Abstract
During the priming step that leaves synaptic vesicles ready for neurotransmitter release, the SNARE syntaxin-1 transitions from a closed conformation that binds Munc18-1 tightly to an open conformation within the highly stable SNARE complex. Control of this conformational transition is key for brain function, but the underlying mechanism(s) is unknown. NMR and fluorescence experiments now show that the Munc13-1 MUN domain, which plays a central role in vesicle priming, dramatically accelerates the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex. This activity depends on weak interactions of the MUN domain with the syntaxin-1 SNARE motif, and probably with Munc18-1. Together with available physiological data, these results provide a defined molecular basis for synaptic vesicle priming, and illustrate how weak protein-protein interactions can play crucial biological roles by promoting transitions between high-affinity macromolecular assemblies.
Keywords: Neurotransmitter release, Munc13, synaptic vesicle priming, syntaxin, Munc18, conformational switch, protein NMR, exocytosis, synaptic transmission
The release of neurotransmitters by Ca2+-triggered synaptic vesicle exocytosis is crucial for interneuronal communication. This process involves docking of synaptic vesicles to specialized sites of the plasma membrane called active zones, priming of the vesicles to a release-ready state, and Ca2+-triggered fusion of the vesicle and plasma membranes1. The core of the sophisticated protein machinery that controls these steps includes Munc18-1 and the SNAREs synaptobrevin, syntaxin-1 and SNAP-252–5. The SNAREs assemble into a highly stable four-helix bundle called the SNARE complex through their SNARE motifs6, 7, which brings the two membranes together and is critical for membrane fusion3, 8. Munc18-1 forms a very tight complex with syntaxin-1 folded into a closed conformation that involves intramolecular binding of its N-terminal Habc domain9 to the SNARE motif and is different from the open conformation of syntaxin-1 in the SNARE complex10, 11 (Figs. 1a,b). Studies using a so called ‘LE mutation’ that helps to open syntaxin-110 showed that the binary Munc18-1-syntaxin-1 interaction gates SNARE complex formation and regulates the vesicular release probability12. Munc18-1 also binds to assembled SNARE complexes containing open syntaxin-113, 14, which is key for exocytosis15, 16 and may enable cooperation between Munc18-1 and the SNAREs in membrane fusion13, 14, 17–19.
Figure 1.
Weak interactions can dramatically accelerate transitions between tight complexes. (a) Domain diagrams of rat syntaxin-1A and Munc13-1. Residue numbers indicate selected domain boundaries. The position of the loop sequence removed in MUN* (residues 1408–1452) is also indicated. (b) Diagrams of the closed syntaxin-1–Munc18-1 complex [Munc18-1 in purple; syntaxin-1 regions colored as in panel a] and the SNARE complex [only the SNARE motifs are shown for synaptobrevin (red) and SNAP-25 (green)]. (c) Free energy diagram illustrating how the energy barrier for the transition from the closed syntaxin-1–Munc18-1 complex to the SNARE complex (with Munc18-1 bound to it) could be decreased through weak interactions with the MUN domain, which might lead to the formation of MUN–SNARE complex–Munc18-1 assemblies.
Synaptic vesicle priming is widely believed to involve opening of syntaxin-1 and partial SNARE complex assembly4, 5, 12, 20, but the molecular mechanism underlying this transition is unknown. Two large proteins from presynaptic active zones, Unc13/Munc13s and Unc10/α-RIMs, are known to play key roles in synaptic vesicle priming. Thus, priming is completely abolished upon genetic ablation of Unc13 in invertebrates or of its homologues Munc13-1 and Munc13-2 in mice21–24, while strong priming defects are also observed in the absence of Caenorhabditis elegans (C. elegans) Unc10 or their mammalian homologues RIM1α and RIM2α25–27. Interestingly, these priming defects were partially rescued by overexpression of the open syntaxin-1 LE mutant in C. elegans Unc13 or Unc10 nulls25, 28, suggesting that Unc13 and Unc10 are somehow involved in opening syntaxin-1. Moreover, reports of Unc13/Munc13 interactions with the syntaxin-1 N-terminal region29–31 suggested that Unc13/Munc13s may open syntaxin-1 directly. However, these interactions required the C2C domain (see domain diagram in Fig. 1a) or were observed with protein fragments that do not correspond to complete folded domains. In contrast, vesicle priming in Munc13-1,2 knockout mice was rescued with an autonomously folded module called MUN domain (without the C2C domain), and no binding of the MUN domain to syntaxin-1 was observed32. Hence, despite the extensive physiological data demonstrating the central role of Unc13/Munc13s in priming, their mechanism of action is highly unclear. Note also that, through modulation by other Unc13/Munc13 domains, and through interactions with RIMs33–35 and other large proteins of presynaptic active zones36, the priming activity of the MUN domain may be the focal point of regulation of neurotransmitter release in a wide variety of presynaptic plasticity processes that mediate diverse forms of information processing in the brain4, 32. Bridging the large gap between the physiological and biochemical characterization of Unc13/Munc13s is thus crucial to understand multiple fundamental aspects of brain function.
In extensive studies performed over several years using gel filtration, isothermal titration calorimetry and pulldown assays, we were unable to obtain convincing evidence for binding of the Munc13-1 MUN domain to recombinant soluble samples of key components of the release machinery such as the SNAREs, Munc18-1, synaptotagmin-1, Rab3A, complexins, or soluble complexes among them (N. Shen, R. Guan, O. Guryev, J. Lu, I. Dulubova and J. Rizo, unpublished data). However, we and others found that the Munc13-1 MUN domain binds to membrane-anchored SNARE complexes37, 38. These findings suggested that the MUN domain interacts weakly with the SNARE complex, and led us to hypothesize that MUN domain function depends critically on weak interactions that are not detectable by the methods mentioned above. Key for this hypothesis was the realization that, whereas the closed syntaxin-1–Munc18-1 complex is very stable [kD 1.3 nM39] and the SNARE complex is even tighter [affinity too high to be measured 40], the transition between the two complexes may not involve strong interactions; thus, weak, transient binding of the MUN domain to the SNAREs and/or Munc18-1 might be sufficient to lower the energy barrier of the transition (Fig. 1c).
To test this hypothesis and shed light on the function of the Munc13-1 MUN domain, we have performed a detailed analysis of its interactions with the SNAREs and Munc18-1, taking advantage of the power of NMR spectroscopy to analyze large macromolecular complexes with a wide range of affinities41, 42. We find that the MUN domain interacts weakly with the SNARE complex, with Munc18-1 and with the syntaxin-1 SNARE motif. Using NMR and fluorescence spectroscopies, we show that the transition from the closed syntaxin-1–Munc18-1 complex to the SNARE complex is dramatically accelerated by the MUN domain. This activity depends critically on the weak interaction of the MUN domain with the syntaxin-1 SNARE motif, and can be bypassed by the LE mutation in syntaxin-1. Together with the available physiological data21–24, 28, 32, these results provide a clear molecular basis for the role of Unc13/Munc13s in synaptic vesicle priming, and illustrate how weak protein interactions can play critical biological functions by accelerating transitions between tight macromolecular complexes.
RESULTS
MUN-SNARE complex-Munc18-1 macromolecular assemblies
The Munc13-1 MUN domain has a tendency to oligomerize that hinders NMR studies and could prevent detection of weak protein-protein interactions. However, we found that removing part of a loop that has variable length and is poorly conserved in MUN domains (residues 1408–1452)32, 43 yields a fragment that is monomeric even at 105 μM concentration (Supplementary Fig. 1).
This fragment was used for most experiments described below (below referred to as MUN*). To analyze interactions of MUN*, we used one- and two-dimensional (1D and 2D) 1H-13C heteronuclear multiple quantum correlation (HMQC) and 1H-15N heteronuclear single quantum correlation (HSQC) spectra of proteins that were uniformly 15N-labeled and/or 1H,13C-labeled at Ile, Leu and Val methyl groups in a perdeuterated background (here referred to as 2H-13CH3 labeling), monitoring the decrease in signal intensities due to binding to unlabeled proteins34, 41.
We first acquired 1H-13C HMQC spectra of 2H-13CH3-MUN* (72 kDa). Addition of Munc18-1 (67 kDa) decreased slightly the cross-peak intensities, while the SNARE complex (55 kDa) caused a more considerable decrease, and an even stronger decrease was observed on addition of both Munc18-1 and SNARE complex (Figs. 2a,b; Supplementary Figs. 2a,b). These results show that MUN* binds very weakly to Munc18-1 and somewhat more strongly to the SNARE complex, and suggest that these interactions cooperate to yield a MUN*-SNARE complex-Munc18-1 macromolecular assembly.
Figure 2.
Formation of MUN*-SNARE complex-Munc18-1 macromolecular assemblies. (a) Superposition of expansions of 1H-13C HMQC spectra of 12 μM 2H-13CH3-MUN* before and after addition of 20 μM Munc18-1 (M18) and 20 μM SNARE complex (SC). The inset was plotted at lower contour levels to show that binding did not induce large cross-peak shifts. (b) Average decrease in the intensities of well-resolved cross-peaks from 1H-13C HMQC spectra of 2H-13CH3-MUN* upon addition of 20 μM Munc18-1, 20 μM SNARE complex or both. (c) Superposition of expansions of 1H-13C HMQC spectra of 15 μM 2H-13CH3-SNARE complex before or after addition of 20 μM Munc18-1 and 30 μM MUN*. The 2H-13CH3-SNARE complex was formed with 2H-Ile-13CH3-syntaxin-1(2–253), 2H-Val,Leu-13CH3-synaptobrevin(29–93), 2H- SNAP-25(11–82) and 2H-SNAP-25(141–203). (d) Average decrease in the intensities of well- resolved cross-peaks from 1H-13C HMQC spectra of 15 μM 2H-13CH3-SNARE complex upon addition of 20 μM Munc18-1, 30 μM MUN*, or both. Color-coded bars represent data obtained for cross-peaks from the syntaxin-1 Habc domain (Habc), from the syntaxin-1 and synaptobrevin SNARE motifs (SNARE motif), or both (all). In (b, d) the error bars denote the standard error calculated from consecutive spectra acquired on the same samples. (e) 1D 1H-15N HSQC spectra of 20 μM 15N-MUN* before and after addition of 40 μM Munc18-1 and SNARE complex. (f) Plots of normalized integrals of the amide region of 1D 1H-15N HSQC spectra of 20 μM 15N-labeled MUN* upon addition of different concentrations of Munc18-1, syntaxin-1(2–253)–Munc18-1 complex (Syx/M18)), SNARE complex and Munc18-1 plus SNARE complex. Curves show the fits obtained with a standard one-to-one protein-ligand binding model.
We also acquired 1H-13C HMQC spectra of 2H-13CH3-SNARE complex where only the syntaxin-1 Ile and the synaptobrevin Leu,Val methyl groups were 1H,13C-labeled. This design, together with the resonance assignments available for the Habc domain9 and additional multidimensional NMR experiments (Supplementary methods), allowed us to distinguish the behavior of cross-peaks from the Habc domain versus those from the four-helix bundle formed by the SNARE motifs (below referred to as SNARE 4-helix bundle) (Fig. 2c; Supplementary Figs. 2c-h). Overall, addition of MUN*, Munc18-1 or both caused progressively stronger decreases in the cross-peak intensities (Fig. 2d, red bars). Munc18-1 caused stronger decreases in the Habc domain cross-peak intensities while MUN* affected more strongly the cross-peaks from the SNARE 4-helix bundle (Fig. 2d, blue and light pink bars). These results are consistent with the finding that Munc18-1 binds preferentially to the syntaxin-1 N-terminal region of soluble SNARE complexes44, and suggest that MUN* binds preferentially to the SNARE 4-helix bundle (Supplementary Note 1). Furthermore, these data reinforce the conclusion that MUN* forms a macromolecular assembly with Munc18-1 and the SNARE complex.
To measure the affinities of these interactions, we monitored the decrease in the integral of the amide region of 1D 1H-15N spectra of 15N-labeled MUN* upon progressive addition of unlabeled proteins (Figs. 2e,f). The titrations showed that MUN* has a relatively weak affinity for the SNARE complex (kD 35 ± 10 μM), and a much weaker affinity for Munc18-1 (estimated kD ≥ 150 μM). The kD measured for binding of MUN* to the Munc18-1–SNARE complex assembly is 13 ± 4 μM, showing that there is some (albeit moderate) cooperativity between the interactions of MUN* with the SNARE complex and Munc18-1.
The MUN domain binds weakly to the syntaxin-1 SNARE motif
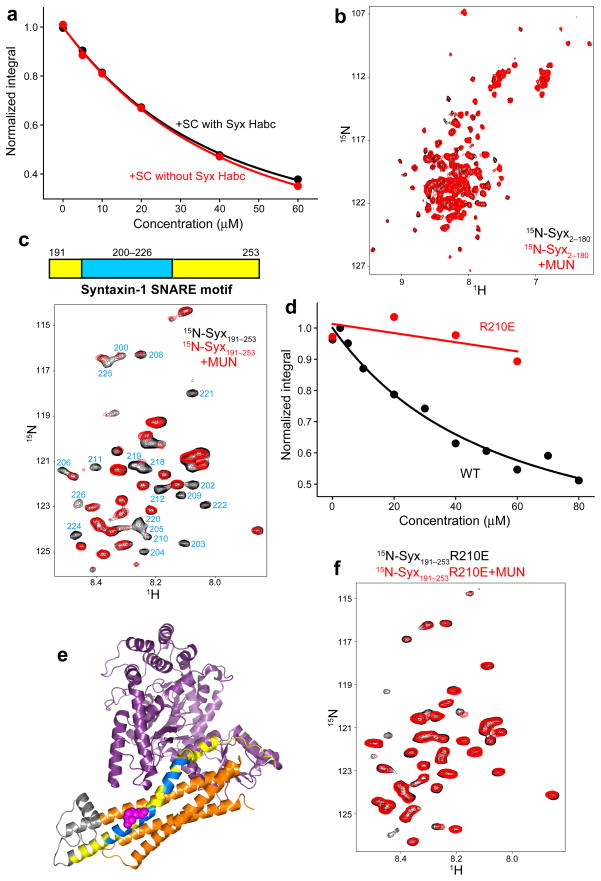
To further characterize which part of the SNARE complex binds to MUN*, we performed titrations of unlabeled MUN* on 15N-labeled SNARE complex containing or lacking the syntaxin-1 N-terminal region. We obtained very similar titration curves (Fig. 3a) and kDs of 26 ± 3 μM and 28 ± 5 μM, which are consistent with those obtained with 15N-MUN* and unlabeled SNARE complex (Fig. 2f), and confirm that MUN* interacts primarily with the SNARE 4-helix bundle rather than the Habc domain. This conclusion was supported by the fact that MUN* caused very few changes on 2D 1H-15N HSQC spectra of 15N-labeled syntaxin-1 N-terminal region (Fig. 3b). Importantly, MUN* caused strong broadening of selected cross-peaks in 2D 1H-15N HSQC spectra of the 15N-syntaxin-1 SNARE motif (Fig. 3c). Based on the available assignments10, all broadened cross-peaks correspond to the region spanning residues 200–226 of syntaxin-1. A titration yielded a kD of 46 ± 20 μM for the MUN*–syntaxin-1 SNARE motif interaction (Fig. 3d), comparable to that of the MUN*–SNARE complex interaction.
Figure 3.
MUN* binds to the syntaxin-1 SNARE motif. (a) Plots of normalized integrals of the amide region of 1D 1H-15N HSQC spectra of 20 μM 15N-SNARE complex containing syntaxin-1(2–253) (SC with Syx Habc) or syntaxin-1(191–253) (SC without Syx Habc) upon addition of different concentrations of MUN*. (b) 2D 1H-15N HSQC spectra of 20 μM 15N-syntaxin-1 N-terminal region (residues 2-180) before or after addition of 30 μM MUN*. (c) 2D 1H-15N HSQC spectra of 20 μM 15N-syntaxin-1 SNARE motif (residues 191–253) before or after addition of 30μM MUN*. Cross-peaks that are strongly broadened by MUN* binding are labeled with their corresponding residue number. The diagram above shows in blue the location of the MUN*-binding region (exhibiting strong broadening) within the syntaxin-1–SNARE motif, including selected residue numbers above. (d) Plots of normalized integrals of the amide region of 1D 1H-15N HSQC spectra of 10 μM wild type (WT) or R210E mutant (R210E) 15N-syntaxin-1 SNARE motif upon addition of different concentrations of MUN*. In (a,d), curves show the fits obtained with a standard one-to-one protein-ligand binding model. (e) Ribbon diagram of the structure of the syntaxin-1 Munc18-1 complex39. Munc18-1 is in purple and syntaxin-1 in orange (Habc domain), gray (linker) and yellow (SNARE motif), except that residues that exhibit strong broadening upon binding of MUN* to the isolated syntaxin-1 SNARE motif (see panel c) are in blue. Arg210 is represented by pink hard spheres. The ribbon diagram was generated with Pymol (DeLano Scientific). (f) 2D 1H-15N HSQC spectra of 20 μM 15N-syntaxin-1-R210E SNARE motif before or after addition of 30 μM MUN*.
The fact that MUN* causes selective broadening of cross-peaks from residues 200–226 arises from the unstructured, flexible nature of the syntaxin-1 SNARE motif10 and provides an internal control for the specificity of the interaction. Nevetherless, this region of syntaxin-1 is highly promiscuous3, 17, which may arise from an abundance of hydrophobic, flexible methionine side chains. To rule out that binding to MUN* merely reflects such promiscuity, we mutated Arg210 to glutamate (R210E mutant), since this conserved residue (Supplementary Fig. 3) is on the polar face of the syntaxin-1 SNARE motif. Moreover, the exposed nature of R210 in the syntaxin-1–Munc18-1 complex (Fig. 3e) makes it an ideal point for an action of MUN* on this complex (see below). Indeed, the R210E mutation strongly impairs binding of MUN* to the syntaxin-1 SNARE motif (Figs. 3d,f).
A new model for the mechanism of action of the MUN domain
Based on the data presented here and previous results32, it became clear that Munc13 function was unlikely to be primarily dictated by the interactions with the syntaxin-1 N-terminal region reported earlier29–31. Instead, our results and energetic arguments (Fig. 1c) suggest a novel model for the role of Munc13-1 in synaptic vesicle priming whereby the MUN domain does accelerate opening of syntaxin-1, but such activity is mediated by interactions with the syntaxin-1 SNARE motif (Fig. 4a). Note that a 1.4 kcal/mol decrease in the transition state energy should accelerate the reaction by a factor of 10, and the MUN–SNARE motif interactions can thus provide ample energy for a strong acceleration of the reaction despite their weak nature (the 46 μM kD measured, Fig. 3d, translates into ca. 6 kcal/mol of binding energy). The model is also based on the realization that the Munc18-1–syntaxin-1 complex can be viewed as a ‘three-layered’ object where the Habc domain is in the middle, sandwiched between Munc18-1 and the SNARE motif (Fig. 4a, left panel); hence, from a mechanistic point of view, it seems difficult to open syntaxin-1 through interactions with the Habc domain. In contrast, the region of the syntaxin-1 SNARE motif that includes the R210 side chain and binds to the MUN domain is more accessible (Fig. 3e and Fig. 4a, left panel), suggesting that a more natural way to open syntaxin-1 involves ‘extraction’ of the SNARE motif from the syntaxin-1–Munc18-1 complex due to binding to the MUN domain (Fig. 4a, middle panel). Such ‘extraction’ might be transient, and would be favored if optimal binding of the MUN domain to the syntaxin-1 SNARE motif is hindered in the syntaxin-1 Munc18-1 complex by steric clashes and/or if the specific conformation adopted by the syntaxin-1 SNARE motif in the syntaxin-1–Munc18-1 complex is different from that required for MUN domain binding.
Figure 4.
The MUN domain accelerates the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex. (a) Proposed model whereby the MUN domain (pink) promotes the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex through its weak interactions with the syntaxin-1 SNARE motif (color-coding as in Fig. 1b). (b) Superposition of expansions of 1H-13C HMQC spectra of 15 μM 2H-Ile-13CH3-syntaxin-1(2–253) bound to Munc18-1 or incorporated into SNARE complex formed with 2H-Val,Leu-13CH3-synaptobrevin(29–93), 2H-SNAP-25(11–82) and 2H-SNAP-25(141–203). The cross-peak assignments that are available are indicated with the residue number; H3 identifies cross-peaks that are known to belong to the SNARE motif but do not have residue-specific assignment; * indicates tentative assignments (Supplementary Methods). (c-d) 1H-13C HMQC spectra of samples of 15 μM 2H-Ile-13CH3-syntaxin-1(2–253) bound to Munc18-1 acquired immediately after addition of 15 μM 2H-Val,Leu-13CH3-synaptobrevin(29–93), 25 μM 2H-SNAP-25(11–82) and 25 μM 2H-SNAP-25(141–203), in the absence (c) or presence of 30 μM MUN* (d). (e-f) The progress of the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex as a function of time was monitored through the intensity of the disappearing Ile203 cross-peak of the syntaxin-1–Munc18-1 complex and the intensity of the appearing Ile203 cross-peak of the SNARE complex in consecutive 1H-13C HMQC spectra (2.3 hr each) obtained after mixing samples as in (c-d), in the absence (e) or presence (f) of 30 μM MUN*. The vertical axis represents the fraction of reaction completed based on normalized cross-peak intensities (see Supplementary Methods). (g) Progress of the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex in the presence of 30 μM MUN* monitored as in panel (f) but acquiring 10 min 1H-13C HMQC spectra. In (e-g), the curves show the fits of the data to an exponential rise to a maximum.
Based on these arguments, our model proposes that, starting with the Munc18-1–syntaxin-1 complex, weak MUN domain–syntaxin-1 SNARE motif interactions mediate the formation of a transient intermediate or transition state (Fig. 4a, middle panel), which lowers the energy barrier for the transition (Fig. 1c) and leads irreversibly to SNARE complex assembly upon binding of SNAP-25 and synaptobrevin to the syntaxin-1 SNARE motif. Note that the syntaxin-1 SNARE motif has an amphipathic nature, with a hydrophobic side that is buried in both the syntaxin-1–Munc18-1 complex and the SNARE complex, and a polar side that binds to the MUN domain (Supplementary Fig. 4). Therefore, we envision that the hydrophobic side of the syntaxin-1 SNARE motif becomes available for binding to SNAP-25 and synaptobrevin when the MUN domain binds to the polar side and releases the SNARE motif from its interactions with the Habc domain and Munc18-1. The MUN*-SNARE complex-Munc18-1 macromolecular assembly identified by our data serves as a natural end point for the transition in vitro (Fig. 4a, right panel), although only partial SNARE complex assembly is expected to occur in vivo during vesicle priming4, 5, 20, since C-terminal assembly is expected to be hindered by repulsion between the vesicle and plasma membranes, and might require additional factors such as synaptotagmin-145.
The MUN domain accelerates opening of syntaxin-1
Our model makes several predictions. First, binding of the MUN domain to the isolated syntaxin-1 SNARE motif should be stronger than binding to the syntaxin-1 closed conformation. Second and most important, the MUN domain should accelerate the rate of the transition from the Munc18-1–closed syntaxin-1 complex to the SNARE complex. Third, this accelerating activity should be impaired by the R210E mutation in syntaxin-1. And fourth, the LE mutation that helps to open syntaxin-110 and partially rescues the Unc13 phenotype28 should at least partially bypass the need for the MUN domain.
To test the first prediction, we performed titrations of 15N-MUN* with syntaxin-1–Munc18-1 complex (95 kDa) and observed very weak binding that paralleled the results obtained in titrations with Munc18-1 alone (Fig. 2f), suggesting that syntaxin-1 contributes little to this already very weak interaction. We also performed titrations of 15N-syntaxin-1 cytoplasmic region (28 kDa) with MUN* and observed only very weak binding (estimated kD ≥200 μM; Supplementary Fig. 5), consistent with previous results that indicated no binding at lower concentrations32. These data and the results of Fig. 3d show that MUN* indeed binds more strongly to the syntaxin-1 SNARE motif than to closed syntaxin-1.
In multiple attempts performed over several years to test the second, central prediction of our model, i.e. that the MUN domain accelerates the transition from the Munc18-1–syntaxin-1 complex to the SNARE complex, we monitored formation of SDS-resistant SNARE complexes, but it was difficult to obtain conclusive results. We finally observed acceleration of SNARE complex formation (Supplementary Fig. 6a) when we used concentrations of MUN* in the 20–30μM range, which correlates with the low affinity of the MUN*–syntaxin-1 SNARE motif interaction. Gel filtration experiments provided further evidence that MUN* accelerates the transition (Supplementary Figs. 6b,c), leading us to design NMR experiments that could provide definitive data and involved a labeling scheme that allowed monitoring the 1H-13C HMQC cross-peaks of syntaxin-1 Ile methyl groups with high sensitivity at 15 μM protein concentration even in the 200 kDa MUN*–SNARE complex–Munc18-1 assembly (Figs. 4b-d). Since several of these cross-peaks have different positions when syntaxin-1 forms the binary complex with Munc18-1 or the SNARE complex (Fig. 4b), these spectra allowed us to unambiguously monitor the transition between the two complexes in real time (note that the positions of the cross-peaks are very similar in the SNARE complex and the MUN*–SNARE complex–Munc18-1 assembly). The transition was also monitored simultaneously through the Leu,Val methyl cross-peaks of synaptobrevin, which change dramatically upon SNARE complex formation (Supplementary Fig. 7).
In reactions started with 2H-Ile-13CH3-syntaxin-1 bound to Munc18-1, consecutive 1H-13C HMQC spectra (2.3 hours duration each) acquired after addition of the synaptobrevin and SNAP-25 SNARE motifs revealed very slow disappearance of the cross-peaks from the Munc18-1–syntaxin-1 complex and concomitant (very slow) appearance of the cross-peaks from the SNARE complex (e.g. Fig. 4c). Kinetic analysis of normalized cross-peak intensities for several cross-peaks (e.g. Fig. 4e) yielded rate constants ranging from 0.021 to 0.16 hr−1 (the large range arises because the reaction is incomplete; Supplementary Note 2). However, both the disappearance of syntaxin-1–Munc18-1 complex cross-peaks and the appearance of SNARE complex cross-peaks were much faster in analogous reactions started in the presence of 30 μM MUN* (e.g. Fig. 4f). The final spectra were analogous to spectra obtained with preformed SNARE complex upon addition of Munc18-1 and MUN* (Fig. 2c; Supplementary Figs. 2e,h), demonstrating that the final product was indeed the MUN*-SNARE complex-Munc18-1 assembly. No reliable rate could be obtained from these data because SNARE complex formation was almost quantitative during acquisition of the first 1H-13C HMQC spectrum (Fig. 4f). However, the high sensitivity of the cross-peak of Ile203 from the SNARE complex and from the syntaxin-1–Munc18-1 complex (see its location in the two complexes in Supplementary Figure 8), as well as the Ile176 and Ile115 cross-peaks from the latter, allowed us to monitor the kinetics of the reaction through 1H-13C HMQC spectra acquired in 10 min each (Fig. 4g). These experiments yielded rate constants ranging from 4.0 to 5.1 hr−1 for the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex formation in the presence of MUN*, indicating that MUN* accelerates the reaction by a factor of 25 to 240 under these conditions (see Supplementary Note 2).
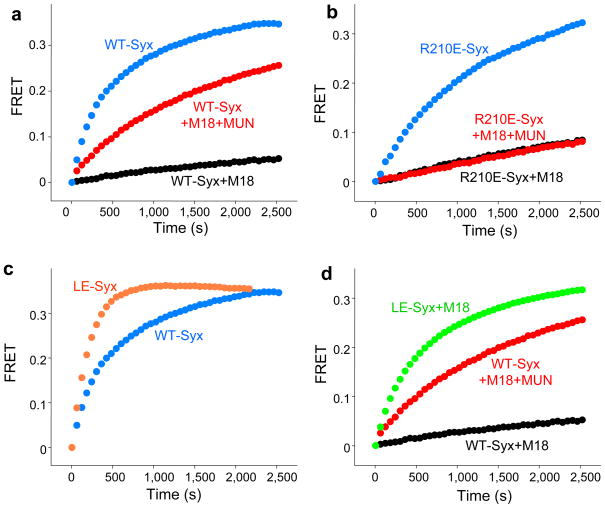
To test the third and forth predictions of our model regarding the effects of the R210E and LE mutations, we turned to fluorescence resonance energy transfer (FRET) experiments. SNARE complex assembly reactions started with isolated syntaxin-1 or the syntaxin-1–Munc18-1 complex (Fig. 5a) illustrated how Munc18-1 dramatically inhibits SNARE complex formation. Addition of MUN* strongly accelerated SNARE complex assembly reactions started with the syntaxin-1–Munc18-1 complex (Fig. 5a), which confirms our NMR results even though the acceleration rate by MUN* is lower (a factor of 11; for a quantitative analysis, see Supplementary Figs. 9a-d and Supplementary Note 2). Importantly, while Munc18-1 binding to the syntaxin-1-R210E mutant also inhibited SNARE complex assembly, MUN* was unable to stimulate SNARE complex formation starting with the syntaxin-1-R210E–Munc18-1 complex (Fig. 5b). These results show that the weak interaction of the MUN domain with the syntaxin-1 SNARE motif is critical for its activity in stimulating the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex, as postulated by our third prediction. Finally, SNARE complex assembly started with isolated syntaxin-1 containing the LE mutation was faster than that observed with wild type (WT)-syntaxin-1 but to a moderate extent (Fig. 5c), consistent with the notion that the closed conformation of isolated syntaxin-1 is not very stable46. However, the LE mutation dramatically accelerated SNARE complex assembly starting with the syntaxin-1-LE–Munc18-1 complex without the need to add MUN* (Fig. 5d). These results verify the fourth prediction of our model and beautifully explain the finding that the LE mutation partially rescues the Unc13 phenotype in C. elegans28.
Figure 5.
MUN* activity depends on interactions with the syntaxin-1 SNARE motif and is bypassed by the syntaxin-1 LE mutation. (a-d) SNARE complex assembly reactions monitored by the time-dependent development of FRET between a BODIPY fluorescence donor on residue 61 of synaptobrevin(29–93) and a Rhodamine fluorescence acceptor on residue 249 of syntaxin-1(2–253). Reactions were started by adding 2 μM labeled synaptobrevin, and 10 μM SNAP-25(11–82) and SNAP-25(141–203) to 10 μM WT syntaxin-1(2–253) or the R210E or LE syntaxin-1(2–253) mutants alone or bound to Munc18-1 (+M18) in the absence or in the presence of 30 μM MUN* (+MUN), as indicated.
In a final set of experiments, we tested whether the intact MUN domain, i.e. without deletion of the loop, is also able to accelerate the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex. We found that the intact MUN domain has a similar stimulation activity as MUN* (Supplementary Fig. 9e), showing that the loop does not alter this activity. We also tested whether MUN* can accelerate SNARE complex assembly starting with isolated syntaxin-1, but comparable rates were obtained in the absence or presence of MUN* (Supplementary Fig. 9f). These data suggest that the syntaxin-1–Munc18-1 complex, rather than isolated syntaxin-1, is the substrate for MUN domain activity, and that this activity is required to release the clamp that Munc18-1 holds on closed syntaxin-1.
DISCUSSION
Identification of several tight protein complexes provided crucial insights into the mechanism of neurotransmitter release2–5, but this mechanism remains poorly understood. Particularly important was to elucidate the function of Unc13/Munc13s in vesicle priming. The work presented here to address this question was driven by the realization that, while tight protein complexes provide ideal starting and end points for biological processes, weak interactions could be critical for rearrangements that occur during highly regulated processes such as neurotransmitter release. Our results provide a compelling illustration of this principle, showing that MUN* accelerates the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex, even though it does not bind tightly to Munc18-1 or the SNAREs. Instead, a weak interaction with the syntaxin-1 SNARE motif is critical for this activity of the MUN domain. These results and our NMR analysis of MUN*–SNARE complex–Munc18-1 interactions provide a well-defined framework to rationalize the physiological data on Unc13/Munc13s and strongly suggest that stimulation of the Munc18-1–syntaxin-1 to SNARE complex transition underlies the central role of Unc13/Munc13s in vesicle priming.
Our data add to increasing evidence revealing weak interactions within the release machinery that may be enhanced by cooperativity effects and/or proper localization, and that are likely critical for release 37, 38, 47, 48. This dependence on weak interactions might be in fact a common feature in biological processes that are tightly regulated. In the case of the MUN*–syntaxin-1 interaction, its weak nature explains the requirement of relatively high concentrations of MUN* for stimulation of SNARE complex formation. It can be expected that localization of Munc13 at the active zone and interactions with the plasma membrane37 dramatically increase its local concentration, favoring syntaxin-1 binding and increasing the speed of MUN domain action. The relevance of our results is also supported by the specificity of the MUN*–syntaxin-1 interaction and the activity of MUN* in stimulating SNARE complex formation, as shown by the data obtained with the syntaxin-1 R210E mutant.
Further research will be required to test and refine several aspects of our model. For instance, although the Munc13-1 C2C domain–syntaxin-1 N-terminus interaction29 is not essential for Munc13 function32, it might cooperate with the MUN domain–syntaxin-1 SNARE motif interaction to enhance Munc13 activity. Moreover, the interplay between the MUN domain, Munc18-1 and the SNAREs is likely complex and needs to be further characterized. Thus, the binary Munc18-1–syntaxin-1 interaction seems to inhibit release, stabilizing the syntaxin-1 closed conformation46 and impairing SNARE complex formation (ref. 39 and Fig. 5a). However, the finding that MUN* accelerates SNARE complex formation starting from the syntaxin-1–Munc18-1–complex (Figs. 4 and 5a) but not from isolated syntaxin-1 (Supplementary Fig. 9f) suggests that Munc18-1 assists in the activity of MUN in opening syntaxin-1, perhaps because weak Munc18-1-MUN* interactions (Fig. 2) help bringing MUN* close to syntaxin-1 and/or cause an allosteric effect. This synergy between Munc18-1 and the MUN domain could be enhanced by interactions with the membranes37, 44, perhaps leading to more efficient SNARE complex assembly than with isolated syntaxin-1. Furthermore, the binary Munc18-1 syntaxin-1 interaction likely plays positive roles by stabilizing both proteins and preventing the formation of unproductive syntaxin-1 complexes12, 49, 50. Munc18-1 also interacts with a syntaxin-1 N-terminal sequence that precedes the Habc domain, and this interaction may also play multiple roles14, 15, 39. In addition, whereas Munc18-1 binding to soluble SNARE complexes involves primarily the syntaxin-1 N-terminal region [Fig. 2d and Ref.44], membrane interactions likely help to translocate Munc18-1 to the SNARE 4-helix bundle after vesicle priming to cooperate with the SNAREs in membrane fusion14, 18, 44, 51. It will be critical to investigate whether the MUN domain competes or cooperates with Munc18-1 for binding to the SNARE 4-helix bundle in the presence of membranes, as well as to study the relation between these interactions and those involving other proteins that also bind to the SNARE 4-helix bundle, such as synaptotagmin-1 and complexins47, 52.
Unc13/Munc13s have additional functions beyond opening syntaxin-1, since the syntaxin-1 LE mutant rescues only some aspects of the Unc13 phenotype in C. elegans28, and does not rescue survival in Munc13-1 knockout mice12. The MUN domain itself could have an additional function downstream of priming, including a direct role in membrane fusion4. Indeed, reconstitution experiments showed that CAPS, which also contains a MUN domain, stimulates liposome docking, trans-SNARE complex formation and fusion53. These data were obtained in the absence of Munc18-1, and CAPS activity required a PH domain that is not present in Unc13/Munc13s. Hence, the relation of these results to our current study is unclear, but the finding that the syntaxin-1 LE mutant can reverse some secretory deficits caused by absence of CAPS54 supports the notion that the functions of Unc13/Munc13s and CAPS are related. Interestingly, the MUN domain was recently shown to exhibit homology to tethering complexes involved in traffic at diverse membrane compartments55 and one of these complexes causes a moderate acceleration of SNARE complex assembly56. Although this acceleration was observed in the absence of a Sec1/Munc18 homologue, it seems likely that some of the interactions of MUN domains and tethering complexes with SNAREs might have a general function in membrane traffic.
Regardless of these possibilities, it is certain that Unc13/Munc13s have multiple functions in diverse presynaptic plasticity processes that govern distinct forms of information processing in the brain4. Such functions are directly mediated by distinct domains of Unc13/Munc13s, and additional roles in presynaptic plasticity are likely played through interactions with αRIMs and other large proteins of the active zone33–36. An intriguing hypothesis is that modulation of MUN-domain activity may be the focal point of these plasticity processes32, which is supported by studies of the Munc13-1 C1 domain57 and by the observation that the syntaxin-1 LE mutation also rescues partially the Unc10 phenotype in C. elegans25. Our discovery that the MUN domain accelerates opening of syntaxin-1 now provides the opportunity to further test this hypothesis and unravel the molecular mechanisms of multiple presynaptic plasticity processes.
ONLINE METHODS
Plasmids and recombinant proteins
Constructs for bacterial expression of full-length rat Munc18-1, squid Munc18-1, or fragments of rat synaptobrevin-2 (residues 29–93), human SNAP25 (residues 11–82 and 141–203), the cytoplasmic domain (residues 2–253) of rat syntaxin-1A, its N-terminal region (residues 1–180) or its SNARE motif (residues 191–253), as well as the syntaxin-1A(2–253) L165E,E166A mutant (LE mutant), were described previously10, 13, 44, 52. The synaptobrevin(29–93) (S61C), syntaxin-1A(191–253) (R210E), syntaxin-1A(2–253) (R210E), syntaxin-1A(2–253) (C145S, S249C), syntaxin-1A(2–253) (L165E,E166A, C145S, S249C) and syntaxin-1A(2–253) (R210E, C145S, S249C) mutants were generated from the corresponding parent fragments using QuickChange site-directed mutagenesis kit (Stratagene). The vector to express rat Munc13-1(859–1407,1453–1531) fragment (MUN*) was prepared from the previously described Munc13-1(859–1531) fragment32 using standard molecular biology techniques. All proteins were expressed as GST fusions, isolated by affinity chromatography, and purified by gel filtration and/or ion exchange chromatography as described10, 13, 32, 52. Isotopic labeling was performed using well-established procedures9, 58. Protein concentrations were determined by UV absorbance. Most experiments involving Munc18-1 were performed with rat Munc18-1. However, because of the limited solubility of rat Munc18-1, binding experiments where isolated Munc18-1 had to be added at concentrations above 20 μM were performed with squid Munc18-1, which is more soluble and has a 66.4% sequence identity with rat Munc18-1, having very similar biochemical properties44.
Pre-assembled SNARE Complexes and Munc18 Syx 2–253 complexes
Pre-assembled SNARE complexes for binding assays were formed by incubating purified synaptobrevin(29–93), syntaxin-1(2–253), SNAP-25(11–82) and SNAP-25(141–203) overnight at 4°C in 20 mM Hepes, pH 7.4, 125 mM NaCl, 2 mM TCEP, in a 1:1:1:1 molar ratio. The assembled SNARE complexes were purified from unassembled fragments by gel filtration on a Superdex 200 column. For the binding experiments in Fig. 3a, SNARE complexes were also formed with syntaxin-1(2–253) replaced by syntaxin-1(191–253), and unassembled fragments were removed by repeated cycles of concentration-dilution with a 30 kDa cutoff52. Munc18-1–syntaxin-1(2–253) complexes were assembled with purified Munc18-1 and WT or mutant syntaxin-1(2–253) in a 1.5:1 molar ratio at room temperature for 1–2 hrs. The assembled complexes were purified by gel filtration on a Superdex 200 column (GE Healthcare).
NMR spectroscopy
1H-13C HMQC spectra were acquired at 25 °C on a Varian INOVA800 spectrometer equipped with a cold probe. Samples contained 12–15 μM 2H-13CH3-labeled proteins alone or with different additions, as indicated in the figure legends, dissolved in 20 mM Tris (pH 8.0), 150 mM NaCl, 2 mM TCEP, using D2O as the solvent. Spectra were generally acquired with a 2.3 hour total acquisition time, except for some of those performed to measure the kinetics of the transition from the syntaxin-1–Munc18-1 complex to the SNARE complex in the presence of MUN* (Fig. 4g), which were acquired with total acquisition time of 10 minutes. The methodology used to quantify the data from these spectra is described in Supplementary Methods. 2D 1H-15N HSQC spectra were acquired at 15 °C or 25 °C on a Varian INOVA600 spectrometer, using H2O/D2O 95:5(v/v) as the solvent. Samples contained 20 μM 15N-syntaxin-1(2–180) or 15N-syntaxin-1(191–253) (either WT or R210E mutant), without or with addition of 30 μM MUN*, and were dissolved in 20 mM HEPES (pH 7.4), 125 mM NaCl, 2 mM TCEP. Total acquisition times were 2 hours. 1D 1H-15N HSQC spectra for binding assays were acquired at 15 °C on Varian INOVA500, INOVA600 or INOVA800 spectrometers equipped with cold probes by running the first trace of a conventional 1H-15N HSQC experiment for 2 hours, using H2O/D2O 95:5(v/v) as the solvent. Samples contained 10 or 20 μM 15N-labeled proteins and variable amounts of unlabeled proteins, dissolved in 20 mM Tris (pH 8.0) containing 120 mM NaCl and 2 mM TCEP (for experiments with 15N-MUN*), or in 20 mM HEPES (pH 7.4), 125 mM NaCl, 2 mM TCEP for all other experiments. Integrals of the amide region of the 1D 1H-15N HSQC spectra were calculated with the Varian VnmrJ software (Agilent Technologies). The titration data were fit with a 1:1 protein-ligand binding model41 using Sigma Plot (Systat Software, Inc.). Note that a 1:1 binding model is the simplest and most natural assumption for the equilibria studied considering that all the data fit well to such model and that all reagents are monomeric and monodisperse based on previous NMR and DLS studies9, 10, 13, 15, 52. However, our data cannot rule out the possibility of more complex stoichiometries. For experiments where binding was very weak and hence we were far from reaching saturation, no reliable kD values could be obtained, and we could only estimate lower limits. 2D NMR data were processed with NMRPipe59 and analyzed with NMRView60.
FRET experiments
Purified synaptobrevin(29–93) (S61C) in PBS buffer (20mM sodium phosphate, pH 7.4, 100 mM NaCl) was labeled by adding 20x molar excess of Bodipy-FL-maleimide (Invitrogen). Purified syntaxin-1(2–253) (C145S, S249C) and its LE and R210E mutants were labeled using a 20x molar excess of tetramethylrhodamine-5-iodoacetamide dihydroiodide (5-TMRIA) (Invitrogen). After incubation at 4°C overnight, the reactions were stopped by adding 10mM dithiothreitol, and free dye was removed by ion exchange chromatography followed by gel filtration on a Superdex 200 column. Labeled proteins were dialyzed against Buffer A (25 mM HEPES pH 7.4, 150 mM KCl, 10 % glycerol (v/v)). FRET experiments were carried out on a Photon Technology Incorporated Spectrofluorometer (PTI, Lawrenceville, NJ). All experiments were performed at 28 °C in Buffer A. In time-based FRET experiments, labeled protein was excited at 485 nm, and the emission intensity was monitored at 513 nm. FRET efficiency was calculated according to E = (F0–Fobs)/F0, where F0 is the fluorescence intensity at time 0, and Fobs is the fluorescence intensity measured as a function of time.
Supplementary Material
Acknowledgments
We thank Yilun Sun for expert technical assistance, Yi Xu and Lijing Su for fruitful discussions, Michael Rosen and Lewis Kay for advice on NMR experiments, and Bill Wickner for insightful comments on the manuscript. This work was supported by a postdoctoral fellowship from the American Heart Association (to Y.X.), by Welch foundation grant I-1304, and by NIH grant NS37200 (to J.R.).
Footnotes
AUTHOR CONTRIBUTIONS
C.M. performed the kinetic studies of SNARE complex formation. C.M., W.L. and J.R. performed the NMR experiments. Y.X. performed initial biochemical studies of the MUN domain. J.R. wrote the paper.
Reference List
- 1.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 2.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 3.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 4.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirier MA, et al. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 7.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 8.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523–535. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez I, et al. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 10.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 12.Gerber SH, et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321:1507–1510. doi: 10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci U S A. 2007;104:2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128:183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 15.Khvotchev M, et al. Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus. J Neurosci. 2007;27:12147–12155. doi: 10.1523/JNEUROSCI.3655-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deak F, et al. Munc18-1 binding to the neuronal SNARE complex controls synaptic vesicle priming. J Cell Biol. 2009;184:751–764. doi: 10.1083/jcb.200812026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizo J, Chen X, Arac D. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release. Trends Cell Biol. 2006;16:339–350. doi: 10.1016/j.tcb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Diao J, et al. Single-Vesicle Fusion Assay Reveals Munc18-1 Binding to the SNARE Core Is Sufficient for Stimulating Membrane Fusion. ACS Chem Neurosci. 2010;1:168–174. doi: 10.1021/cn900034p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carr CM, Rizo J. At the junction of SNARE and SM protein function. Curr Opin Cell Biol. 2010;22:488–495. doi: 10.1016/j.ceb.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sorensen JB. Synaptobrevin N-terminally bound to syntaxin-SNAP-25 defines the primed vesicle state in regulated exocytosis. J Cell Biol. 2010;188:401–413. doi: 10.1083/jcb.200907018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richmond JE, Davis WS, Jorgensen EM. UNC-13 is required for synaptic vesicle fusion in C. elegans Nat Neurosci. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Augustin I, Rosenmund C, Sudhof TC, Brose N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature. 1999;400:457–461. doi: 10.1038/22768. [DOI] [PubMed] [Google Scholar]
- 23.Varoqueaux F, et al. Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming. Proc Natl Acad Sci U S A. 2002;99:9037–9042. doi: 10.1073/pnas.122623799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aravamudan B, Fergestad T, Davis WS, Rodesch CK, Broadie K. Drosophila UNC-13 is essential for synaptic transmission. Nat Neurosci. 1999;2:965–971. doi: 10.1038/14764. [DOI] [PubMed] [Google Scholar]
- 25.Koushika SP, et al. A post-docking role for active zone protein Rim. Nat Neurosci. 2001;4:997–1005. doi: 10.1038/nn732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schoch S, et al. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature. 2002;415:321–326. doi: 10.1038/415321a. [DOI] [PubMed] [Google Scholar]
- 27.Schoch S, et al. Redundant functions of RIM1alpha and RIM2alpha in Ca(2+)-triggered neurotransmitter release. EMBO J. 2006;25:5852–5863. doi: 10.1038/sj.emboj.7601425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richmond JE, Weimer RM, Jorgensen EM. An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming. Nature. 2001;412:338–341. doi: 10.1038/35085583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betz A, Okamoto M, Benseler F, Brose N. Direct interaction of the rat unc-13 homologue Munc13-1 with the N terminus of syntaxin. J Biol Chem. 1997;272:2520–2526. doi: 10.1074/jbc.272.4.2520. [DOI] [PubMed] [Google Scholar]
- 30.Stevens DR, et al. Identification of the minimal protein domain required for priming activity of Munc13-1. Curr Biol. 2005;15:2243–2248. doi: 10.1016/j.cub.2005.10.055. [DOI] [PubMed] [Google Scholar]
- 31.Madison JM, Nurrish S, Kaplan JM. UNC-13 interaction with syntaxin is required for synaptic transmission. Curr Biol. 2005;15:2236–2242. doi: 10.1016/j.cub.2005.10.049. [DOI] [PubMed] [Google Scholar]
- 32.Basu J, et al. A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol. 2005;12:1017–1018. doi: 10.1038/nsmb1001. [DOI] [PubMed] [Google Scholar]
- 33.Betz A, et al. Functional interaction of the active zone proteins Munc13-1 and RIM1 in synaptic vesicle priming. Neuron. 2001;30:183–196. doi: 10.1016/s0896-6273(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 34.Dulubova I, et al. A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J. 2005;24:2839–2850. doi: 10.1038/sj.emboj.7600753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu J, et al. Structural Basis for a Munc13-1 Homodimer to Munc13-1/RIM Heterodimer Switch. PLoS Biol. 2006;4:e192. doi: 10.1371/journal.pbio.0040192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, et al. A protein interaction node at the neurotransmitter release site: domains of Aczonin/Piccolo, Bassoon, CAST, and rim converge on the N-terminal domain of Munc13-1. J Neurosci. 2009;29:12584–12596. doi: 10.1523/JNEUROSCI.1255-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan R, Dai H, Rizo J. Binding of the Munc13-1 MUN Domain to Membrane-Anchored SNARE Complexes. Biochemistry. 2008;47:1474–1481. doi: 10.1021/bi702345m. [DOI] [PubMed] [Google Scholar]
- 38.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory Proteins Stabilize the Acceptor Complex for Synaptobrevin, the 1:1 Syntaxin/SNAP-25 Complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27:923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fasshauer D, Antonin W, Subramaniam V, Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat Struct Biol. 2002;9:144–151. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- 41.Arac D, Murphy T, Rizo J. Facile detection of protein-protein interactions by one-dimensional NMR spectroscopy. Biochemistry. 2003;42:2774–2780. doi: 10.1021/bi0272050. [DOI] [PubMed] [Google Scholar]
- 42.Ruschak AM, Kay LE. Methyl groups as probes of supra-molecular structure, dynamics and function. J Biomol NMR. 2010;46:75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- 43.Brose N, Hofmann K, Hata Y, Sudhof TC. Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem. 1995;270:25273–25280. doi: 10.1074/jbc.270.42.25273. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y, Su L, Rizo J. Binding of Munc18-1 to Synaptobrevin and to the SNARE Four-Helix Bundle. Biochemistry. 2010;49:1568–1576. doi: 10.1021/bi9021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008;15:1160–1168. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Lu J, Dulubova I, Rizo J. NMR analysis of the closed conformation of syntaxin-1. J Biomol NMR. 2008;41:43–54. doi: 10.1007/s10858-008-9239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai H, Shen N, Arac D, Rizo J. A Quaternary SNARE-Synaptotagmin-Ca(2+)-Phospholipid Complex in Neurotransmitter Release. J Mol Biol. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue M, et al. Binding of the complexin N terminus to the SNARE complex potentiates synaptic-vesicle fusogenicity. Nat Struct Mol Biol. 2010;17:568–575. doi: 10.1038/nsmb.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verhage M, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science. 2000;287:864–869. doi: 10.1126/science.287.5454.864. [DOI] [PubMed] [Google Scholar]
- 50.Medine CN, Rickman C, Chamberlain LH, Duncan RR. Munc18-1 prevents the formation of ectopic SNARE complexes in living cells. J Cell Sci. 2007;120:4407–4415. doi: 10.1242/jcs.020230. [DOI] [PubMed] [Google Scholar]
- 51.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190:55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X, et al. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 53.James DJ, Kowalchyk J, Daily N, Petrie M, Martin TF. CAPS drives trans-SNARE complex formation and membrane fusion through syntaxin interactions. Proc Natl Acad Sci U S A. 2009;106:17308–17313. doi: 10.1073/pnas.0900755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, et al. Two distinct secretory vesicle-priming steps in adrenal chromaffin cells. J Cell Biol. 2010;190:1067–1077. doi: 10.1083/jcb.201001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pei J, Ma C, Rizo J, Grishin NV. Remote homology between Munc13 MUN domain and vesicle tethering complexes. J Mol Biol. 2009;391:509–517. doi: 10.1016/j.jmb.2009.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren Y, et al. A structure-based mechanism for vesicle capture by the multisubunit tethering complex Dsl1. Cell. 2009;139:1119–1129. doi: 10.1016/j.cell.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basu J, Betz A, Brose N, Rosenmund C. Munc13-1 C1 domain activation lowers the energy barrier for synaptic vesicle fusion. J Neurosci. 2007;27:1200–1210. doi: 10.1523/JNEUROSCI.4908-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tugarinov V, Sprangers R, Kay LE. Line narrowing in methyl-TROSY using zero-quantum 1H-13C NMR spectroscopy. J Am Chem Soc. 2004;126:4921–4925. doi: 10.1021/ja039732s. [DOI] [PubMed] [Google Scholar]
- 59.Delaglio F, et al. Nmrpipe - A Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 60.Johnson BA, Blevins RA. Nmr View - A Computer-Program for the Visualization and Analysis of Nmr Data. Journal of Biomolecular Nmr. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.