Abstract
Chagas' disease, caused by the hemoflagellate protozoan Trypanosoma cruzi, affects millions of people in South and Central America. Chronic chagasic cardiomyopathy, the most devastating manifestation of this disease, occurs in approximately one-third of infected individuals. Events associated with the parasite's tropism for and invasion of cardiomyocytes have been the focus of intense investigation in recent years. In the present study, we use murine microarrays to investigate the cellular response caused by invasion of primary murine cardiomyocytes by T. cruzi trypomastigotes. These studies identified 353 murine genes that were differentially expressed during the early stages of invasion and infection of these cells. Genes associated with the immune response, inflammation, cytoskeleton organization, cell-cell and cell-matrix interactions, apoptosis, cell cycle, and oxidative stress are among those affected during the infection. Our data indicate that T. cruzi induces broad modulations of the host cell machinery in ways that provide insight into how the parasite survives, replicates, and persists in the infected host and ultimately defines the clinical outcome of the infection.
INTRODUCTION
Chagas' disease, caused by the intracellular flagellate protozoan Trypanosoma cruzi, is a major public health problem in Latin America. According to the World Health Organization, approximately 13 million Latin Americans are currently infected by the parasite and another 90 million are at risk for infection (58). Chronic chagasic cardiomyopathy (CCC) is the most devastating manifestation of Chagas' disease. Affecting approximately one-third of infected individuals, CCC represents the major cause of cardiomyopathy in Latin America. Despite the obvious clinical importance, the pathogenesis of CCC remains poorly understood (24).
Intracellular pathogens like T. cruzi exhibit a broad range of strategies to guarantee their establishment and persistence in the host. These strategies depend on the ability of the microorganisms to take control of the host cell machinery and undermine host defense mechanisms (21). Two such strategies, i.e., appropriation of components of the cytoskeleton to enhance invasion and intracellular motility and subversion of pathways for signal transduction and apoptosis, are well-documented examples of mechanisms by which some intracellular parasites promote their invasion, replication, completion of their cell cycle, and, ultimately, survival in the infected host (9). Infection of mammalian host cells by T. cruzi is a multistep process that requires activation of multiple signal transduction pathways in both the host and the parasite that lead to parasite entry (6, 15). Upon completion of a replicative cycle as intracellular amastigotes, the parasites escape from the cell as trypomastigotes, infect neighboring cells, and are eventually disseminated throughout the body, leading to the establishment of the systemic infection.
It has been shown that T. cruzi invading mammalian cells binds to the TrkA receptor, the receptor tyrosine kinase widely expressed in the mammalian nervous system, activating TrkA-dependent survival mechanisms and facilitating its adherence, invasion, and survival (40). This binding is mediated by the parasite-derived neurotrophic factor (PDNF), a trans-sialidase located on the surface of the parasite. PDNF in the cytosol of the host cell apparently activates Akt signaling, leading to a suppression of apoptosis (10). Furthermore, T. cruzi trans-sialidase binds to endothelial cells, triggering activation of NF-κB and leading to protection against apoptosis caused by growth factor deprivation (13).
Although T. cruzi trypomastigotes are broadly dispersed among many different organs in the mammalian host, cardiac tissue represents an important target for the parasite and the T. cruzi-cardiomyocyte interaction has been the subject of intense investigation. Several molecules at the cardiomyocyte surface, including carbohydrates, fibronectin, and heparan sulfate proteoglycans, are involved in the parasite-host cell recognition and mediate the invasion process (3, 4, 7, 8, 45). Structural changes in cardiomyocytes observed during intracellular development of the parasite may contribute to a reduction of transmission of the contractile force and lead to alterations in function of the heart in Chagas' disease (1, 37, 38, 39, 46). Finally, T. cruzi-infected cardiomyocytes elicit a strong immune response, characterized by the production of the chemokines RANTES and macrophage inflammatory protein 2 and the cytokines gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α), which triggers a potent nitric oxide-dependent trypanocidal activity (33). As part of this response, infected cardiomyocytes produce interleukin-1β (IL-1β), which has been reported to be associated with the hypertrophic effect observed in these cells (47).
Several recent studies have applied broad-based gene expression analysis in myocardial tissue derived from infected animals to gain insight into the molecular events associated with CCC. Garg et al. (16), in a time course experiment (up to 100 days postinfection), showed that the initial cellular response to T. cruzi invasion is characterized by (i) an upregulation of genes associated with inflammation and interferon-induced immune response; (ii) expression of extracellular matrix proteins (ECMs), suggestive of active reparative and remodeling reactions following injury to the myocardium; and (iii) a generalized depression of mitochondrial function during the progression to chronic disease, indicative of a deficiency of mitochondrial oxidative phosphorylation in T. cruzi-infected murine hearts. Mukherjee et al. (44), using a single time point (100 days postinfection), reported that the infected heart showed symptoms of chronic inflammation, vasculitis, and fibrosis and, as previously reported by Garg et al. (16), an upregulation of atrial natriuretic peptide precursor, a strong indicator of cardiac pathogenesis (57). These authors also found that the ECM genes, especially those associated with fibrosis, were upregulated in the chronic disease model, similar to that observed at day 37 postinfection (16), suggesting different kinetics of infection. Although these studies provided important insights into the molecular events associated with the pathogenesis of CCC at the organ level, the specific cellular response of the cardiomyocytes cannot be separated from the organismal response in these experiments. The global effect of the infection by T. cruzi in cardiomyocytes was characterized for the first time by Goldenberg et al. (20) using a single time point (48 h postinfection). These studies revealed that alterations in cardiac gene expression in Chagas' disease are the consequence of both direct infection of cardiomyocytes and the presence of other cell types in the myocardium.
In the present study, we use microarrays to characterize the global response of murine cardiomyocytes after infection by trypomastigotes in a carefully controlled progression. In contrast to previous reports, our results indicate that T. cruzi induces a broad-based global modulation of genes associated with many pathways and processes in the infected host cell. This global response includes but is not limited to the immune response, inflammation, cell cycle, apoptosis, stress response, and redox homeostasis and disrupts the cytoskeleton and tissue architecture. These effects are generally conducive to the replication and survival of T. cruzi in this hostile intracellular environment. Together, our data provide significant new insight into the early events in the infection of cardiomyocytes that lead to the replication and survival of intracellular T. cruzi after infection by trypomastigotes and the events that may eventually lead to the development of CCC.
MATERIALS AND METHODS
Parasites.
T. cruzi trypomastigotes were obtained from the supernatant of Vero cells infected with the Dm28c clone as previously described (45). Trypomastigotes liberated 96 h after invasion of the Vero cells were collected, washed by centrifugation in serum-free medium, and used in our invasion experiments.
Primary murine cardiomyocyte isolation, culture, and infection assay.
Hearts of 18-day-old embryos of Swiss mice were submitted to mechanical and enzymatic dissociation using 0.05% trypsin and 0.01% collagenase in phosphate-buffered saline at 37°C, following the method previously described (36). The ventricular heart muscle cells were plated at a density of 2.5 × 106 cells in 60-mm sterile plastic petri dishes. Cultures were maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum, 2.5 mM CaCl2, 1 mM l-glutamine, 2% chicken embryo extract, 1,000 U/ml penicillin, and 50 μg/ml streptomycin.
After 24 h, cardiomyocyte monolayers were exposed to culture-derived trypomastigotes at a ratio of 10 parasites per host cell. Free trypomastigotes were removed after 6 h of interaction by washing the cultures with Ringer's solution (154 mM NaCl, 56 mM KCl, 17 mM Na2HPO4, pH 7.0), and fresh medium was added to the culture. The time course of infection was interrupted after 1, 2, 4, 6, 12, 24, and 48 h. Uninfected cardiomyocytes or cardiomyocytes incubated with epimastigotes were used as controls.
RNA extraction.
Total RNA was extracted from uninfected and T. cruzi-infected cardiomyocytes using an RNeasy minikit (Qiagen Inc., Valencia, CA), according to the manufacturer's protocol, and stored in liquid nitrogen until use. RNA was extracted after 1, 2, 4, 6, 12, 24, and 48 h of infection.
Target preparation and microarray hybridization.
Labeling of target RNA and hybridization were performed following the manufacturer's instructions (Affymetrix). Briefly, double-stranded cDNA was synthesized directly from total RNA using a Superscript Choice system (Invitrogen Life Technologies) and the primer 5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG(dT)24-3′. Biotin-labeled cRNA was synthesized with a BioArray high-yield RNA transcript labeling kit (Enzo Biochemicals). After purification on RNeasy columns (Qiagen), 15 μg of fragmented cRNA was hybridized to an MG_U74Av2 array (Affymetrix), and the chips were washed and stained with streptavidin-phycoerythrin on a Fluidics station, as described by the manufacturer. The arrays were scanned at 570 nm at a resolution of 3 μm/pixel, using a GeneArray scanner (Agilent Technologies).
Identification of differentially expressed genes (DEGs) during infection. (i) Image analysis.
Images resulting from scanning of each of the hybridized GeneChips were processed using MAS (version 5.0) software (Affymetrix), and the resulting CEL files were analyzed using the robust multiarray average (RMA) method (27), incorporating the Affymetrix package (17) included in Bioconductor software (18). All hybridizations were analyzed in one batch to maximize their positive correlation.
(ii) Statistical determination of DEGs.
The data resulting from the RMA analysis were submitted to the significance analysis of microarrays method (55) by a two-class unpaired test with 2,000 permutations, using false discovery rate and fold change thresholds of 0.05 and 1.5, respectively, providing us with the list of DEGs.
Verification by real-time PCR.
Genes that were identified to be differentially expressed during the infection in our microarray analysis were verified by SYBR green-based quantitative reverse transcription-PCR (RT-PCR). Specific primers for selected differentially expressed genes (see Table S1 in the supplemental material) were designed using Primer Select (Applied Biosystems). In brief, total RNA from samples was reversed transcribed, and the cDNA synthesized was used in assays performed and quantified using an ABI Prism 7900HT sequence detection system (Applied Biosystems). The results for each gene were normalized to transcripts from the 18S rRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes, which are constitutively expressed during these infections.
Functional classification of DEGs.
Differentially expressed gene lists were classified for gene ontology and function using Ingenuity Pathways Analysis (IPA) software (Ingenuity Systems), which looks for both known canonical pathways and networks of genes. The analyses with IPA software identified the biological functions and/or diseases that were most significant to the data set. Genes from the data set that met the cutoff of 0.05 and that were associated with molecular and cellular functions in the Ingenuity Pathway Knowledge Base were considered for the analysis.
RESULTS
Analysis of the temporal changes in the transcriptome of cardiomyocytes infected with T. cruzi reveals a global modulation of the host cell response.
The gene expression profiles of the infected murine cardiomyocytes were obtained by comparison of RNA from T. cruzi-infected and uninfected cardiomyocytes hybridized to an Affymetrix MG_U74Av2 GeneChip. As a negative control, cardiomyocytes were exposed to noninvasive T. cruzi epimastigotes. Minimal transcriptional changes were observed in these controls (data not shown). The experiment resolved into three distinct empirically defined phases: early (1, 2, 4, 6 h), intermediate (12 h), and late (24, 48 h). These phases loosely represent three distinct biologically relevant stages in the intracellular cycle of the parasite. Thus, microscopic examination of cardiomyocytes exposed to trypomastigotes for 2 h showed that approximately 35% of the mammalian cells were infected, and this remained stable through the 6-h time point (data not shown). However, the number of intracellular amastigotes increased rapidly at approximately 12 h of infection, because, unlike some other strains of T. cruzi, the intracellular cycle of T. cruzi Dm28c is characterized by a rapid transition to the amastigote stage after invasion (data not shown). Thus, our results indicate that, after an initial early largely nonreplicative phase (e.g., 1 to 6 h), trypomastigotes differentiate into amastigotes and replicate profusely. The 6-h time point likely marks the exit of trypomastigotes from the parasitophorous vacuole (PV) and the beginning of the replicative process of amastigotes in the cytoplasm of the cell. Cell lysis begins at later time points (24 to 48 h), loosely marking the third phase of the infection.
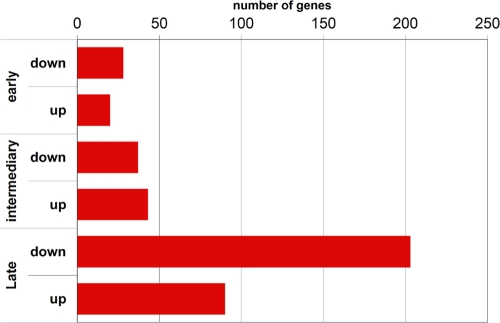
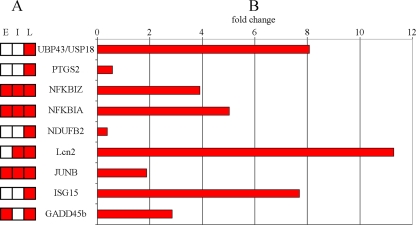
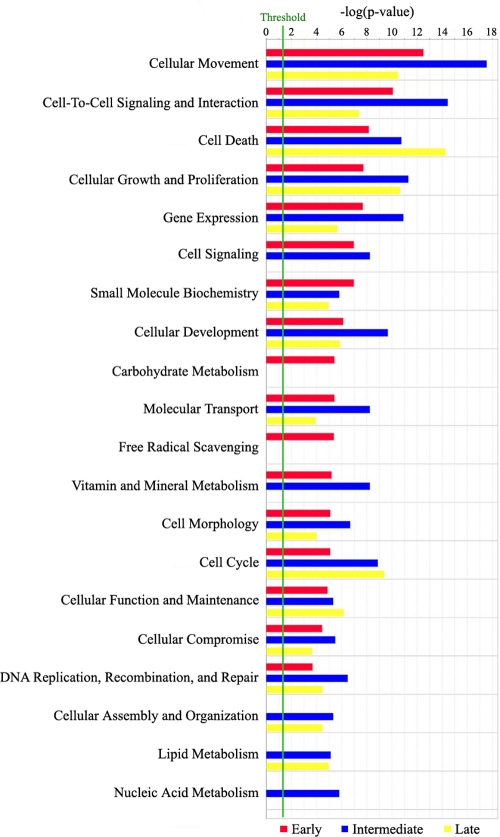
Our analysis of the gene expression profiles at these time points revealed 353 genes that were differentially expressed during at least one of the three different phases of infection (see Table S2 in the supplemental material). Of the 353 DEGs, 111 were upregulated and 242 were downregulated at one or more time points during the experiment. The overall number of DEGs increased with time during the infection, peaking in the late 24- to 48-h stage (Fig. 1). Confirmation of the DEG results was obtained by RT-PCR of a fraction of the genes detected (Fig. 2). Examination of the 353 DEGs showed that a broad range of major functional processes was affected (Fig. 3).
Fig. 1.
Number of DEGs at each time point. Each bar indicates the number of down- or upregulated genes for each time point.
Fig. 2.
Confirmation of microarray results by RT-PCR. Nine DEGs and their expression profiles are shown. The time points at which the differential expression was detected are shown schematically for each gene (A), and the RT-PCR result for each gene is represented as fold change (B). The RT-PCR analyses were performed on RNA extracted from infections in the late time period.
Fig. 3.
Functional analysis of DEGs during the infection of cardiomyocytes with Trypanosoma cruzi. The functional analysis identified the biological functions that were most significant to the data set. Genes from the data set associated with biological functions and/or diseases in the Ingenuity Pathways Knowledge Base (see Materials and Methods) were considered for the analysis. Fisher's exact test was used to calculate a P value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone.
T. cruzi infection induces a broad inflammatory response.
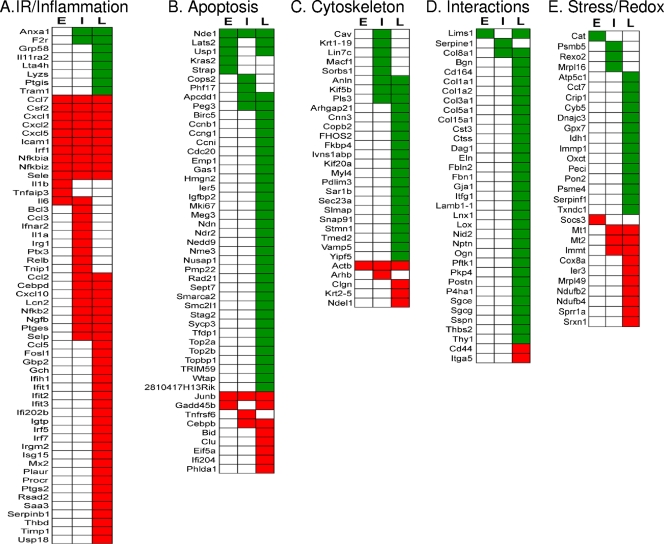
Infection of mammalian cells by T. cruzi has previously been shown to elicit a strong immune response characterized by the production of cytokines, including tumor necrosis factor, gamma interferon, and a potent nitric oxide-dependent trypanocidal activity (33). In our experiments, several immune response- and inflammation-associated genes represented a subset of the most strongly differentially expressed genes across all of the time points of the infection (Fig. 4A). These genes can be generally grouped according to their functional roles, i.e., recruitment of immune cells, interferon signaling, and mediators of inflammation. Among genes associated with recruitment of immune cells, CCL3, CXCL5, NGFB, selectins P and E, CSF-2, and PLAUR, which are key factors for leukocyte recruitment in inflammatory processes (23, 31, 42, 49, 53), were upregulated during the infection (Fig. 4A). Interestingly, the ISG15 gene, a member of the ubiquitin-like protein superfamily of proteins which is strongly induced by viral infection or exposure to type I IFN (11), was also upregulated in the late phase of the infection (Fig. 4A). In addition, the UBP43 gene (also known as USP18), an ISG15-specific protease that belongs to the ubiquitin-specific protease family and that is also strongly activated by IFN and lipopolysaccharide (35), is also upregulated in the late phase of the infection. Furthermore, other genes associated with the production of mediators of inflammation were differentially expressed. For example, PTGES and PTGS2, two genes strongly associated with prostaglandin synthesis, were upregulated in the later stages of the cardiomyocyte infections.
Fig. 4.
Heat map representations of DEGs associated with T. cruzi infection of primary murine cardiomyocytes. (A) DEGs associated with immune response (IR) and inflammation; (B) DEGs associated with cell cycle and apoptosis; (C) DEGs associated with cytoskeleton and intracellular trafficking; (D) DEGs associated with cell-cell and cell-matrix interactions; (E) DEGs associated with stress response and redox homeostasis. DEGs were classified according to their functions and the time point (E, early; I, intermediary; L, late) at which the gene was selected. Green squares, downregulation; red squares, upregulation.
A delicate balance in apoptosis might determine the fate of the infected cardiomyocyte.
Exploitation of apoptotic pathways is a common strategy for survival and/or dissemination used by many intracellular organisms (9). In our infections of T. cruzi in murine cardiomyocytes, we observed upregulation of two classical proapoptotic genes associated with activation of the intrinsic and extrinsic apoptotic pathways; i.e., BID, the BH3-interacting domain death agonist gene, and TNFRSF6 (or FAS), the TNF receptor superfamily gene (Fig. 4B). In addition, the GADD45B gene, a member of the GADD45 family of inducible factors associated with cell cycle control and DNA repair (61), was also upregulated during the infection. GADD45B has been implicated in stress signaling, resulting in either cell cycle arrest, DNA repair, cell survival, or apoptosis (32). Moreover, the PHLDA1 gene (also known as TDAG51), a member of the pleckstrin homology-related domain family, and the interferon-induced helicase IFIH1 (also known as MDA-5), both associated with programmed cell death in human cells (19, 25, 29), were upregulated. In contrast to the proapoptosis pathways that are induced, our data confirm the upregulation of the NF-κB pathway, which plays a pivotal role in modulating the resistance of T. cruzi-infected cardiomyocytes to apoptosis (48) (Fig. 4A). Interestingly, several negative regulators of this pathway, including NF-κBIA, NF-κBIZ, and the TNIP1 gene, are upregulated throughout the infection (Fig. 4A). Other genes associated with cell survival, including those encoding autotaxin and clusterin (CLU), are also upregulated in the late phase of the cardiomyocyte infection (Fig. 4B).
T. cruzi invasion leads to disruption of the host cell cytoskeleton, intracellular trafficking, and tissue architecture.
The cytoskeleton of the host cell has been shown to play an important role during pathogen invasion, establishment, and replication in the cytoplasm (43). We identified a number of genes associated with the host cell cytoskeleton and intracellular trafficking that were differentially expressed primarily during the late phase of the T. cruzi infection of murine cardiomyocytes (Fig. 4C). Thus, several myosin-associated genes (MYL1 and MYL4) and cytoskeleton dynamic and organization-associated genes were downregulated. In addition to the effect of the infection on the cytoskeleton, we observed a downregulation of several genes associated with intracellular trafficking and endocytosis at late time points of the infection. The most affected function is associated with endoplasmic reticulum-Golgi and vesicular trafficking (i.e., the COPB2, SAR1B, SEC23A, TEMD2, and VAMP5 genes) (Fig. 4C).
Disruption of the cytoskeletal architecture is likely to impact interactions between cells, interactions between the cells and the ECM, the integrity of tissue, and intercellular communication. In this context, CCC is characterized by inflammation, infiltration, and myocardial fibrosis to replace the damaged tissue (54). Our analysis revealed that several genes (Fig. 4D) associated with cell-cell and cell-ECM interactions and matrix remodeling were largely downregulated during the infection. Thus, both FBN1 and ITM2A, which are associated with cell-cell and cell-ECM interactions, are downregulated in the late phase of the infection. Unexpectedly, several collagen-associated genes (COL1A1, COL1A2, COL3A1, COL5A1, COL8A1, and COL15A1; collagen is a main component of the basement membrane that is reportedly highly expressed during CCC [2, 8]) are downregulated during the late stages of the infection. In addition, integrin signaling-associated genes were also affected, including LIMS1, NEDD9, CYR61, and ITGA5, which were downregulated. Finally, the interaction between cardiomyocytes and the basement membrane is also compromised in these infections. Thus, the NID2 gene, which is a basement membrane glycoprotein that plays a crucial role in assembly, organization, and integration of the basal membrane (30), is downregulated late in infection.
T. cruzi infection modulates the oxidative and cellular stress response and induces profound changes in the expression of mitochondrion-associated proteins.
T. cruzi invasion and intracellular replication lead to profound molecular perturbations that alter the intracellular homeostasis. It is generally accepted that, in CCC, a sustained production of reactive oxygen species due to the parasite infection coupled with an insufficient antioxidant response leads to long-term oxidative stress in the heart. Thus, the severity of chagasic cardiomyopathy is associated with injury due to increased oxidative stress (22). Our analysis indicated that several genes classically associated with the oxidative stress response, CAT, GPX7, PON2, and TXNDC1, are downregulated early in the infection (Fig. 4E). In addition, two genes of the proteasome-ubiquitin pathway (UBE2E1 and PSMB5), which is associated with the response to cellular stress, are also downregulated. In addition to the above genes associated with oxidative stress, we identified several other genes related to mitochondrial function that were differentially regulated at different stages of the T. cruzi infection (Fig. 4E). Seven genes associated with metabolic processes performed by the mitochondria were downregulated, suggesting an early impairment of critical metabolic functions associated with this organelle in infected cardiomyocytes.
DISCUSSION
In this study, we characterized the temporal changes in the transcriptome of primary murine cardiomyocytes infected with T. cruzi. Our results, which reveal a global cellular response to T. cruzi infection, are in contrast to previous reports that suggest that infection and invasion of mammalian organs and cell lines by T. cruzi induce only a very limited modulation of the host cell transcriptome (16, 26, 44, 50, 56). In fact, our results demonstrate for the first time an intense modulation of gene expression beginning at the early stages of infection (Fig. 3). This is likely the result of the initial interaction of the parasite with cellular receptors that lead to the activation of several signaling cascades as well as the presence of the parasite inside the cell. In addition, our data confirm observations of Goldenberg et al. (20), related to the late stages of cardiomyocyte infection, indicating that these broad gene expression changes are a direct consequence of the invasion of the cardiomyocytes by the parasite. We believe that the differences between our observations and those of the previous studies are due to the challenges of identifying subtle changes in gene expression in whole-animal models, use of different mammalian cell targets, use of different T. cruzi strains, or other differences in experimental design. In fact, clone Dm28c, used in this study, is a TCI lineage isolate, whereas the strains Y and Brazil, which were used in the previous experiments cited, are TCII lineage isolates. Although TCI and TCII isolates cause similar diseases, the diseases that they cause exhibit many biological, physiological, and pathogenic differences, including different host cell responses (14), and they have different mechanisms of invasion (23, 58).
Inflammation is one of the signature events during chagasic cardiomyopathy. Our data are largely consistent with what is known about the inflammatory processes exhibited by experimental models of CCC. However, our work significantly extends these previous observations and provides an integrated view of the genes and proteins involved. Thus, genes associated with recruitment of immune cells, the IFN response, and specific mediators of inflammation are strongly upregulated during the infections. Interestingly, chemokines not previously associated with the response to T. cruzi infection were also identified in our study. Thus, chemokine CXCL5, which is induced by IL-1 or TNF-α and is generally associated with recruitment of immune cells, is upregulated early during the infection. Since it was previously reported that T. cruzi-infected cardiomyocytes secrete IL-1 (47), it is possible that these chemokines (CXCL10, CCL3, and CXCL5) play a protective role in the infected tissue and are responsible for early induction of the inflammatory processes associated with infection of cardiac tissue. In addition, nerve growth factor beta (NGFB), a neurotrophic factor that is responsible for survival of specific populations of neurons in the peripheral and central nervous system, is also upregulated during these stages of the infection. Recent work suggests that NGFB is associated with inflammation by inducing the influx of inflammatory cells; i.e., it is chemotactic for leukocytes and elevated levels are observed at sites of inflammation (49). NGFB is also considered to be a stress response gene that, when it is upregulated, leads to the expression of intercellular adhesion molecule 1 (ICAM-1), an adhesion molecule previously implicated in recruitment after T. cruzi infection (41). The upregulation of ISG15, a protein associated with ISGylation (52), and UBP43 suggests an involvement of protein ISGylation in the regulation of the JAK-STAT pathway (34) and opens the question of the role of JAK-STAT during the infection. These observations clearly show that many IFN pathway-related genes that were previously not known to be associated with T. cruzi infection are strongly induced in these T. cruzi-infected cardiomyocytes.
The apoptotic response of the T. cruzi-infected host cell, including cardiomyocytes, remains controversial, and the differences observed in the literature may suggest that the degree of the response varies with the host cell type and the parasite strain (9). Some reports suggest that infected cardiomyocytes undergo apoptosis in a parasite strain-dependent fashion (12), whereas others suggest that activation of NF-κB prevents apoptosis in these infected cells (48). Again, it is important to note that these studies examined strains from different lineages of T. cruzi. In the present study, we used T. cruzi Dm28c, an important and well-characterized member of the TCI major lineage. Our analysis identified several genes associated with either apoptosis or survival of the infected cells that were significantly affected during the infection (Fig. 4B). These apparently contradictory observations suggest a delicate balance between these cellular states. Thus, to help to clarify previous observations of apoptosis in cardiomyocytes infected with the Dm28c clone (12) and the activation of a series of survival genes reported here, a detailed temporal analysis of the change of the genes identified here and by others could provide more definitive answers about the fate of infected cardiomyocytes.
Our study provides further evidence on the extent of the disruption of the host cell cytoskeleton, including alterations in genes associated with the organization and dynamics of the host cytoskeleton during infection. These observations could provide further understanding of the molecular basis for the disturbances in the contractile force of the heart muscle that is observed in CCC. Our observations are also in agreement with previous suggestions that disarray of the cytoskeleton in T. cruzi-infected cells is a necessity for and/or a consequence of the establishment and subsequent replication of the parasite in the cytoplasm. For example, it was recently demonstrated that the alterations in the cytoskeletal organization of infected cardiomyocytes could be reverted by treatment with trypanocidal drugs (51). Our data identified dramatic changes in the expression of genes associated with the tissue architecture, providing the molecular bases for the disruption observed in CCC. Interestingly, several genes associated with remodeling of the extracellular matrix are modulated during the infection. Thus, cathepsin S (CTSS), a papain-like cysteine protease that has been implicated in tissue destruction caused by lung disease and that may have a role in the response to oxidative stress (59), is consistently downregulated in response to T. cruzi invasion (Fig. 4D). In addition, the neutrophil serine protease inhibitor SERPINB1A is upregulated later in the infection (Fig. 4D). Interestingly, SERPINB1A has been associated with maintenance of lung defense functions in Pseudomonas infections by disabling critical components of the host response to bacterial infection (5). Moreover, TIMP1, an inhibitor of the metalloproteinase that is responsible for the degradation of the ECM (60), is upregulated late in the infection (Fig. 4D). These observations may be interpreted in the context of an effort by the parasite to delay the arrival of immune cells to the site of the infection or as a host cell response intended to limit the large inflammatory response observed in CCC.
The changes in the expression profiles of several stress-related genes (Fig. 4E) provide further molecular explanation for the association of oxidative stress-related injury with CCC and provide novel insight into the role of the ubiquitin-dependent catabolism pathway in the pathology of CCC. Thus, the parasite infection both induces the oxidative response and suppresses the ability of the cells to modulate and control it, thus leading to survival of the parasite and to a permanent damage of the heart by reactive species.
Interestingly, complex I-associated genes NDUFB2 and NDUFB4, together with CYP1B1, were upregulated during the infection. These results are in contrast to previous reports that showed a downregulation of OXPHOS complexes; i.e., complex I (NADH-ubiquinone oxidoreductase) and complex IV (cytochrome c oxidase) were reported to be dramatically decreased in the myocardium in late stages of CCC in an in vivo murine model (16). This observation suggests that at early stages of infection, represented by the work described herein, there may be an attempt by the cell to compensate for the loss of mitochondrial function. This hypothesis is supported by the upregulation of IMMT, a gene that is essential for the normal mitochondrial function (28). Later in the infection, represented by the previous studies (22), the cell may no longer be able to sustain a compensatory effort, thus explaining the apparently conflicting results.
Summary and conclusions.
We have examined the gene expression profile of primary cardiomyocytes after infection with T. cruzi trypomastigotes using murine microarrays. In contrast to previous reports, which indicate a modest modulation of gene expression in response to T. cruzi infection, our results suggest a very broad-based modulation of gene expression. At least 353 genes in many relevant pathways and networks are apparently differentially expressed in these infected cell lines. The difference between our results and those of previous investigators can be attributed to the different infected mammalian cell lines used, to the use of whole-animal models and different parasite strains, or to the experimental design. Our results do clearly reflect three specific stages in the process of cellular invasion by the parasite (entry into the cell, differentiation of trypomastigotes into amastigotes in the parasitophorous vacuole, followed by escape of the latter to the cell cytoplasm and rapid multiplication, and finally, cellular lysis). Moreover, we have clearly identified differentially expressed genes associated with many relevant cellular processes, including but not limited to inflammation and cytokine production, apoptosis, disruption of the cellular cytoskeleton, and the oxidative and cellular stress response, confirming a broad-based global cellular response to invasion. Finally, our observations are consistent with the hypothesis that the parasite is exerting control over the mammalian cell processes, e.g., apoptosis, and that this control is optimizing conditions for the parasite to replicate and persist in its infection. Further study will be required to confirm and extend these hypotheses.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI050196 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
Microarray analysis, DNA synthesis, and RT-PCR were performed in the Nucleic Acids Research Facilities at Virginia Commonwealth University.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 22 February 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Adesse D., et al. 2008. Trypanosoma cruzi induces changes in cardiac connexin43 expression. Microbes Infect. 10:21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrade S. G., Grimaud J. A., Stocker-Guerret S. 1989. Sequential changes of the connective matrix components of the myocardium (fibronectin and laminin) and evolution of cardiac fibrosis in mice infected with Trypanosoma cruzi. Am. J. Trop. Med. Hyg. 40:252–260 [DOI] [PubMed] [Google Scholar]
- 3. Barbosa H. S., Meirelles M. N. 1993. The role of RCA-binding sites in the adhesion of Trypanosoma cruzi to heart muscle cells, as revealed by electron spectroscopic imaging. J. Submicrosc. Cytol. Pathol. 25:47–51 [PubMed] [Google Scholar]
- 4. Barbosa H. S., Meirelles M. N. 1992. Ultrastructural detection in vitro of WGA-, RCA I-, and ConA-binding sites involved in the invasion of heart muscle cells by Trypanosoma cruzi. Parasitol. Res. 78:404–409 [DOI] [PubMed] [Google Scholar]
- 5. Benarafa C., Priebe G. P., Remold-O'Donnell E. 2007. The neutrophil serine protease inhibitor serpinb1 preserves lung defense functions in Pseudomonas aeruginosa infection. J. Exp. Med. 204:1901–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burleigh B. A., Woolsey A. M. 2002. Cell signalling and Trypanosoma cruzi invasion. Cell. Microbiol. 4:701–711 [DOI] [PubMed] [Google Scholar]
- 7. Calvet C. M., Toma L., De Souza F. R., Meirelles M. N., Pereira M. C. 2003. Heparan sulfate proteoglycans mediate the invasion of cardiomyocytes by Trypanosoma cruzi. J. Eukaryot. Microbiol. 50:97–103 [DOI] [PubMed] [Google Scholar]
- 8. Calvet C. M., Meuser M., Almeida D., Meirelles M. N., Pereira M. C. 2004. Trypanosoma cruzi-cardiomyocyte interaction: role of fibronectin in the recognition process and extracellular matrix expression in vitro and in vivo. Exp. Parasitol. 107:20–30 [DOI] [PubMed] [Google Scholar]
- 9. Carmen J. C., Sinai A. P. 2007. Suicide prevention: disruption of apoptotic pathways by protozoan parasites. Mol. Microbiol. 64:904–916 [DOI] [PubMed] [Google Scholar]
- 10. Chuenkova M. V., Perrin M. P. 2008. Trypanosoma cruzi targets AKt in host cells as an intracellular antiapoptotic strategy. Sci. Signal 17:97ra74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. D'Cunha J., Knight E., Jr., Haas A. L., Truitt R. L., Borden E. C. 1996. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc. Natl. Acad. Sci. U. S. A. 93:211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Souza E. M., et al. 2003. Host and parasite apoptosis following Trypanosoma cruzi infection in in vitro and in vivo models. Cell Tissue Res. 314:223–235 [DOI] [PubMed] [Google Scholar]
- 13. Dias W. B., et al. 2008. Endothelial cell signalling induced by trans-sialidase from Trypanosoma cruzi. Cell. Microbiol. 10:88–99 [DOI] [PubMed] [Google Scholar]
- 14. Fernandes O., et al. 1999. Populational heterogeneity of Brazilian Trypanosoma cruzi isolates revealed by the mini-exon and ribosomal spacers. Mem. Inst. Oswaldo Cruz 94(Suppl. 1):195–197 [DOI] [PubMed] [Google Scholar]
- 15. Ferreira D., Cortez M., Atayde V. D., Yoshida N. 2006. Actin cytoskeleton-dependent and -independent host cell invasion by Trypanosoma cruzi is mediated by distinct parasite surface molecules. Infect. Immun. 74:5522–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garg N., Popov V. L., Papaconstantinou J. 2003. Profiling gene transcription reveals a deficiency of mitochondrial oxidative phosphorylation in Trypanosoma cruzi-infected murine hearts: implications in chagasic myocarditis development. Biochim. Biophys. Acta 1638:106–120 [DOI] [PubMed] [Google Scholar]
- 17. Gautier L., Cope L., Bolstad B. M., Irizarry R. A. 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315 [DOI] [PubMed] [Google Scholar]
- 18. Gentleman R. C., et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gitlin L., et al. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldenberg R. C., et al. 2009. Transcriptomic alterations in Trypanosoma cruzi-infected cardiac myocytes. Microbes Infect. 11:1140–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gruenheid S., Finlay B. B. 2003. Microbial pathogenesis and cytoskeletal function. Nature 422:775–781 [DOI] [PubMed] [Google Scholar]
- 22. Gupta S., Wen J. J., Garg N. J. 2009. Oxidative stress in Chagas disease. Interdiscip. Perspect. Infect. Dis. 2009:190354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamilton N. H., Mahalingam S., Banyer J. L., Ramshaw I. A., Thomson S. A. 2004. A recombinant vaccinia virus encoding the interferon-inducible T-cell alpha chemoattractant is attenuated in vivo. Scand. J. Immunol. 59:246–254 [DOI] [PubMed] [Google Scholar]
- 24. Higuchi M. L., Benvenuti L. A., Martins Reis M., Metzger M. 2003. Pathophysiology of the heart in Chagas' disease: current status and new developments. Cardiovasc. Res. 60:96–107 [DOI] [PubMed] [Google Scholar]
- 25. Hossain G. S., et al. 2003. TDAG51 is induced by homocysteine, promotes detachment-mediated programmed cell death, and contributes to the development of atherosclerosis in hyperhomocysteinemia. J. Biol. Chem. 278:30317–30327 [DOI] [PubMed] [Google Scholar]
- 26. Imai K., Mimori T., Kawai M., Koga H. 2005. Microarray analysis of host gene-expression during intracellular nests formation of Trypanosoma cruzi amastigotes. Microbiol. Immunol. 49:623–631 [DOI] [PubMed] [Google Scholar]
- 27. Irizarry R. A., et al. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 28. John G. B., et al. 2005. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16:1543–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang D. C., et al. 2004. Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene 23:1789–1800 [DOI] [PubMed] [Google Scholar]
- 30. Kohfeldt E., Sasaki T., Gohring W., Timpl R. 1998. Nidogen-2: a new basement membrane protein with diverse binding properties. J. Mol. Biol. 282:99–109 [DOI] [PubMed] [Google Scholar]
- 31. Langer H. F., Chavakis T. 2009. Leukocyte-endothelial interactions in inflammation. J. Cell. Mol. Med. 13:1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liebermann D. A., Hoffman B. 2007. Gadd45 in the response of hematopoietic cells to genotoxic stress. Blood Cells Mol. Dis. 39:329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Machado F. S., et al. 2000. Trypanosoma cruzi-infected cardiomyocytes produce chemokines and cytokines that trigger potent nitric oxide-dependent trypanocidal activity. Circulation 102:3003–3008 [DOI] [PubMed] [Google Scholar]
- 34. Malakhova O. A., et al. 2003. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 17:455–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Malakhova O., Malakhov M., Hetherington C., Zhang D. E. 2002. Lipopolysaccharide activates the expression of ISG15-specific protease UBP43 via interferon regulatory factor 3. J. Biol. Chem. 277:14703–14711 [DOI] [PubMed] [Google Scholar]
- 36. Meirelles M. N., de Araujo-Jorge T. C., Miranda C. F., de Souza W., Barbosa H. S. 1986. Interaction of Trypanosoma cruzi with heart muscle cells: ultrastructural and cytochemical analysis of endocytic vacuole formation and effect upon myogenesis in vitro. Eur. J. Cell Biol. 41:198–206 [PubMed] [Google Scholar]
- 37. Melo T. G., Almeida D. S., Meirelles M. N., Pereira M. C. 2006. Disarray of sarcomeric alpha-actinin in cardiomyocytes infected by Trypanosoma cruzi. Parasitology 133:171–178 [DOI] [PubMed] [Google Scholar]
- 38. Melo T. G., Almeida D. S., Meirelles M. N., Pereira M. C. 2004. Trypanosoma cruzi infection disrupts vinculin costameres in cardiomyocytes. Eur. J. Cell Biol. 83:531–540 [DOI] [PubMed] [Google Scholar]
- 39. Melo T. G., Meirelles M. N., Pereira M. C. 2008. Trypanosoma cruzi alters adherens junctions in cardiomyocytes. Microbes Infect. 10:1405–1410 [DOI] [PubMed] [Google Scholar]
- 40. Melo-Jorge M., Perrin M. P. 2007. The Chagas' disease parasite Trypanosoma cruzi exploits nerve growth factor receptor Trka to infect mammalian hosts. Cell Host Microbe 1:251–261 [DOI] [PubMed] [Google Scholar]
- 41. Michailowsky V., et al. 2004. Intercellular adhesion molecule 1 deficiency leads to impaired recruitment of T lymphocytes and enhanced host susceptibility to infection with Trypanosoma cruzi. J. Immunol. 173:463–470 [DOI] [PubMed] [Google Scholar]
- 42. Mondino A., Blasi F. 2004. uPA and uPAR in fibrinolysis, immunity and pathology. Trends Immunol. 25:450–455 [DOI] [PubMed] [Google Scholar]
- 43. Mott A., Lenormand G., Costales J., Fredberg J. J., Burleigh B. A. 2009. Modulation of host cell mechanics by Trypanosoma cruzi. J. Cell. Physiol. 218:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mukherjee S., et al. 2003. Microarray analysis of changes in gene expression in a murine model of chronic chagasic cardiomyopathy. Parasitol. Res. 91:187–196 [DOI] [PubMed] [Google Scholar]
- 45. Oliveira F. O., Jr., et al. 2008. Trypanosoma cruzi heparin-binding proteins and the nature of the host cell heparan sulfate-binding domain. Microb. Pathog. 44:329–338 [DOI] [PubMed] [Google Scholar]
- 46. Pereira M. C., Costa M., Chagas Filho C., Meirelles M. N. 1993. Myofibrillar breakdown and cytoskeletal alterations in heart muscle cells during invasion by Trypanosoma cruzi: immunological and ultrastructural study. J. Submicrosc. Cytol. Pathol. 25:559–569 [PubMed] [Google Scholar]
- 47. Petersen C. A., Burleigh B. A. 2003. Role for interleukin-1 beta in Trypanosoma cruzi-induced cardiomyocyte hypertrophy. Infect. Immun. 71:4441–4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petersen C. A., Krumholz K. A., Carmen J., Sinai A. P., Burleigh B. A. 2006. Trypanosoma cruzi infection and nuclear factor kappa B activation prevent apoptosis in cardiac cells. Infect. Immun. 74:1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raychaudhuri S. K., Raychaudhuri S. P., Weltman H., Farber E. M. 2001. Effect of nerve growth factor on endothelial cell biology: proliferation and adherence molecule expression on human dermal microvascular endothelial cells. Arch. Dermatol. Res. 293:291–295 [DOI] [PubMed] [Google Scholar]
- 50. Shigihara T., Hashimoto M., Shindo N., Aoki T. 2008. Transcriptome profile of Trypanosoma cruzi-infected cells: simultaneous up- and down-regulation of proliferation inhibitors and promoters. Parasitol. Res. 102:715–722 [DOI] [PubMed] [Google Scholar]
- 51. Silva D. T., Meirelles M. N. S. L., Almeida D., Urbina J. A., Pereira M. C. 2006. Cytoskeleton reassembly in cardiomyocytes infected by Trypanosoma cruzi is triggered by treatment with ergosterol biosynthesis inhibitors. Int. J. Antimicrob. Agents 27:530–537 [DOI] [PubMed] [Google Scholar]
- 52. Staub O. 2004. Ubiquitylation and isgylation: overlapping enzymatic cascades do the job. Sci. STKE 2004:pe43. [DOI] [PubMed] [Google Scholar]
- 53. Talvani A., et al. 2000. Kinetics of cytokine gene expression in experimental chagasic cardiomyopathy: tissue parasitism and endogenous IFN-gamma as important determinants of chemokine mRNA expression during infection with Trypanosoma cruzi. Microbes Infect. 2:851–866 [DOI] [PubMed] [Google Scholar]
- 54. Tanowitz H. B., et al. 2009. Perspectives on Trypanosoma cruzi-induced heart disease (Chagas disease). Prog. Cardiovasc. Dis. 51:524–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tusher V. G., Tibshirani R., Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vaena de Avalos S., Blader I. J., Fisher M., Boothroyd J. C., Burleigh B. A. 2002. Immediate/early response to Trypanosoma cruzi infection involves minimal modulation of host cell transcription. J. Biol. Chem. 277:639–644 [DOI] [PubMed] [Google Scholar]
- 57. Vikstrom K. L., Bohlmeyer T., Factor S. M., Leinwand L. A. 1998. Hypertrophy, pathology, and molecular markers of cardiac pathogenesis. Circ. Res. 82:773–778 [DOI] [PubMed] [Google Scholar]
- 58. WHO 2002. The world health report, 2002. WHO, Geneva, Switzerland [Google Scholar]
- 59. Williams A. S., et al. 2009. Role of cathepsin S in ozone-induced airway hyperresponsiveness and inflammation. Pulm. Pharmacol. Ther. 22:27–32 [DOI] [PubMed] [Google Scholar]
- 60. Woessner J. F., Jr 2002. MMPs and TIMPs—an historical perspective. Mol. Biotechnol. 22:33–49 [DOI] [PubMed] [Google Scholar]
- 61. Zhan Q., et al. 1999. Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene 18:2892–2900 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.