Abstract
The miRNA Registry provides a service for the assignment of miRNA gene names prior to publication. A comprehensive and searchable database of published miRNA sequences is accessible via a web interface (http://www.sanger.ac.uk/Software/Rfam/mirna/), and all sequence and annotation data are freely available for download. Release 2.0 of the database contains 506 miRNA entries from six organisms.
INTRODUCTION
MicroRNAs (miRNAs) are a class of non-coding RNA gene whose products are ∼22 nt sequences that play important roles in the regulation of translation and degradation of mRNAs through base pairing to partially complementary sites in the untranslated regions (UTRs) of the message. Since the discovery of the founding members of the class, let-7 and lin-4 miRNAs in Caenorhabditis elegans (reviewed in 1), more than 300 miRNAs have been found in animals and plants (2–19). In animals, the expression of miRNAs has been shown to involve at least two processing steps (20). miRNAs are transcribed as long primary transcipts (pri-miRNAs), which may contain more than one miRNA. The primary transcript is processed in the nucleus to give one or more hairpin precursor sequences (pre-miRNAs). This processing step defines one end of the mature miRNA sequence, which is contained in one arm of the hairpin precursor. The hairpin precursor is exported to the cytoplasm where the mature miRNA is excised by the RNase III-like enzyme Dicer, suggesting a relationship with RNA interference (RNAi) (21–23). The criteria for distinguishing miRNAs from other classes of RNA, such as small interferring RNAs (siRNAs) have recently been agreed by a number of miRNA scientists (24).
The rapid rate of miRNA gene discovery has led to two basic needs for the miRNA community. To avoid inadvertant overlap, it is important for miRNA researchers to have access to an independent arbiter of gene names. In addition, a comprehensive and up-to-date repository for published miRNA sequences and annotation greatly facilitates the rapid development of computational approaches for the prediction of miRNA genes and targets, as well as aiding sequence and genome annotation. Several groups have recently published work on prediction of miRNAs in C.elegans (14,16) and human (17), and reports of prediction and verification of the mRNA targets of a number of miRNAs have started to emerge (12,25).
AIMS OF THE miRNA REGISTRY
The primary aims of the miRNA Registry are two-fold. The first is to assign unique names to distinct miRNAs prior to publication of their discovery. A web interface has been developed to facilitate the submission of miRNA sequences for naming. To avoid accidental overlap of gene names, and to minimize ‘pre-booking’ of assignments, the Registry will assign a name only after a paper describing the sequence has been accepted for publication. Authors are advised to use temporary names in initial submission of articles to journals for peer-review. On acceptance, final names are discussed and agreed with the corresponding author. The miRNA Registry maintains complete confidentiality for pre-publication data.
miRNAs are given numerical identifiers based on sequence similarity. At the time of writing, the last assigned name is miR-318 from Drosophila melanogaster. The next miRNA with no similarity to previously identified sequences will receive the name miR-319. It is desirable for homologues in different organisms to receive the same name. Names are based on the similarity of the excised ∼22 nt sequence to previously identified miRNAs. Identical mature sequences are assigned the same name—if they originate from seperate genomic loci in a given organism they are given numberical suffixes, such as mir-6-1 and mir-6-2 from D.melanogaster (4). Sequences with one or two base changes are assigned suffixes of the form miR-181a and miR-181b (17). Homologous sequences with more base differences may be suggested by sequence similarity in the hairpin portion of the primary transcript, and such cases are discussed and names agreed with the corresponding author. Some miRNA hairpin precursors give rise to two excised miRNAs, one from each arm. Different naming conventions have been used to describe these sequences. Where cloning studies have allowed researchers to determine which arm of the precursor gives rise to the predominantly expressed miRNA, an asterisk has been used to denote the less predominant form, as in miR-56 and miR-56* from C.elegans (2). Previous reports have also denoted miRNAs from opposite arms of the hairpin precursor as, for example, miR-142-s (5′ arm) and miR-142-as (3′ arm) (5). Current opinion favours using names of the form miR-142-5p and miR-142-3p to designate miRNAs from the 5′ and 3′ arms, respectively, until the data are sufficient to confirm which is predominantly expressed (T. Tuschl and D. Bartel, personal communication). Capitalisation of names should not be relied upon to confer meaning, but historically, mir-16 has been used to designate the gene (and also the predicted stem–loop portion of the primary transcript), whereas miR-16 signifies the excised ∼22 nt sequence. Plant gene names follow a slightly different convention—of the form MIR156 (10).
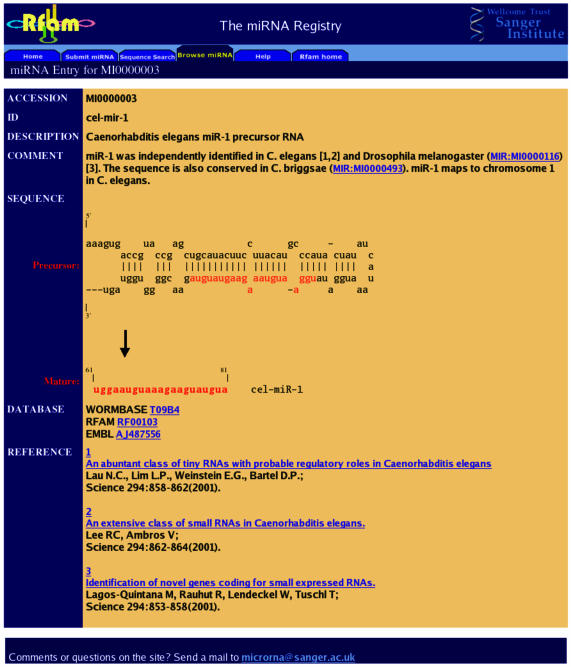
The second aim of the miRNA Registry is to provide a comprehensive and searchable database of all published miRNA sequences. To this end, submitted sequences are moved to the public sections of the database on their publication. The website includes a browsable list of miRNA entries, name, keyword and publication searches, and allows the user to search a sequence against the database of predicted hairpins and mature miRNAs. Each database entry represents a predicted stem–loop containing the miRNA, with the bounds of the excised sequence(s) reported. The publication describing the discovery of the miRNA is cited as the primary reference. A brief description of the genomic location, homologous sequences and possible targets is provided, with links to literature references for more information. Cross-links to nucleotide databases, model organism databases and RNA family databases are given. Hairpin base-paired structures are depicted as predicted by the RNAfold program from the ViennaRNA package (26). A typical entry page is shown in Figure 1.
Figure 1.
The entry for mir-1 from C.elegans. The predicted stem–loop portion of the primary transcript and the excised miRNA sequence are depicted. Links to other data sources, references and annotation are also shown.
A commitment to the long-term curation of the miRNA Registry ensures the rapid dissemination of new sequence data and annotation. Each database entry is identified by a stable accession number in addition to the miRNA gene name. This enables the rationalisation of gene names as more data become available, whilst maintaining information for tracking changes from initial published names and descriptions. At the time of writing, the database contains only published miRNA loci, but miRNA annotation guidelines allow for the computational identification of homologues of validated miRNA sequences (24). The size of the database is likely to increase significantly as such sequences are curated by us and others. As more information becomes available about the biogenesis of miRNAs, we predict that it will become desirable to curate sequence information for the primary transcipt and the hairpin precursor, as well as the excised mature miRNA. Close integration with the Rfam database (27) facilitates the classification of related miRNA sequences into families.
AVAILABILITY
The database is hosted by the Rfam (UK) website at http://www.sanger.ac.uk/Software/Rfam/mirna/ and is freely available to all. Predicted stem–loop and mature miRNA sequence data are also available for download from the FTP site in FASTA format, and complete with annotation in EMBL format. Release 2.0 of the database (July 2003) contains 506 entries from C.elegans, Caenorhabditis briggsae, D.melanogaster, human, mouse and Arabidopsis thaliana. Queries and feedback, including data revisions are welcomed by email to microrna@sanger.ac.uk.
Acknowledgments
ACKNOWLEDGEMENTS
I would like to thank Mhairi Marshall for web design and database support, and Simon Moxon for annotating many entries in the database. I am grateful to David Bartel, Tom Tuschl, Victor Ambros, Sean Eddy and Alex Bateman for their support and useful discussions, and David Bartel for critical manuscript reading.
REFERENCES
- 1.Pasquinelli A.E. and Ruvkun,G. (2002) Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol., 18, 495–513. [DOI] [PubMed] [Google Scholar]
- 2.Lau N.C., Lim,L.P., Weinstein,E.G. and Bartel,D.P. (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science, 294, 858–862. [DOI] [PubMed] [Google Scholar]
- 3.Lee R.C. and Ambros,V. (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science, 294, 862–864. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M., Rauhut,R., Lendeckel,W. and Tuschl,T. (2001) Identification of novel genes coding for small expressed RNAs. Science, 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M., Rauhut,R., Yalcin,A., Meyer,J., Lendeckel,W. and Tuschl,T. (2002) Identification of tissue-specific microRNAs from mouse. Curr. Biol., 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 6.Llave C., Xie,Z., Kasschau,K.D. and Carrington,J.C. (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science, 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- 7.Mette M.F., van der Winden,J., Matzke,M. and Matzke,A.J. (2002) Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol., 130, 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mourelatos Z., Dostie,J., Paushkin,S., Sharma,A., Charroux,B., Abel,L., Rappsilber,J., Mann,M. and Dreyfuss,G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev., 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park W., Li,J., Song,R., Messing,J. and Chen,X. (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol., 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinhart B.J., Weinstein,E.G., Rhoades,M.W., Bartel,B. and Bartel,D.P. (2002) MicroRNAs in plants. Genes Dev., 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ambros V., Lee,R.C., Lavanway,A., Williams,P.T. and Jewell,D. (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol., 13, 807–818. [DOI] [PubMed] [Google Scholar]
- 12.Brennecke J., Hipfner,D.R., Stark,A., Russell,R.B. and Cohen,S.M. (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell, 113, 25–36. [DOI] [PubMed] [Google Scholar]
- 13.Dostie J., Mourelatos,Z., Yang,M., Sharma,A. and Dreyfuss,G. (2003) Numerous microRNPs in neuronal cells containing novel microRNAs. RNA, 9, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grad Y., Aach,J., Hayes,G.D., Reinhart,B.J., Church,G.M., Ruvkun,G. and Kim,J. (2003) Computational and experimental identification of C. elegans microRNAs. Mol. Cell, 11, 1253–1263. [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M., Rauhut,R., Meyer,J., Borkhardt,A. and Tuschl,T. (2003) New microRNAs from mouse and human. RNA, 9, 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim L.P., Lau,N.C., Weinstein,E.G., Abdelhakim,A., Yekta,S., Rhoades,M.W., Burge,C.B. and Bartel,D.P. (2003) The microRNAs of Caenorhabditis elegans. Genes Dev., 17, 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim L.P., Glasner,M.E., Yekta,S., Burge,C.B. and Bartel,D.P. (2003) Vertebrate microRNA genes. Science, 299, 1540. [DOI] [PubMed] [Google Scholar]
- 18.Sempere L.F., Sokol,N.S., Dubrovsky,E.B., Berger,E.M. and Ambros,V. (2003) Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and Broad-Complex gene activity. Dev. Biol., 259, 9–18. [DOI] [PubMed] [Google Scholar]
- 19.Lai E.C., Tomancak,P., Williams,R.W. and Rubin,G.M. (2003) Computational identification of Drosophila microRNA genes. Genome Biol., 4, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee Y., Jeon,K., Lee,J.T., Kim,S. and Kim,V.N. (2002) MicroRNA maturation: stepwise processing and subcellular localization. EMBO J., 21, 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grishok A., Pasquinelli,A.E., Conte,D., Li,N., Parrish,S., Ha,I., Baillie,D.L., Fire,A., Ruvkun,G. and Mello,C.C. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell, 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 22.Ketting R.F., Fischer,S.E., Bernstein,E., Sijen,T., Hannon,G.J. and Plasterk,R.H. (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev., 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutvagner G. and Zamore,P.D. (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- 24.Ambros V., Bartel,B., Bartel,D.P., Burge,C.B., Carrington,J.C., Chen,X., Dreyfuss,G., Eddy,S.R., Griffiths-Jones,S., Marshall,M. et al. (2003) A uniform system for microRNA annotation. RNA, 9, 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhoades M.W., Reinhart,B.J., Lim,L.P., Burge,C.B., Bartel,B. and Bartel,D.P. (2002) Prediction of plant microRNA targets. Cell, 110, 513–520. [DOI] [PubMed] [Google Scholar]
- 26.Hofacker I.L. (2003) Vienna RNA secondary structure server. Nucleic Acids Res., 31, 3429–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S., Bateman,A., Marshall,M., Khanna,A. and Eddy,S.R. (2003) Rfam: an RNA family database. Nucleic Acids Res., 31, 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]