inferring human TRNs.38,39 The method relies on MI to detect regulatory interactions, but carries out an additional step to remove indirect connections from the network, which often appear when similarity measures are used to evaluate dependency between expression profiles. For ARACNe and many other methods, indirect links in the resulting TRN are undesired, because they are assumed not to represent physical interactions. ARACNe uses the data processing inequality, a property of information flow in cascades, to test whether a link is indirect, and therefore should be removed from the network. The method was used to reverse-engineer the B cell network, by predicting both protein–DNA and protein–protein interactions from a panel of normal and transformed human B cell lines, encompassing 336 microarray samples.
The context likelihood of relatedness (CLR) method40 also relies on an approach to minimize spurious or indirect relationships retrieved using MI. To evaluate whether a given TF–target gene interaction is significant, the method constructs two background distributions, and a given TF–target gene relationship is double background-corrected to assure that the high MI is meaningful when compared to other possible interactions. This double background correction scheme conferred superior performance to CLR compared to ARACNe and other NI methods40 when applied to reconstruct the E. coli TRN. Even though the proportion of recovered interactions from the known E. coli TRN was apparently small (2%–10%, depending on the false positive rate), in reality the method identified many new direct physical regulatory interactions, 21 of which were experimentally validated.
Synergy-augmented CLR (sa-CLR) is similar to CLR, except it also considers synergy in addition to the MI when computing the relatedness of gene expression profiles.41 Synergy (Box 1) is the opposite of the three-way mutual information between three gene expression profiles, and represents the information that is common to all three genes but is absent from the pairwise combination of any two genes. Including synergy in the similarity measure further improved the performance of CLR.41
Other NI methods
While relevance networks are relatively accessible and easy to implement, coexpression does not always imply coregulation: strong coexpression can appear by chance if the number of microarrays is small, or can result from a common environmental input to two distinct pathways. Conversely, lack of coexpression between mRNA expression profiles does not always imply lack of regulation, because the DNA-binding activity of many TFs depends on post-transcriptional modifications not reflected in microarray data. Many other types of NI methods exist, some of which may be able to circumvent these problems by the integration of diverse data types or by using more complex models of gene regulation.
A second class of NI methods relies on the assumption that TRNs can be described by a set of differential equations, with the expression of each gene dependent on a certain number of “inputs” from other genes. One early example is network identification by multiple regression (NIR), which approximates network dynamics through a set of linear differential equations near steady state. A perturbation (overexpression) is then applied to each gene in the network, resulting in a system of equations that can be robustly solved by linear regression.42 Inferelator is a more recent method unique in the sense that it combines steady state and time-course data into a common modeling framework (non-linear differential equations with a piecewise-linear activation function),43 and that it operates on top of the network response identification algorithm cMonkey44 (see below). The model allows for efficient estimation of the regulatory effect of TFs and environmental factors on individual genes or modules predetermined using cMonkey, as demonstrated by the reconstructed TRN of the archaeon Halobacterium salinarum NRC-1, containing 1431 interactions that the authors used to predict the expression of most modules in conditions that were not used for NI. It is yet unclear how the computational time required for these approaches will scale as the number of interactions expands, for example in highly complex mammalian TRNs.
Bayesian networks represent a third type of NI methods that have been used extensively to infer the connectivity and regulatory effects present in TRNs.45 The goal of these methods is to identify directed acyclic graphs and the corresponding conditional probability structures likely to have generated the gene expression or signaling data, based on the concept of conditional independence (two genes becoming independent when the third is fixed).46 This is a computationally expensive problem operating on large datasets, which requires limiting the search space and usually involves heuristic search algorithms. Integration of diverse data types was shown to improve the accuracy of Bayesian NI.47
Supervised and semi-supervised learning is a fourth NI approach that relies on existing, known TRNs to infer new regulatory interactions. For example, the machine learning method SEREND expands an existing bibliomic network11 after integrating it with microarray data and TF–promoter binding scores obtained from known position weight matrices.48 After training two logistic regression classifiers using the microarray data and motif scores, respectively, SEREND revisits TF–target gene pairs without evidence of regulation, and applies semi-supervised learning to infer additional target genes for each TF.
Network response identification
Modular network response
While TRN inference is a crucial step in the systems biology workflow (Fig. 1), the reconstructed network provides only a global summary of the multitude of molecular interactions that can take place in the organism. A major challenge is to discover the subset of interactions active in a particular subcellular component and/or condition. Accumulating evidence indicates that various parts of TRNs are utilized in an environment-dependent fashion,3,12,49 due to the subcellular localization, membership in molecular complexes, or chemical specificity of biological components.4 For example, if two proteins are not expressed simultaneously in a cell or cellular component, they cannot interact with each other, and if a TF is absent or inactive in a certain condition, then it cannot regulate its target genes. In multicellular organisms, network utilization is tissue-dependent and customized to perform tissue-specific functions.50 Such condition-dependent network utilization (modular network response) may be an evolved property of biological networks3 and must be better understood if we are to apply the workflow shown in Fig. 1 to eradicate microbial infections and interfere with cancer progression.
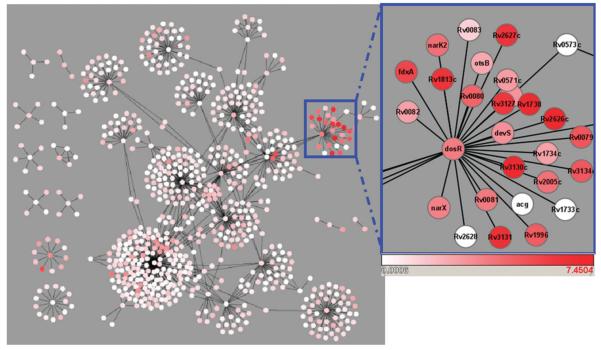
Two extreme views of network response called “molecular autocracy” and “molecular democracy” were proposed recently.51 “Molecular autocracy” means that a single gene regulates part of the genome directly or indirectly in response to environmental or developmental stimuli. On the other hand, “molecular democracy” corresponds to a general response, with all genes exerting regulatory influence on all other genes, leading to stability and homeostasis in the face of fluctuations. Modular network response corresponds to the former, autocratic scenario, when specialized sentinel sensors regulate the expression of quasi-disjoint gene groups immediately after an environmental perturbation, possibly preparing the network for a subsequent democratic response. For example, a well-defined subnetwork (controlled by the regulator dosR) initiates network response in M. tuberculosis in hypoxia (Fig. 3).
Fig. 3.
Modular response of the M. tuberculosis TRN at day 4 in hypoxia. DosR is a transcriptional regulator shared by a pair of two-component systems that sense the environmental signal and activate DosR, which then triggers a specific, modular network response by regulating its target genes.
The concept of modular network response stems from the assumption that network modules are utilized dynamically, “just-in-time”,52,53 ensuring that the cost of protein expression is compensated by the benefit conferred by the module's expression in a varying environment.54 Modularity in TRNs and other biological systems may have been evolutionarily selected due to its role in adaptation to a changing environment,55,56 or may be a byproduct of network growth.57 In either case, modularity seems to be a universal theme in living systems, in part supported by the success of the methods described below.
Two types of approaches are available for network response identification. Data-centric methods (such as biclustering and singular value decomposition) rely primarily on large-scale datasets to learn modular network response, and do not require the knowledge of an underlying network. By contrast, network-centric approaches overlay large-scale data on a known TRN to identify significantly affected modules in various conditions. Below we briefly describe some examples of data-centric and network-centric approaches.
Data-centric methods
Clustering was probably one of the first data-centric tools widely accepted in the systems biology community.58-60 The goal of clustering is to identify groups of genes with similar behavior across conditions based on some similarity measure.61 For example, hierarchical clustering agglomerates genes iteratively into clusters based on the similarity of their expression profiles (measured by cross-correlation or some other metric). This method coupled with a heatmap representation has successfully identified gene expression signatures of cancer subtypes,59 as well as the genome-wide stress response and cell cycle regulation of yeast genes.62,63 In these studies the resulting gene lists were not analyzed in the context of a large-scale network, but they can still be considered early examples of network response identification. Nowadays, gene clusters and expression data are frequently overlaid on TRNs to visualize active network regions and thereby understand context-dependent network utilization.64
With the explosive increase of microarray data covering several organisms and a wide spectrum of conditions, it became clear that genes cluster differently in different experiments. This fact prompted the development of novel “biclustering” methods able to group genes and conditions simultaneously along both dimensions (genes and conditions) represented in gene expression data. The first biclustering algorithm65 was followed by numerous newer versions, and the resulting biclusters were soon proposed to represent network modules.66 For example, the algorithm SAMBA implements a generalized biclustering method that works across multiple “dimensions” (data types), integrating gene expression, TF binding, knockout phenotypes, and protein interactions into the analysis.67 cMonkey is a more recent network response identification method that integrates biclustering with the co-occurrence of putative TF binding sites in promoters and the presence of highly connected subgraphs in metabolic, signaling, protein-protein, and comparative genomics networks.44 Similar to traditional clustering approaches, biclustering can be followed by network visualization to identify a context-dependent network. Biclustering remains a booming area of bioinformatics research today, including recent efforts to cross-compare and evaluate the performance of various biclustering approaches.68
Matrix algebra offers a powerful toolbox for data-centric identification of network response. For example, singular value decomposition (SVD) can be applied as a network response identification method to transform gene expression data into a space of mutually orthogonal eigengenes and eigenarrays, each being a linear combination of the original gene expression profiles and microarrays, respectively.69 SVD was applied to expression data collected during the yeast cell cycle, and remarkably found that two of the eigengenes and eigenarrays captured most of the biological expression changes during the yeast cell cycle, displaying periodic oscillations with different phase lags.
Network-centric methods
Network-centric approaches represent another family of network response identification methods that take a known regulatory network and large-scale data as inputs, and produce a list of subnetworks significantly affected in specific conditions.21,49,70,71 By analogy, if the known TRN corresponds to the roadmap of a city and gene expression data corresponds to city-wide traffic information on various days of the week, then network-centric approaches would identify the neighborhoods most affected by traffic on Tuesday morning, Saturday evening, and so on. Similar to data-centric methods, network-centric approaches identify TRN regions (modules) active in specific conditions and thus reveal context-dependent network utilization.
Ideker et al. pioneered an approach for identifying active regions in large-scale networks.71 They studied how gene deletions in the galactose utilization pathway affect a large-scale yeast TRN assembled from known transcriptional regulatory and protein–protein interactions.72 The method consisted of (i) z-scoring individual genes by their expression change; (ii) defining a background-corrected “subnetwork z-score”; and (iii) using simulated annealing to identify the top-scoring connected region (subnetwork). Background correction of subnetwork scores was performed through comparison with subnetworks of identical connectivity obtained after randomizing the network. An improved version of this method was able to identify “network signatures” of breast cancer metastasis (network regions that are significantly different in metastatic vs. non-metastatic tumors) by overlaying breast cancer gene expression data on top of a protein–protein interaction network.70 Network signatures identified from two different breast cancer datasets were more reproducible than single gene-based signatures, emphasizing current challenges with cancer signatures and the need for independent validation by innovative approaches.
Studying modular network response is highly facilitated if network modules are predefined from the start, based on network topology. This way, the computationally expensive task of mapping and scoring subnetworks (resolved by simulated annealing in71) is simplified to scoring a set of topology-based network modules. For example, the responses of the E. coli TRN11 to various environments have been studied using predefined modules (origons) that comprise the group of operons regulated directly or indirectly by a common TF. Microarray data collected in well-defined environments were then superimposed on the known TRN to identify origons significantly responsive to specific conditions, based on the covariance between an environment-dependent input signal and each gene's expression profile. The approach was later applied to reveal the context-dependent utilization of the yeast TRN73 and the response of the E. coli TRN to crp deletion in cell lines evolved over 20 000 generations;74 and was further developed to uncover the temporal succession of subnetworks responding to oxygen deprivation in Mycobacterium tuberculosis.21 This latter version called NetReSFun successfully recovered network regions induced by hypoxia, such as the dosR origon (Fig. 3) and made experimentally testable predictions about transcriptional modules affected by hypoxic stress. However, these methods are limited by the accuracy and extent of the information base used to predefine the network modules.
Once network regions utilized in specific contexts have been identified, the molecular function of the active subnetworks can be studied by enrichment analysis tools, which integrate gene lists with gene ontology information. A prominent example is DAVID75 that, given a list of genes (subnetworks), outputs the enriched molecular functions by retrieving data from several biological databases. Alternative methods are: BINGO, which identifies GO terms statistically over-represented in a subnetwork;76 ClueGO, which quantifies the association with different GO terms using kappa statistic;77 and PIPE, an in silico annotation network based on pathways and protein interaction data for mapping the gene list of interest.78
Outlook: what is still missing?
With the accelerating pace of biological discovery and technological development new opportunities arise for inferring TRNs and studying their response to perturbations. In this final section we mention some of the emerging opportunities and challenges that the systems biology research community is currently facing.
Nowadays, genome annotation as well as every task related to sequence analysis such as genomic sequence assembly, sequence comparison, and genome rearrangement detection can be performed entirely in silico. Clearly, the theoretical framework and corresponding computational tools for sequence analysis are sufficiently developed to be applied in an automated and trustful manner. Similarly, the next step on the road to successfully execute the research pipeline shown in Fig. 1 will be to improve the accuracy and reliability of NI and network response identification methods to a similar level. Presently, some of the best performing NI methods are still far from robust, and only recover a modest portion (up to 12%) of experimentally determined regulatory interactions as measured by precision-recall curves.40,79 Further, since no molecular network is known completely, it is not entirely clear how susceptible each of the approaches are to false discovery. What may be the obstacles that need to be overcome, and how can NI and network response identification methods be further improved?
One opportunity is to utilize current knowledge regarding the topological properties of TRNs.6 Most current NI methods learn regulatory interactions de novo, without any underlying assumptions about the network topology. However, the structure of TRNs is far from random: similar in- and out-degree distributions have been observed in various organisms,21 and TRNs are enriched in specific small subgraphs called network motifs.80 As general properties of network structure are being elucidated, they can be used to constrain the search space of future NI methods. For example, a novel class of NI methods relies on the fact that large parts of microbial TRNs are aggregates of network motifs.81 For instance, a large number of feed-forward (FF) motifs and bi-fan (BF) motifs are present in the E. coli TRN. Veiga et al.82 designed FF and BF predictors, using artificial neural networks that classify whether a group of genes are likely to form a motif. These motif predictors were applied to identify new TFs regulating transport proteins acting in efflux pumps linked to multidrug resistance in E. coli. In a similar approach, recurrent neural networks that mimic the topology and model the expression dynamics of motifs in yeast were used to identify novel interactions between TFs and a cluster of genes involved in the cell cycle.83
More fundamentally, the models underlying NI and network response identification approaches should be developed to incorporate additional layers of complexity in gene expression control. Current models operate based on an oversimplified view of gene expression, largely emphasizing biological events involved in transcriptional regulation, assuming that a gene's expression can be predicted by a linear or non-linear function of its activator and suppressor TFs. However, TRNs involve additional transcriptional and post-transcriptional regulation mechanisms that have rarely been considered. For instance, small, non-coding RNAs (sRNAs) play an important role in regulating mRNA transcription and translation initiation, both in bacteria and eukaryotes. In bacteria, especially in E. coli, hundreds of sRNAs have been found and classified according to their mechanisms of action,84 including trans-encoded sRNAs that act by basepairing and repressing translation of target mRNAs, in a mechanism similar to miRNAs in eukaryotes.85 The coordination of rapid, integrative responses of specific gene groups to environmental changes and stress conditions may be the reason why this type of regulation is frequently found in all domains of life. Given the importance of regulatory RNAs in affecting the dynamics of TRNs, future NI and network response identification methods could integrate this layer of information for generating more accurate context-dependent networks, and for learning more regulatory interactions from data. Other types of molecules that could be included in future models of gene regulation (and NI algorithms) are post-translational modifications and small metabolite signals, including alarmones and autoinducers. This will require the simultaneous collection of diverse data types and the integration of different molecular interaction networks.86
The rapid increase of completely sequenced genomes offers an excellent opportunity for learning networks by comparative genomics. For example, TRNs have been inferred in 175 prokaryotes based on the known TRN of E. coli.87 To generate these networks, a TF and a gene were linked if both were bidirectional best hits of a corresponding TF–target gene pair in E. coli. While this method should be applied with caution due to the rapid evolutionary divergence of bacterial gene regulation,88 a refined approach based on phylogenetic trees might be a promising strategy to quickly infer TRNs in closely related organisms.19 Such ab initio methods predicting novel TF binding sites by scanning promoter regions using position weight matrices or simply consensus TF binding sites89,90 will surely benefit from the increasing number and availability of completely sequenced genomes and experimentally validated TF binding sequences.
Synthetic biology offers a novel way to study biological networks, aiming to develop well-characterized molecular building blocks, followed by their assembly into increasingly complex synthetic networks with prescribed behavior, such as toggle switch,91 oscillators92,93 and linearizer.94 Using these circuits to deliver well-defined perturbations into specific regions of natural networks will likely improve NI and network response identification by eliminating most of the pleiotropic effects of natural perturbations. Recently, it has become possible to cross-compare the performance of various NI methods on a synthetic five-gene network assembled in budding yeast.95
Gene expression in single cells (measurable by flow cytometry or fluorescence microscopy) is often different from the population average (as measured by microarrays), and these deviations can have major biological implications (reviewed in96,97) due to their impact on cell survival98-100 and cell fate decisions.101-103 As a consequence, NI methods should ideally operate at the single cell level, accounting for expression differences between members of a cell population. However, technological limitations restrict our current ability to quantify the expression of more than a handful of molecules in single cells. Remarkably, Bayesian networks and multicolor flow cytometry have been combined to reverse-engineer small signaling networks in cancer cells.104 Moreover, gene expression measurements at the single cell level coupled with mathematical modeling were able to distinguish between active and inactive gene regulatory interactions in both synthetic and natural gene networks.105 Future development in this area will be very interesting to follow.
Network inference and network response identification are quickly developing research areas with new methods proposed at an accelerating pace. They represent crucial steps in a systems biology research workflow that we propose as a dependable strategy for converting various types of genomic data into biomedical applications. Future advance in these areas will require increasingly collaborative data collection efforts and improved experimental design to go hand in hand with these newly proposed approaches. While it might be tempting to ignore these or other steps in the systems biology research workflow shown in Fig. 1 (for example, by trying to predict drug targets directly from the annotated sequence or microarray data), such “shortcuts” will most likely increase the chance of false predictions and will result in waste of effort. In fact, additional tiers of knowledge generation may need to be included for multicellular eukaryotes, due to their more complex regulation operating across several organizational scales.
Acknowledgements
We thank Boris Hayete, Dmitry Nevozhay, Rhys Adams, James J. Collins, Gordon Mills and Marila Gennaro for comments. We acknowledge funding support from the TB PAN-NET consortium funded by the European Commission's Seventh Framework Programme for Research (FP7) to DFTV and GB; from the CAPES/Fulbright Program (Brazil) to DFTV; from the National Institutes of Health through the NIH Director's New Innovator Award Program, 1-DP2-OD006481-01 to GB and from the NIH-funded UCSF siRNA therapeutics consortium to BD.
Abbreviations
- BF
Bi-fan motifs
- CLR
Context likelihood of relatedness
- FF
Feed-forward motifs
- MI
Mutual information
- NI
Network inference
- PCor
Partial correlation
- TF
Transcription factor
- TRN
Transcriptional regulatory network
Biographies
 Diogo F. T. Veiga received his education in computer science at the Federal University of Santa Catarina, Brazil. During his undergraduate studies, he worked in the Genomics Engineering Group, in a highly interdisciplinary environment where he was introduced to applied computational biology problems. He spent one year at the National Laboratory for Scientific Computing studying in silico methods for reconstruction of prokaryotic regulatory networks using gene expression data. Currently, he is a CAPES/Fulbright PhD student at the University of Texas-Graduate School of Biomedical Sciences in Houston, investigating the molecular basis of therapy resistance in tuberculosis strains through network and comparative genomics approaches.
Diogo F. T. Veiga received his education in computer science at the Federal University of Santa Catarina, Brazil. During his undergraduate studies, he worked in the Genomics Engineering Group, in a highly interdisciplinary environment where he was introduced to applied computational biology problems. He spent one year at the National Laboratory for Scientific Computing studying in silico methods for reconstruction of prokaryotic regulatory networks using gene expression data. Currently, he is a CAPES/Fulbright PhD student at the University of Texas-Graduate School of Biomedical Sciences in Houston, investigating the molecular basis of therapy resistance in tuberculosis strains through network and comparative genomics approaches.
 Bhaskar Dutta was first introduced to scientific research as an undergraduate student at the Indian Institute of Technology (IIT) where he decided to pursue a career in science. He joined the University of Maryland as a graduate student in 2002 and started working in the field of systems biology. In collaboration with the Institute for Genomic Research (TIGR) he studied the dynamic transcriptomic and metabolomic response of Arabidopsis thaliana exposed to combinatorial environmental stress. During his postdoctoral training at the University of Texas M. D. Anderson Cancer Center he was involved in analyzing high-throughput siRNA screening data, network reconstruction, and developing approaches for overlaying data with large-scale networks. Currently he is a staff scientist at the Department of Defense, working in the field of network medicine.
Bhaskar Dutta was first introduced to scientific research as an undergraduate student at the Indian Institute of Technology (IIT) where he decided to pursue a career in science. He joined the University of Maryland as a graduate student in 2002 and started working in the field of systems biology. In collaboration with the Institute for Genomic Research (TIGR) he studied the dynamic transcriptomic and metabolomic response of Arabidopsis thaliana exposed to combinatorial environmental stress. During his postdoctoral training at the University of Texas M. D. Anderson Cancer Center he was involved in analyzing high-throughput siRNA screening data, network reconstruction, and developing approaches for overlaying data with large-scale networks. Currently he is a staff scientist at the Department of Defense, working in the field of network medicine.
 Gábor Balázsi has completed his undergraduate education in physics at the Babeş-Bolyai University of Cluj, Romania. He obtained his PhD in physics from the University of Missouri-St. Louis, where he worked in the Center for Neurodynamics, analyzing perturbation propagation in noisy excitable systems and studying the stochastic synchronization of calcium fluctuations in glial cell cultures. Following his graduation, he spent three years as a post-doctoral researcher at Northwestern University in Chicago, analyzing the modular response of transcriptional regulatory networks to environmental perturbations. Then he moved to the Applied BioDynamics Laboratory at the Department of Biomedical Engineering at Boston University, where he started working in the field of synthetic biology to engineer stochastic gene expression and demonstrate the impact of gene expression noise on the fitness of cell populations during drug treatment. Currently, he is Assistant Professor of Systems Biology at the University of Texas M. D. Anderson Cancer Center in Houston, Texas, USA.
Gábor Balázsi has completed his undergraduate education in physics at the Babeş-Bolyai University of Cluj, Romania. He obtained his PhD in physics from the University of Missouri-St. Louis, where he worked in the Center for Neurodynamics, analyzing perturbation propagation in noisy excitable systems and studying the stochastic synchronization of calcium fluctuations in glial cell cultures. Following his graduation, he spent three years as a post-doctoral researcher at Northwestern University in Chicago, analyzing the modular response of transcriptional regulatory networks to environmental perturbations. Then he moved to the Applied BioDynamics Laboratory at the Department of Biomedical Engineering at Boston University, where he started working in the field of synthetic biology to engineer stochastic gene expression and demonstrate the impact of gene expression noise on the fitness of cell populations during drug treatment. Currently, he is Assistant Professor of Systems Biology at the University of Texas M. D. Anderson Cancer Center in Houston, Texas, USA.
References
- 1.Kyrpides NC. Fifteen years of microbial genomics: meeting the challenges and fulfilling the dream. Nat. Biotechnol. 2009;27:627–632. doi: 10.1038/nbt.1552. [DOI] [PubMed] [Google Scholar]
- 2.Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, Longabaugh W, Vuthoori M, Whitehead K, Madar A, Suzuki L, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 3.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 5.Szent-Gyorgyi A. The living state and cancer. Proc. Natl. Acad. Sci. U. S. A. 1977;74:2844–2847. doi: 10.1073/pnas.74.7.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albert R. Scale-free networks in cell biology. J. Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- 7.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, Fraenkel E, Bell GI, Young RA. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo M, Feng H, Zhang J, Wang W, Wang Y, Li Y, Gao C, Chen H, Feng Y, He ZG. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009;19:1301–1308. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, McCartney RR, Schmidt MC, Rachidi N, Lee SJ, Mah AS, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 11.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, Bonavides-Martinez C, Abreu-Goodger C, Rodriguez-Penagos C, Miranda-Rios J, Morett E, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luscombe NM, Babu MM, Yu H, Snyder M, Teichmann SA, Gerstein M. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 13.Mathivanan S, Periaswamy B, Gandhi TK, Kandasamy K, Suresh S, Mohmood R, Ramachandra YL, Pandey A. An evaluation of human protein-protein interaction data in the public domain. BMC Bioinformatics. 2006;7(suppl 5):S19. doi: 10.1186/1471-2105-7-S5-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat. Rev. Genet. 2006;7:119–129. doi: 10.1038/nrg1768. [DOI] [PubMed] [Google Scholar]
- 15.Albert R, DasGupta B, Dondi R, Kachalo S, Sontag E, Zelikovsky A, Westbrooks K. A novel method for signal transduction network inference from indirect experimental evidence. J. Comput. Biol. 2007;14:927–949. doi: 10.1089/cmb.2007.0015. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual JF, Dricot A, Vazquez A, Murray RR, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J. Mol. Biol. 2006;360:213–227. doi: 10.1016/j.jmb.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 18.Makita Y, Nakao M, Ogasawara N, Nakai K. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res. 2004;32:75D–77D. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumbach J. CoryneRegNet 4.0—A reference database for corynebacterial gene regulatory networks. BMC Bioinformatics. 2007;8:429. doi: 10.1186/1471-2105-8-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brinkrolf K, Brune I, Tauch A. The transcriptional regulatory network of the amino acid producer Corynebacterium glutamicum. J. Biotechnol. 2007;129:191–211. doi: 10.1016/j.jbiotec.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Balazsi G, Heath AP, Shi L, Gennaro ML. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol. Syst. Biol. 2008;4:225. doi: 10.1038/msb.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol. Syst. Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonneau R. Learning biological networks: from modules to dynamics. Nat. Chem. Biol. 2008;4:658–664. doi: 10.1038/nchembio.122. [DOI] [PubMed] [Google Scholar]
- 24.Cuccato G, Della Gatta G, di Bernardo D. Systems and synthetic biology: tackling genetic networks and complex diseases. Heredity. 2009;102:527–532. doi: 10.1038/hdy.2009.18. [DOI] [PubMed] [Google Scholar]
- 25.Butte AJ, Kohane IS. Unsupervised knowledge discovery in medical databases using relevance networks. Proc. AMIA Symp. 1999:711–715. [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput. Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter SL, Brechbuhler CM, Griffin M, Bond AT. Gene co-expression network topology provides a framework for molecular characterization of cellular state. Bioinformatics. 2004;20:2242–2250. doi: 10.1093/bioinformatics/bth234. [DOI] [PubMed] [Google Scholar]
- 28.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 29.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 30.de la Fuente A, Bing N, Hoeschele I, Mendes P. Discovery of meaningful associations in genomic data using partial correlation coefficients. Bioinformatics. 2004;20:3565–3574. doi: 10.1093/bioinformatics/bth445. [DOI] [PubMed] [Google Scholar]
- 31.Wille A, Zimmermann P, Vranova E, Furholz A, Laule O, Bleuler S, Hennig L, Prelic A, von Rohr P, Thiele L, Zitzler E, Gruissem W, Buhlmann P. Sparse graphical Gaussian modeling of the isoprenoid gene network in Arabidopsis thaliana. GenomeBiology. 2004;5:R92. doi: 10.1186/gb-2004-5-11-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Veiga DF, Vicente FF, Grivet M, de la Fuente A, Vasconcelos AT. Genome-wide partial correlation analysis of Escherichia coli microarray data. Genet. Mol. Res. 2007;6:730–742. [PubMed] [Google Scholar]
- 33.Arkin A, Shen P, Ross J. A test case of correlation metric construction of a reaction pathway from measurements. Science. 1997;277:1275–1279. [Google Scholar]
- 34.Qian J, Dolled-Filhart M, Lin J, Yu H, Gerstein M. Beyond synexpression relationships: local clustering of time-shifted and inverted gene expression profiles identifies new, biologically relevant interactions. J. Mol. Biol. 2001;314:1053–1066. doi: 10.1006/jmbi.2000.5219. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt WA, Jr., Raab RM, Stephanopoulos G. Elucidation of gene interaction networks through time-lagged correlation analysis of transcriptional data. Genome Res. 2004;14:1654–1663. doi: 10.1101/gr.2439804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steuer R, Kurths J, Daub CO, Weise J, Selbig J. The mutual information: detecting and evaluating dependencies between variables. Bioinformatics. 2002;18(suppl 2):S231–S240. doi: 10.1093/bioinformatics/18.suppl_2.s231. [DOI] [PubMed] [Google Scholar]
- 37.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pac. Symp. Biocomput. 2000;5:418–429. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 38.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat. Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 39.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watkinson J, Liang KC, Wang X, Zheng T, Anastassiou D. Inference of regulatory gene interactions from expression data using three-way mutual information. Ann. N. Y. Acad. Sci. 2009;1158:302–313. doi: 10.1111/j.1749-6632.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- 42.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 43.Bonneau R, Reiss DJ, Shannon P, Facciotti M, Hood L, Baliga NS, Thorsson V. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. GenomeBiology. 2006;7:R36. doi: 10.1186/gb-2006-7-5-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reiss DJ, Baliga NS, Bonneau R. Integrated biclustering of heterogeneous genome-wide datasets for the inference of global regulatory networks. BMC Bioinformatics. 2006;7:280. doi: 10.1186/1471-2105-7-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J. Comput. Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- 46.Pe'er D. Bayesian network analysis of signaling networks: a primer. Science's STKE. 2005;2005:pl4. doi: 10.1126/stke.2812005pl4. [DOI] [PubMed] [Google Scholar]
- 47.Bernard A, Hartemink AJ. Informative structure priors: joint learning of dynamic regulatory networks from multiple types of data. Pac. Symp. Biocomput. 2005. 2005;10:459–470. [PubMed] [Google Scholar]
- 48.Ernst J, Beg QK, Kay KA, Balazsi G, Oltvai ZN, Bar-Joseph Z. A semi-supervised method for predicting transcription factor-gene interactions in Escherichia coli. PLoS Comput. Biol. 2008;4:e1000044. doi: 10.1371/journal.pcbi.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balazsi G, Barabasi AL, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183:1654–1655. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 51.Bar-Yam Y, Harmon D, de Bivort B. Systems biology. Attractors and democratic dynamics. Science. 2009;323:1016–1017. doi: 10.1126/science.1163225. [DOI] [PubMed] [Google Scholar]
- 52.Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science. 2001;292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- 53.Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U. Just-in-time transcription program in metabolic pathways. Nat. Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
- 54.Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- 55.Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat. Rev. Genet. 2007;8:921–931. doi: 10.1038/nrg2267. [DOI] [PubMed] [Google Scholar]
- 56.Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13773–13778. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sole RV, Valverde S. Spontaneous emergence of modularity in cellular networks. J. R. Soc. Interface. 2008;5:129–133. doi: 10.1098/rsif.2007.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 60.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 61.D'Haeseleer P. How does gene expression clustering work? Nat. Biotechnol. 2005;23:1499–1501. doi: 10.1038/nbt1205-1499. [DOI] [PubMed] [Google Scholar]
- 62.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ganter B, Giroux CN. Emerging applications of network and pathwayanalysis in drug discovery and development. Curr. Opin. Drug Discovery Dev. 2008;11:86–94. [PubMed] [Google Scholar]
- 65.Cheng Y, Church GM. Biclustering of expression data. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2000;8:93–103. [PubMed] [Google Scholar]
- 66.Ihmels J, Bergmann S, Barkai N. Defining transcription modules using large-scale gene expression data. Bioinformatics. 2004;20:1993–2003. doi: 10.1093/bioinformatics/bth166. [DOI] [PubMed] [Google Scholar]
- 67.Tanay A, Sharan R, Kupiec M, Shamir R. Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2981–2986. doi: 10.1073/pnas.0308661100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prelic A, Bleuler S, Zimmermann P, Wille A, Buhlmann P, Gruissem W, Hennig L, Thiele L, Zitzler E. A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics. 2006;22:1122–1129. doi: 10.1093/bioinformatics/btl060. [DOI] [PubMed] [Google Scholar]
- 69.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(suppl 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- 72.Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 73.Farkas IJ, Wu C, Chennubhotla C, Bahar I, Oltvai ZN. Topological basis of signal integration in the transcriptional-regulatory network of the yeast, Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:478. doi: 10.1186/1471-2105-7-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cooper TF, Remold SK, Lenski RE, Schneider D. Expression profiles reveal parallel evolution of epistatic interactions involving the CRP regulon in Escherichia coli. PLoS Genet. 2008;4:e35. doi: 10.1371/journal.pgen.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 76.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 77.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramos H, Shannon P, Aebersold R. The protein information and property explorer: an easy-to-use, rich-client web application for the management and functional analysis of proteomic data. Bioinformatics. 2008;24:2110–2111. doi: 10.1093/bioinformatics/btn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michoel T, De Smet R, Joshi A, Van de Peer Y, Marchal K. Comparative analysis of module-based versus direct methods for reverse-engineering transcriptional regulatory networks. BMC Syst. Biol. 2009;3:49. doi: 10.1186/1752-0509-3-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 81.Dobrin R, Beg QK, Barabasi AL, Oltvai ZN. Aggregation of topological motifs in the Escherichia coli transcriptional regulatory network. BMC Bioinformatics. 2004;5:10. doi: 10.1186/1471-2105-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Veiga DF, Vicente FF, Nicolas MF, Vasconcelos AT. Predicting transcriptional regulatory interactions with artificial neural networks applied to E. coli multidrug resistance efflux pumps. BMC Microbiol. 2008;8:101. doi: 10.1186/1471-2180-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang Y, Xuan J, de los Reyes BG, Clarke R, Ressom HW. Network motif-based identification of transcription factor-target gene relationships by integrating multi-source biological data. BMC Bioinformatics. 2008;9:203. doi: 10.1186/1471-2105-9-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009;4:85–103. doi: 10.2217/17460913.4.1.85. [DOI] [PubMed] [Google Scholar]
- 85.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeger-Lotem E, Margalit H. Detection of regulatory circuits by integrating the cellular networks of protein-protein interactions and transcription regulation. Nucleic Acids Res. 2003;31:6053–6061. doi: 10.1093/nar/gkg787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 88.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 2007;3:1739–1750. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Perez AG, Angarica VE, Vasconcelos AT, ColladoVides J. Tractor_DB (version 2.0): a database of regulatory interactions in gamma-proteobacterial genomes. Nucleic Acids Res. 2007;35:D132–D136. doi: 10.1093/nar/gkl800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodionov DA. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 2007;107:3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 92.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 93.Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nevozhay D, Adams RM, Murphy KF, Josic K, Balazsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5123–5128. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cantone I, Marucci L, Iorio F, Ricci MA, Belcastro V, Bansal M, Santini S, di Bernardo M, di Bernardo D, Cosma MP. A yeast synthetic network for in vivo assessment of reverse-engineering and modeling approaches. Cell. 2009;137:172–181. doi: 10.1016/j.cell.2009.01.055. [DOI] [PubMed] [Google Scholar]
- 96.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 97.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bayer TS, Hoff KG, Beisel CL, Lee JJ, Smolke CD. Synthetic control of a fitness tradeoff in yeast nitrogen metabolism. J. Biol. Eng. 2009;3:1. doi: 10.1186/1754-1611-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blake WJ, Balazsi G, Kohanski MA, Isaacs FJ, Murphy KF, Kuang Y, Cantor CR, Walt DR, Collins JJ. Phenotypic consequences of promoter-mediated transcriptional noise. Mol. Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 100.Smith MC, Sumner ER, Avery SV. Glutathione and Gts1p drive beneficial variability in the cadmium resistances of individual yeast cells. Mol. Microbiol. 2007;66:699–712. doi: 10.1111/j.1365-2958.2007.05951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 104.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 105.Dunlop MJ, Cox RS, 3rd, Levine JH, Murray RM, Elowitz MB. Regulatory activity revealed by dynamic correlations in gene expression noise. Nat. Genet. 2008;40:1493–1498. doi: 10.1038/ng.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Daub CO, Steuer R, Selbig J, Kloska S. Estimating mutual information using B-spline functions—an improved similarity measure for analysing gene expression data. BMC Bioinformatics. 2004;5:118. doi: 10.1186/1471-2105-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]