Abstract
The RNA-binding protein Hfq is recognized as an important regulatory factor in a variety of cellular processes, including stress resistance and pathogenesis. Hfq has been shown in several bacteria to interact with small regulatory RNAs and act as a post-transcriptional regulator of mRNA stability and translation. Here we examined the impact of Hfq on growth, stress tolerance, and gene expression in the intracellular pathogen Francisella novicida. We present evidence of Hfq involvement in the ability of F. novicida to tolerate several cellular stresses, including heat-shock and oxidative stresses, and alterations in hfq gene expression under these conditions. Furthermore, expression of numerous genes, including several associated with virulence, is altered in a hfq mutant strain suggesting they are regulated directly or indirectly by Hfq. Strikingly, we observed a delayed entry into stationary phase and increased biofilm formation in the hfq mutant. Together, these data demonstrate a critical role for Hfq in F. novicida growth and survival.
Introduction
Francisella tularensis is a gram-negative facultative intracellular pathogen and the etiologic agent of tularemia. Inhalation of as few as 10 organisms can result in development of pneumonic tularemia, a form of the disease associated with the highest morbidity and mortality. Due to its high infectivity and relative ease of dissemination, F. tularensis has been designated a category A select agent by the Centers for Disease Control and Prevention [1]. In humans, the most common form of tularemia is an ulceroglandular disease resulting from the bite of a bacteria-carrying arthropod vector or exposure to the infected blood of an animal through an abrasion in the skin [2]. Numerous arthropod vectors have been implicated in the transmission of tularemia and a wide range of animal hosts, including small mammals, birds, and humans, are susceptible to infection by F. tularensis [3]. Besides possessing a wide range of hosts, the pathogen is also quite resilient to environmental conditions and has been found to survive in mud or water for over a year [4].
Several studies have shown that F. tularensis pathogenicity is dependent on its ability to persist and replicate inside host cells, especially macrophages [1], [5]. Several of the genes required for virulence are located within the Francisella pathogenicity island (FPI), though additional genes are likely to be required [6]. A number of genes in the FPI, such as pdpA and pdpB, have been shown to be required for intracellular bacterial growth and survival [7], [8]. Genes outside the FPI have also been implicated in aspects of F. tularensis virulence and dissemination through the utilization of genome-wide screens [8], [9]. While a limited number of F. tularensis regulators are known, those shown to be involved in virulence include MglA, SspA, FevR, and RelA [10], [11], [12]. Recently, the F. tularensis subspecies holarctica strain LVS Hfq protein was reported to play a role in virulence, possibly by negatively regulating the pdp operon of the FPI [13].
The RNA-binding protein Hfq is an important regulator of gene expression in a number of bacterial pathogens including Yersinia pseudotuberculosis, Neisseria meningitidis, and Salmonella enterica [14], [15], [16]. The Hfq protein was first identified as a host factor required for synthesis of bacteriophage Qβ RNA in Escherichia coli and belongs to the Sm and Sm-like family of proteins found in both prokaryotes and eukaryotes [17], [18]. Hfq is present in many bacterial species and hfq mutants often exhibit pleiotropic phenotypes including defects in virulence, quorum sensing, growth rate, and stress tolerance [19], [20], [21], [22], [23].
Recent studies suggest that Hfq acts primarily as a post-transcriptional regulator that facilitates RNA-RNA interactions between small noncoding RNAs (sRNAs) and their mRNA targets [18], [24]. Small RNAs are a novel group of gene regulators that typically act by base pairing with target mRNAs resulting in either the up- or down-regulation of protein synthesis. These sRNAs are often encoded in trans and commonly rely on the Hfq protein for mediating the sRNA-mRNA interaction [25].
Because the genomes of Francisella species encode only one alternative sigma factor, σ32, and one complete two-component regulatory system [26], [27], the Hfq protein of the mouse pathogen F. novicida was targeted in an effort to discover new regulatory mechanisms employed by Francisella for stress survival. F. novicida is closely related to F. tularensis but is unable to cause disease in healthy individuals, though it does cause a disease in mice that is very similar to human tularemia [7]. As such, it provides an alternative model system for tularemia work. Here we report the characterization of two separate F. novicida hfq transposon insertion mutants, a complete hfq deletion mutant, and the heterologous expression of the F. novicida Hfq in an E. coli hfq mutant. We demonstrate that the N-terminal half of the protein is most critical to heat stress tolerance and that the entire protein is unable to complement the phenotype of an E. coli hfq mutant. Not only does Hfq play a role in the response to salt and heat shock, as reported for F. tularensis subspecies holarctica [13], it also plays a role in pH tolerance, peroxide stressors, entry into stationary phase, and biofilm formation in F. novicida. We also report the expression profile of a subset of genes previously identified by Meibom and coworkers to be regulated by Hfq in F. tularensis subspecies holarctica [13]. Thus Hfq and likely sRNAs appear to play a role in regulating global gene expression, stress resistance, and formation of biofilms in F. novicida.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1. F. novicida strain U112 and all its derivatives were routinely grown at 37°C in Tryptic Soy (TS) broth (Difco) supplemented with 0.1% L-cysteine or on TS agar plates supplemented with 0.1% cysteine, unless otherwise noted. E. coli strains were grown in Luria-Bertani (LB) medium or on LB agar plates. When required, media was supplemented with chloramphenicol (50 µg/ml), erythromycin (250 µg/ml), kanamycin (50 µg/ml), or tetracycline (10 µg/ml).
Table 1. Bacterial strains and plasmids used in this study.
| Strains | Genotype or description | Source or reference |
| F. novicida | ||
| U112 | Wild-type F. tularensis subsp. novicida strain U112 | [56] |
| U112 Δhfq | U112 Δhfq, EmR | This study |
| U112 Tnhfq1 | U112 hfq :: EZ-Tn5 <KAN-2> at ORF position 229 of 327 bp | [37] |
| U112 Tnhfq2 | U112 hfq :: EZ-Tn5 <KAN-2> at ORF position 133 of 327 bp | [37] |
| E. coli | ||
| MC4100 | araD139 Δ(argF-lac)205 flb-5301 pstF25 rpsL150 deoC1 relA1 | [24] |
| GS081 | MC4100 hfq-1 :: Ω(CmR) | [24] |
| Plasmids | ||
| pKK214gfp | Low-copy-number expression vector with groEL promoter of F. tularensis LVS, Δcat::gfp | [30] |
| pKK214hfq | hfq ORF and 191 bp-upstream region inserted into pKK214gfp | This study |
| pACYC177 | Low-copy-number cloning vector, AmpR, KmR | New England Biolabs |
| pACYCMchfq | MC4100 hfq ORF and 163 bp upstream region inserted into pACYC177 | This study |
| pACYCFnhfq | U112 hfq ORF and MC4100 hfq promoter region region inserted into pACYC177 | This study |
| pIDN4 | Source of erythromycin gene cassette | [29] |
| pMAL-c4X | Vector for protein purification containing maltose binding protein | New England Biolabs |
Strain construction
To construct a non-polar F. novicida hfq deletion mutant, a 3,033 bp DNA cassette containing an ermC ORF flanked by 1,170 bp upstream and 1,128 bp downstream of the hfq (FTN_1051) ORF was constructed via fusion of three separate PCR products [28]. Primers for the cassette contained overlapping sequences and are listed in Table 2. The following PCR reaction mixture was used to amplify the upstream (primers FnHfqKO-upF/FnHfqKO-upR) and downstream (primers FnHfqKO-downF/FnHfqKO-downR) regions flanking the hfq ORF from F. novicida as well as the 735 bp ermC ORF from pIDN4 [29] (primers FnHfqKO-ermF/FnHfqKO-ermR): 1× PfuUltra™ II Buffer (Stratagene), 0.25 mM each dNTP, 2.5 pmol each primer, 1 µl PfuUltra™ II Fusion HS DNA polymerase (Stratagene), and 1 µl template DNA in a total volume of 50 µl. Reactions were cycled according to the following program: 95°C denaturation for 2 min; followed by 6 cycles consisting of 95°C for 30 sec, 45°C for 30 sec, and 72°C for 70 sec; then 25 cycles consisting of 95°C for 30 sec, 54°C for 30 sec, and 72°C for 70 sec; and ending with a 6 min extension at 72°C. The resulting PCR products were gel purified using the QiaQuick Gel Extraction kit (Qiagen).
Table 2. Primers used in this study.
| Primer name | Primer sequence (5′ - 3′) |
| FN_hfqProm-F-SmaI | TTTCCCGGGATGGAGGACGAAACTAAAGAGTTAGATAT |
| FN-hfq.orf-EcoRI-R | CGGAATTCTTACTCGTGAATATTACCTTCATTC |
| FN_hfq-R-PstI | GCAATCTGCAGTTACTCGTGAATATTA |
| FN_hfq-22F | CAAGACCCGTTCTTAAATGC |
| FN_hfq-326R | TCGTGAATATTACCTTCATTCTC |
| FN_hfq-151R | AGCTGGAACTATAGTAGAAATAGCATGT |
| FN-hfq-ATG-F | ATGTCAAGAATATCATCTTTACAAGACC |
| FN-hfq-330R-PstI | GCAATCTGCAGTTACTCGTGAATATTA |
| FnHfqKO-ermF | TAGATTCGAGGTACGGCTACAGTCTTTTGGCTAACACACACGCCATTC |
| FnHfqKO-ermR | CGGTCTATGAACTTAGTGAGCGGATTAGTTTATGCATCCCTTAACTTACTTA |
| FnHfqKO-upF | CTATGGTTTGGCTGGACCAACAGCTTC |
| FnHfqKO-upR | AAGACTGTAGCCGTACCTCGAATCTATGTCTCACTTCCTTTTAA |
| FnHfqKO-downF | AATCCGCTCACTAAGTTCATAGACCGAGTTCCATTGTGGAGTAATATTAG |
| FnHfqKO-downR | TCCTCAAGAGGCACGAAACTTGGC |
| FN_hflX-373F | AAACTTCAGGTTGAGTTGGCGCAG |
| FN_hflX-1013R | TCAAGAGGCACGAAACTTGGCTTG |
| FN_hflX-501R | TATCTCAAGCTGTGTCTCACCAGGTC |
| FN_miaA-25F | GCTGGACCAACAGCTTCAGGTAAA |
| FN_miaA-274F | GGAAGAGAAGTCTTACTTGTTGGTGGG |
| FN_miaA-833F | CATGGATTCGTAATTGGCAGAG |
| Q_FN-bioD-111F | ACCTGTAGCGTCTGGACAAAGTCA |
| Q_FN-bioD-241R | GAGGAGCAACTGCTTGATTGAATG |
| Q_FN-dnaG-859F | GCTGCAGTAGCTACATTAGGCACA |
| Q_FN-dnaG-966R | TTGCCCTGCTTCATCACCATCA |
| Q_Fn_hfq-84F | CGGGATCAAACTACAAGGTCAAGT |
| Q_Fn_hfq-195R | AGCTGGAACTATAGTAGAAATAGCATGT |
| Q_FN-katG-798F | AACTCATGGTGCAGTTCCAGAGGA |
| Q_FN-katG-895R | TATTGTGCCAGCCTAGACCTTGCT |
| Q_FN_mglA-156F | ACCTACGCTTAGCACAGATGA |
| Q_FN_mglA-249R | TGGAAACATCGGAGGAAAGGGA |
| Q_FN-pdpA-692F | ACGGCATAAACGGCTGGTTAACTT |
| Q_FN-pdpA-787R | CAAGCAATATGGGTTGATTTGGGC |
| Q_FN-pdpB-2681F | ACTCGGCTGCAACAAATGAAGC |
| Q_FN-pdpB-2767R | GTGGAGATAGCTGCTCTATAAATCCAGAGT |
| Q_FN-pyrF-485F | ATGTCCCAGGTGTAAGGCTTGAGA |
| Q_FN-pyrF-604R | CGCGTAATAGCGGCCTACCTACAATA |
| Q_FN-relA-1042F | ACAGTTGTCAAAGTTGGCGAGCAG |
| Q_FN-relA-1148R | CCTTCTTTATAACGCCAATGTGCCGC |
| Q_FN-uvrD-140F | GTCGTGATAAAGGCGTATCTGTGG |
| Q_FN-uvrD-226R | TTTCTACACGCTGCTGGATCTC |
| Q_FN-yhbG-183F | AGTACGCATGGGTCAAGAGGATGT |
| Q_FN-yhbG-273R | AACCGAAGCCTCCTGAGGCAAATA |
| Ec_hfq-R-PstI | GCAATCTGCAGTTATTCGGTTTCTTCGCTG |
| Ec_hfq-up-F-BamHI | CTTGGATCCTTCACTGGCTTGACAGTGAAAAACCAGAAC |
| EcFnHfq-FusF | GCATATAAGGAAAAGAGAGAATGTCAAGAATATCATCTTT |
| EcHfqProm-FusR | AAAGATGATATTCTTGACATTCTCTCTTTTCCTTATATGC |
For the initial fusion reaction, 100 ng of each purified product was added to a PCR mixture as described above except no primers were added. The products themselves were used as primers. This initial fusion mixture was cycled according to the following program: 95°C denaturation for 15 sec; followed by 5 cycles consisting of 95°C for 15 sec, 55°C for 60 sec, and 72°C for 210 sec. The final fusion reaction contained 20 µl of the initial fusion product, 2.5 pmol each of primers FnHfqKO-upF and FnHfqKO-downR, 1× PfuUltra™ II Polymerase Buffer, 0.25 mM each dNTP, and 1 µl PfuUltra™ II Fusion HS DNA polymerase. This reaction was cycled using the following parameters: 25 cycles consisting of 95°C for 15 sec, 55°C for 60 sec, and 72°C for 210 sec; and ending with a 8 min extension at 72°C. Sequence analysis of the final fusion product was used to verify lack of sequence errors and that the hfq ORF was replaced with the ermC ORF.
To construct the complementing plasmid pKK214hfq, the hfq coding region and 191 bp directly upstream were PCR amplified from F. novicida using primers FN_hfqProm-F-SmaI and FN-hfq.orf-EcoRI-R using a standard reaction mixture and cycling parameters. The product was digested with SmaI and EcoRI and ligated into SmaI-EcoRI-digested pKK214gfp [30], producing pKK214hfq. All constructs were verified by DNA sequence analysis and introduced into the appropriate F. novicida strains by electroporation [31]. Transformants were screened and confirmed by PCR analysis and DNA sequencing, and the resulting mutant strains were designated F. novicida Δhfq and complemented strain Δhfq/pKK214hfq.
To complement an E. coli hfq mutant with the F. novicida Hfq protein under control of the E. coli hfq promoter, the wild-type F. novicida hfq ORF was PCR amplified using primers EcFnHfq-FusF and FN_hfq-R-PstI (Table 2) in a standard reaction mixture. The 163 bp promoter region of the native E. coli hfq ORF was amplified with primers Ec_hfq-up-F-BamHI and EcHfqProm-FusR. The two PCR products were ligated together using a PCR fusion reaction as described above. The PCR product was digested with PstI and BamHI and ligated into pACYC177 that had been digested with the same restriction enzymes, and then transformed into the E. coli hfq mutant strain GS081 [24]. Kanamycin-resistant transformants were screened for the presence of F. novicida hfq linked to the E. coli hfq promoter region by PCR and analyzed by DNA sequence analysis. One transformant was selected for further characterization; its plasmid designated pACYC177Fnhfq. The presence of F. novicida Hfq protein was verified by Western blot analysis using the method described below. Construction of the control plasmid pACYC177Mchfq containing the native E. coli hfq ORF and 163 bp upstream was performed as described above using primers Ec_hfq-R-PstI and Ec_hfq-up-F-BamHI. Complementation was determined by phenotypic analysis at 37°C in LB plus appropriate antibiotics as described above.
Phenotypic analyses
Strains were grown overnight at 37°C in TS broth supplemented with 0.1% cysteine and the appropriate antibiotic. The cultures were diluted 1000-fold in fresh TS broth with cysteine and appropriate antibiotics, supplemented with the appropriate stressor and grown at 37°C (42°C for heat shock) with shaking at 200 rpm. Optical density was measured at 600 nm. For stress tolerance tests the following conditions were used: 1% and 2% additional NaCl, medium adjusted to pH 5, 0.0015% hydrogen peroxide, and heat shock at 42°C. All phenotypic analyses were performed at least two times and the results of a single representative experiment are presented.
Antibody production and immunoblot analysis
The coding sequence for Hfq was PCR amplified from F. novicida using PfuUltra II Fusion HotStart Polymerase (Agilent Technologies) and primers FN-hfq-ATG-F and FN-hfq-330R-PstI (Table 2) at 50°C annealing. The resulting blunt-ended PCR product was digested with PstI, gel purified, and cloned into the XmnI and PstI sites of the expression vector pMAL-c4X (New England BioLabs), which contains a cleavable N-terminal maltose-binding protein ORF. The tagged Hfq was expressed in DE3 cells via IPTG induction, purified using amylose resin, and cleaved from the maltose-binding protein by Factor Xa as described in the pMAL Protein Fusion and Purification System manual (New England BioLabs). An equal volume of SDS sample buffer (40 mM Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol, 10% glycerol, 0.4 mg/ml bromophenol blue) was added to the resulting digest, boiled at 95°C for five minutes, and separated by SDS-PAGE (12% gel). A ∼13 kD band corresponding to the Hfq protein was excised and sent for commercial anti-sera production (Cocalico Biologicals).
For immunoblot analysis, F. novicida and E. coli strains were grown to exponential phase in TS broth supplemented with 0.1% cysteine and LB, respectively, with appropriate antibiotics at 37°C; equivalent units of optical density at 600 nm (OD600 nm) were taken from each culture, resuspended in SDS sample buffer, boiled at 95°C for five minutes, and separated by SDS-PAGE (12% gel). After electrophoresis, proteins were transferred to a PVDF membrane (Millipore) as described by the manufacturer using a Trans-Blot apparatus (Bio-Rad) at 350 mA for 1 h. The resulting membrane was blocked for 1 h and then incubated overnight with F. novicida Hfq antiserum. After washing and blocking, a horseradish peroxidase-linked secondary antibody (Thermo Scientific) was added and additional washing steps were performed. Final binding was detected using SuperSignal West Pico Chemiluminescent substrate (Thermo Scientific).
Microtiter plate biofilm production assay
Crystal violet assaying for biofilm formation was performed as previously described [32]. Francisella strains were grown overnight at 37°C in either Mueller-Hinton (MH) broth or TS broth supplemented with cysteine. Cultures were diluted 1∶100 in fresh medium and vortexed. After vortexing, 150 µl volumes were aliquoted per well in a 96-well polystyrene plate, previously rinsed with 70% ethanol and air-dried. The bacteria were grown statically at 37°C for 48 hours. Wells were washed three times with distilled water, allowed to dry, and then 200 µl of 0.1% crystal violet was added to each well for 45 min of incubation. Wells were washed five times with distilled water, allowed to dry, and the remaining biomass that absorbed crystal violet was solubilized with 95% ethanol. Staining was then quantified by obtaining the OD590 nm using a Nano-Drop spectrophotometer (Thermo Scientific) [33].
RNA isolation and techniques
Cultures of U112 and its Δhfq derivative were grown to exponential (OD600 nm 0.3–0.6) and stationary phase (OD600 nm>0.8) in TS broth supplemented with cysteine. Prior to nucleic acid extraction, a 1/5 volume of ethanol/5% phenol at −20°C was added as a stop solution to prevent transcriptional changes during cell harvesting. Total RNA was isolated using TRI Reagent (Ambion) followed by treatment with TURBO DNase (Ambion) and precipitation with 3 M sodium acetate (pH 5.2) and 100% ethanol. To perform reverse transcription (RT)-PCR, 10 pmol of corresponding reverse primer (Table 2) was allowed to anneal to 1 µg of RNA and heated for 5 min at 70°C. To initiate cDNA synthesis, the following was added to the above mixture to yield a 20 µl (total volume) reaction: 1 mM mixture of dNTPs, 40 U of Ribolock RNase Inhibitor (Fermentas), 3 mM MgCl2, 1 µl ImProm-II reverse transcriptase (Promega), and 5× reaction buffer. The reaction was then heated for 5 min at 25°C, 60 min at 42°C, and 15 min at 70°C. Then 5 µl of the cDNA reaction mixture was used as a template in a 50 µl PCR amplification reaction mixture with corresponding forward and reverse primers (Table 2) and GoTaq DNA polymerase (Promega), as described by the supplier. For control reactions, RNA without reverse transcriptase or chromosomal DNA was used as a template.
5′ RACE was performed on 5 µg DNase-treated RNA using the GeneRacer Kit (Invitrogen) according to the manufacturer's instructions except for dephosphorylation with calf intestinal phosphatase, which was omitted. RT-PCR was performed using the FN_hfq-326R primer (Table 2) and AMV reverse transcriptase as described above. The cDNA was then amplified by PCR using GoTaq DNA Polymerase (Promega) along with FN_hfq-326R (Table 2) and the provided 5′ GeneRacer primer. PCR products were cloned into pGEM-T (Promega) and the resulting plasmids were transformed into Top10 cells. At least five transformants harboring cDNA-containing plasmid were analyzed by DNA sequencing.
Quantitative RT-PCR (qRT-PCR)
F. novicida and its Δhfq derivative were grown to exponential phase in TS broth supplemented with cysteine at 37°C. A portion of culture was removed for RNA extraction as described above and used as time point zero. The remaining culture was divided in half. One portion was grown in TS broth supplemented with cysteine at 37°C, while the other portion was grown at particular stress conditions as described above. Portions of both cultures were removed at 20 minutes, one hour, and four hours and RNA was extracted as described above. One microgram of DNase-treated RNA was reverse transcribed using SuperScript III First-strand synthesis SuperMix (Invitrogen) according to the protocol provided by the manufacturer. Quantitative RT-PCR was performed with SYBR green dye (Quanta Biosciences) in a MiniOpticon thermocycler (Bio-Rad) using gene-specific primers (Table 2). To calculate reaction efficiency of each gene-specific primer set, a standard curve using a series of diluted cDNA was generated. Expression of hfq was determined by comparing samples grown in the presence and absence of a specified stress condition. To compare transcript levels, the amounts of transcript were normalized using the DNA helicase gene uvrD (FTN_1594) (Fig. S1) and fold changes were calculated using the Pfaffl method in CFX Manager software (Bio-Rad) [34].
Results
Transcriptional analysis of hfq ORF
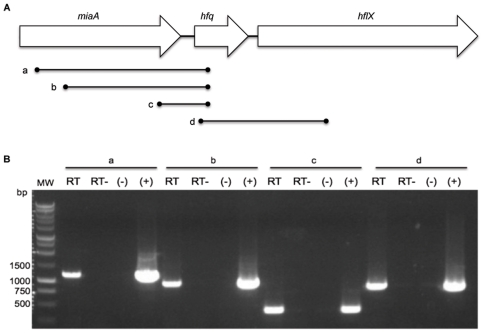
While the F. tularensis LVS hfq gene locus, promoter region, and corresponding protein sequence have previously been described [13], it was unknown if the F. novicida homolog possessed the same characteristics. The F. novicida hfq gene (FTN_1051) is located on the negative strand at bps 1110271–1110600 of the genome. The gene encodes a 109 amino acid protein and is 99.39% identical to its F. tularensis LVS homolog at the nucleotide level and 100% at the amino acid level. In both species, the hfq gene is flanked by the miaA gene upstream [encoding tRNA delta(2)-isopentenylpyrophosphate transferase] and hflX downstream [encoding a GTP-binding protein] (Fig. 1A) [13], a configuration found in a number of bacteria such as E. coli that may form part of an operon [35].
Figure 1. Transcription of the hfq locus.
(A) Genetic organization of the hfq locus. Lines a, b, c, and d indicate regions of the locus amplified by PCR as shown in panel B. (B) RT-PCR of the hfq locus. Lanes 1–4 corresponds to line a, lanes 5–8 to line b, lanes 9–12 to line c, and lanes 13–16 to line d. Within each set of 4 wells, PCR to amplify lines a–d was performed with RNA after RT, RNA without RT as a control, PCR without DNA template as a control, and PCR with genomic DNA from U112 as a positive control. MW denotes molecular weight marker (1 kb BenchTop marker, Promega).
To test if F. novicida hfq was part of an operon we performed RT-PCR experiments targeting the intergenic regions directly up- and downstream of hfq. Transcription of hfq in E. coli has been shown to initiate from multiple upstream promoter sequences, including two located within the miaA gene [36]. To test if hfq transcription in F. novicida originated from other promoters besides the single one identified 65 bp upstream of the F. holarctica hfq gene [13], RT-PCR was performed using reverse primer FN-hfq-151R and forward primers FN_miaA-25F, FN_miaA-274F, and FN_miaA-833F (designated lines a, b, and c respectively) internal to miaA (Fig. 1A). The resulting PCR products (1164 bp, 915 bp, and 357 bp) indicated co-transcription of hfq and miaA throughout the length of the miaA ORF, suggesting at least one additional promoter region is responsible for hfq transcription (Fig. 1B). 5′ RACE experiments placed the start site of hfq transcription −65 bases relative to its ATG start codon, which matches similar work performed in F. tularensis LVS [13]. However, 5′ RACE did not indicate additional transcription start sites further upstream. In order to ascertain if hfq is co-transcribed with the downstream gene hflX, RT-PCR using primers FN_hfq-22F and FN_hflX-501R (line d) was performed (Fig. 1A). An 853 bp product resulted, suggesting co-transcription of hfq with the downstream gene hflX (Fig. 1B).
F. novicida Hfq is unable to complement an E. coli hfq mutation
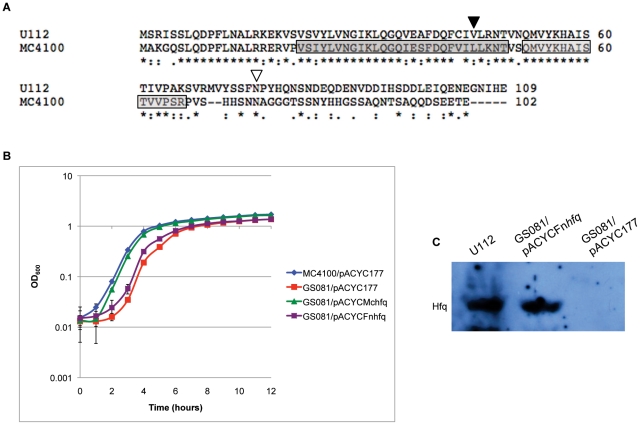
The F. novicida Hfq protein is slightly larger than the Hfq protein of E. coli strain MC4100 (109 a.a. versus 102 a.a.) and is 39% and 49% identical at the nucleotide and amino acid level respectively (Fig. 2A). To investigate the functionality of F. novicida Hfq in the E. coli hfq mutant strain GS081, a region spanning the hfq gene was ligated to the E. coli hfq promoter region, cloned into plasmid pACYC177 and transformed into GS081 [24] yielding strain GS081/pACYCFnhfq. A region containing the native E. coli strain MC4100 hfq gene and promoter region was also cloned into pACYC177 as a control yielding pACYCMchfq, which was transformed into GS081. These two transformants, along with the E. coli parent strain MC4100 and E. coli hfq mutant strain GS081, were each grown in LB broth plus appropriate antibiotics at 37°C to allow comparison of their growth rates. Additionally, MC4100 and GS081 were each grown harboring empty pACYC177 as a control, both of which displayed the same growth rate as the parent strains (data not shown). The E. coli hfq mutant GS081 showed a modest growth defect relative to the parent strain. Expression of E. coli Hfq, under the control of its native promoter, almost fully restored normal growth. However, the strain containing F. novicida hfq exhibited a slower growth rate than the strain containing native MC4100 hfq and did not appear to complement the hfq mutant phenotype (Fig. 2B). Non-quantitative immunoblot analysis verified expression of F. novicida Hfq protein in the E. coli mutant strain GS081 (Fig. 2C).
Figure 2. Complementation of E. coli hfq mutant with F. novicida Hfq.
(A) Alignment of the Hfq proteins from F. novicida strain U112 and E. coli strain MC4100 using the ClustalW program. Periods designate semiconserved substitutions, colons designate conserved substitutions, and asterisks designate residues found in both strains. A dark gray and light gray box indicates the location of E. coli Sm1 and Sm2 sequence motifs respectively. Inverted arrows designate the location of transposon insertion within F. novicida hfq. The arrow at position 45 a.a. (▾) corresponds to strain Tnhfq2 insertion, and the arrow at position 76 a.a. (▿) corresponds to strain Tnhfq 1 insertion. (B) Growth of E. coli strains MC4100/pACYC177, GS081/pACYC177, GS081/pACYCMchfq, and GS081/pACYCFnhfq in LB broth at 37°C with appropriate antibiotics. (C) Detection of F. novicida Hfq assessed by immunoblot analysis in F. novicida strain U112, GS081/pACYCFnhfq, and GS081/pACYC177 using anti-Hfq antisera.
Phenotypic analysis of hfq transposon insertion mutants
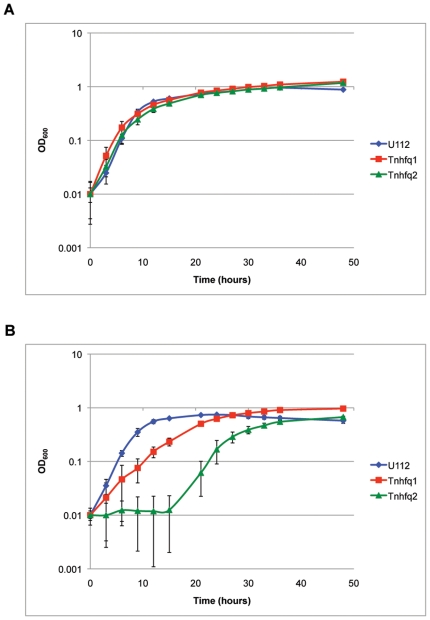
To determine what effect interruption of the hfq gene has on the F. novicida phenotype, two separate transposon insertion mutants were obtained from a F. novicida library created by Gallagher and co-workers [37]. Locations of the EZ-Tn5<KAN-2> insertions mapped to positions 229 and 133 bp relative to the 327 bp hfq ORF for strains Tnhfq1 and Tnhfq2, respectively (Fig. 2A). Phenotypic analysis of these two hfq insertion mutants compared to U112 at 37°C indicated no growth difference for Tnhfq1 or Tnhfq2 (Fig. 3A). Further analysis using 42°C heat stress showed a slower rate of growth for Tnhfq1 compared to U112 at 42°C and its own growth phenotype at 37°C. A more severe defect in growth rate was observed for Tnhfq2, which took over fifteen hours to exit lag phase (Fig. 3B). The different phenotypes observed between Tnhfq1 and Tnhfq2, suggests that the N-terminal half of the F. novicida Hfq is more important for growth during growth in certain stress conditions than the C-terminal half.
Figure 3. Growth characterization of hfq transposon strains.
(A) F. novicida strains U112, Tnhfq1, and Tnhfq2 were cultured in TS broth supplemented with cysteine and appropriate antibiotics at 37°C and the OD600 nm of each culture was measured at select time points. (B) Growth characteristics in TS broth supplemented with cysteine at 42°C.
Construction of F. novicida hfq deletion mutant and growth in vitro
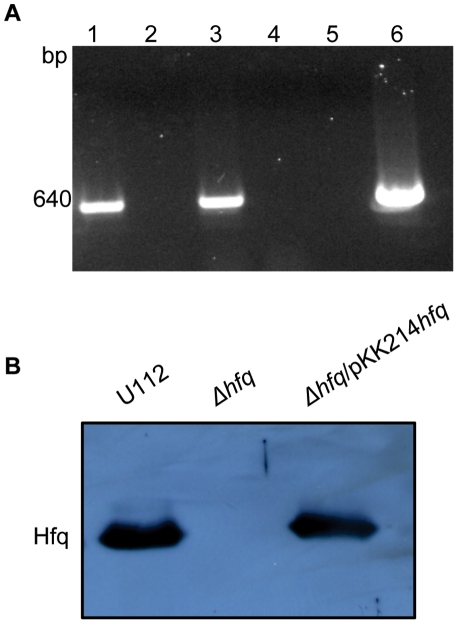
Homologous recombination was used to create a deletion mutant in which the F. novicida hfq ORF was replaced by an ermC ORF. After verifying that marker exchange had occurred by PCR and Southern blotting (data not shown), RT-PCR using primers FN_hflX-373F and FN_hflX-1013R internal to hflX verified that deletion of hfq did not have a polar effect on transcription of the downstream gene hflX (Fig. 4A). We then used the low copy-number vector pKK214gfp to generate a plasmid-based complementing clone of hfq under the control of its native promoter, creating strain Δhfq/pKK214hfq. Subsequent non-quantitative immunoblot analysis verified the production of Hfq protein in both the wild-type U112 and the complemented strain, and the loss of Hfq in the mutant (Fig. 4B). Immunoblot analysis of the hfq mutant strain harboring empty pKK214 as a control also exhibited a loss of Hfq (data not shown).
Figure 4. Affect of hfq deletion on hflX expression and Hfq protein expression in F. novicida strains.
(A) RT-PCR using primers internal to hflX was performed with RNA isolated from U112 with RT (lane 1), RNA from U112 without RT (lane 2), RNA isolated from the hfq mutant with RT (lane 3), RNA from the hfq mutant without RT (lane 4), PCR with no DNA template as a negative control (lane 5), and PCR with DNA isolated from U112 as a positive control (lane 6). (B) Immunoblot analysis of U112, the hfq mutant, and Δhfq/pKK214hfq strains using anti-Hfq antiserum.
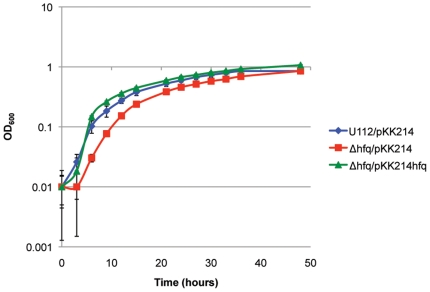
To examine if the entire Hfq protein is important for F. novicida growth, we compared the growth of U112, Δhfq, and Δhfq/pKK214hfq strains in liquid medium at 37°C. Both the wild-type and hfq mutant were grown with empty pKK214 vector to account for any plasmid maintenance effects in the complemented strain. The deletion strain, Δhfq/pKK214, exhibited a longer lag phase compared to wild-type U112/pKK214. The complemented strain, Δhfq/pKK214hfq, exhibited a similar growth compared to U112/pKK214 but eventually reached a higher final optical density (Fig. 5).
Figure 5. Growth characteristics of the U112 hfq mutant.
F. novicida strains U112/pKK214, Δhfq/pKK214, and Δhfq/pKK214hfq were cultured in TS broth supplemented with cysteine and appropriate antibiotics at 37°C and the OD600 nm of each culture was measured over the course of time.
Hfq is important for certain F. novicida stress responses
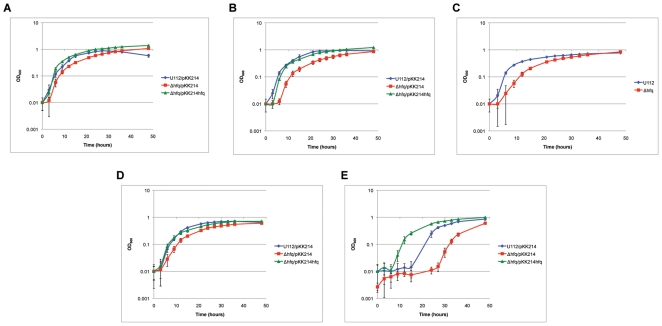
Francisella possesses a diverse lifestyle and like most pathogens is exposed to various stressors. We therefore examined if Hfq is important in the stress resistance of F. novicida by subjecting U112/pKK214, Δhfq/pKK214, and Δhfq/pKK214hfq strains to certain stress conditions during growth in liquid medium. When adding 1% NaCl to the medium, the U112/pKK214 strain exhibited an enhanced growth rate and reached a higher final optical density than in medium lacking this supplement, however a rapid decrease in cell density quickly followed. Growth of the Δhfq/pKK214 strain was also slightly enhanced with additional 1% NaCl compared to growth in medium without this supplement (Figs. 5 and 6A). In medium containing 2% NaCl, Δhfq/pKK214 exhibited a prolonged lag phase relative to medium containing 1% NaCl (Fig. 6B). Upon entry into stationary phase, U112/pKK214 cell density decreased while the hfq mutant remained unchanged compared to growth in unadjusted medium (data not shown). The growth of the strains was also compared in medium adjusted to pH 5. Strain U112 exhibited a slightly slower rate of growth and reached a lower final optical density compared to unadjusted medium. The Δhfq strain also showed a slower growth rate and prolonged lag phase in the acidic medium compared to unadjusted medium but still reached a higher density than U112 (Fig. 6C). Growth of Δhfq/pKK214hfq and pKK214-containing control strains exhibited little or no growth during the initial 24 hours of incubation at acidic pH (data not shown). At 42°C, the U112/pKK214 rate of growth was unchanged but the final density was lower compared to growth at 37°C. The Δhfq/pKK214 strain exhibited a slightly slower rate of growth at 42°C. The Δhfq/pKK214 strain also reached a lower final optical density and never surpassed the density of U112/pKK214 (Fig. 6D). Addition of 0.0015% hydrogen peroxide led to a prolonged lag phase for both U112/pKK214 and Δhfq/pKK214, after which both strains exhibited a rate of growth similar to incubation in unadjusted medium. However, the complemented strain Δhfq/pKK214hfq was able to exit lag phase sooner than even U112/pKK214 (Fig. 6E).
Figure 6. Stress resistance of the U112 hfq mutant.
Growth characteristics of F. novicida strain U112/pKK214, Δhfq/pKK214, and Δhfq/pKK214hfq strains in TS broth supplemented with cysteine and appropriate antibiotics under the following conditions: (A) additional 1% NaCl at 37°C, (B) additional 2% NaCl at 37°C, (C) adjusted to pH 5 at 37°C, (D) at 42°C, and (E) addition of 0.0015% H2O2 at 37°C.
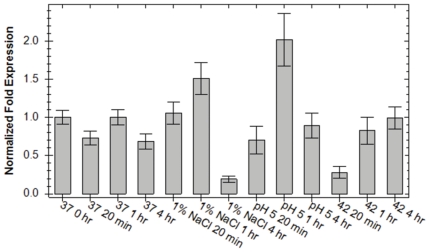
To further examine the role of Hfq in the stress response of F. novicida we monitored hfq expression using qRT-PCR experiments. Cultures of U112 were grown to exponential phase, then separated into two portions as described in the methods. One portion was allowed to continue growth at 37°C while the other was incubated with additional 1% NaCl, in medium adjusted to pH 5, or in unadjusted medium at 42°C. Expression of hfq was quantified (using primers Q_Fn_hfq-84F/Q_Fn_hfq-195R) from RNA extracted at 20 minutes, one hour, and four hours post-separation and compared to hfq levels from time 0 (pre-separation). Measurements of culture optical density at these time points indicate an approximate correlation to exponential, early stationary, and late stationary phases respectively. Expression of hfq exhibited a 5.21-fold decrease after prolonged growth (4 hr) in medium containing additional 1% NaCl. A similar decrease (3.55-fold) in hfq expression was observed shortly after incubation at 42°C. Conversely, a 2.02-fold increase in expression was observed after one hour of growth in acidic medium. We did not observe significant changes in hfq expression for the other measured time points (Fig. 7).
Figure 7. Quantification of hfq expression during stress exposure using quantitative real-time PCR.
Normalized fold expression of hfq transcripts in U112 grown in TS broth supplemented with cysteine at 37°C, at 37°C with the addition of 1% NaCl, at 37°C in medium adjusted to pH 5, and in unadjusted medium at 42°C. Transcript levels were normalized to the level of DNA helicase gene expression (FTN_1594).
Hfq promotes formation of biofilms in F. novicida
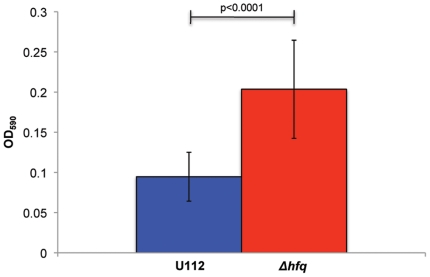
Biofilm growth has been shown to confer both enhanced survival during stress conditions and environmental persistence. Because Francisella has been shown to produce biofilms [10], [33], [38], we decided to test biofilm formation by U112 and the Δhfq strain. Cultures were grown statically in microtiter plates at 37°C for 48 hours followed by staining with 0.1% crystal violet and measurement of the OD590 nm. Both F. novicida strain U112 and the hfq mutant showed similar crystal violet staining when grown in TS broth supplemented with cysteine (data not shown). However, the hfq mutant exhibited increased crystal violet staining compared to wild-type when grown in MH broth indicating increased accumulation of adherent biomass (Fig. 8).
Figure 8. Biofilm formation by F. novicida U112 and Δhfq strains.
Both strains were assayed for biofilm formation using crystal violet staining after 48 hours of static growth in MH broth at 37°C. Culture OD590 nm was determined for multiple replicates. U112 demonstrated decreased crystal violet straining compared to Δhfq (p<0.0001).
Loss of Hfq results in expression changes for a variety of genes
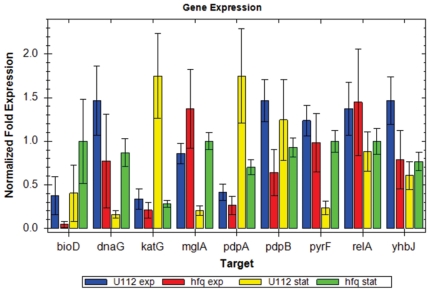
From microarray analysis of a F. tularensis LVS hfq mutant, numerous genes were identified as being regulated by Hfq [13]. We selected genes from that study which exhibited at least a 2-fold change in expression to profile in F. novicida strains U112 and Δhfq. Quantitative real-time PCR was performed on RNA extracted from cultures of U112 and Δhfq grown to exponential and stationary phase. During exponential phase, the expression of two genes decreased significantly (bioD [FTN_0812] and pdpB [FTN_1310]). One of these genes, pdpB, is part of the Francisella pathogenicity island [7]. Four genes displayed significantly altered expression between U112 and Δhfq during growth in stationary phase. Two of these genes were upregulated in the hfq mutant (dnaG [FTN_0914] and pyrF [FTN_0035]), while the other two genes were downregulated (katG [FTN_0633] and pdpA [FTN_1309]). Additionally, we examined expression of the mglA gene (FTN_1290) which encodes a global transcriptional regulator previously shown to regulate hfq [39]. While no significant change was found during exponential phase, expression of mglA was upregulated during stationary phase in the hfq mutant.
Discussion
The ability to monitor environmental changes within the host organism and adjust the expression of stress- and virulence-associated genes accordingly is critical for the growth and survival of pathogenic bacteria. The importance of the small RNA chaperone Hfq in modulating such genes has been demonstrated in a variety of both intra- and extracellular bacterial pathogens [19], [20], [40]. This study demonstrates the role Hfq plays in stress tolerance, gene expression, and biofilm formation in the intracellular pathogen Francisella novicida.
Previously, it was shown that hfq transcription in F. tularensis LVS is controlled by a single promoter located in between the miaA and hfq ORFs [13]. Interestingly, we found that while hfq is transcribed from an identical promoter region in F. novicida, it is also co-transcribed with the upstream gene miaA. Thus expression of the locus in F. novicida appears to differ from that of F. tularensis LVS. Alternatively, this conflicting data could be explained by differences in experimental design. In E. coli, multiple upstream promoters, including two within miaA, drive transcription of hfq, which is located within the miaA-hfq-hflX operon [36]. This genomic locus is conserved in F. novicida, making it tempting to speculate that hfq expression is also under the control of multiple promoters both within and upstream of miaA. However, because our results only indicate a minimum of co-transcription with the majority of the miaA transcript (Fig. 1), the exact location and number of additional promoter regions remain to be determined. Despite the shared operon structure, Hfq in F. novicida is unable to complement an E. coli hfq mutant (Fig. 2B). This inability to complement an E. coli hfq mutant is likely due to basic structural differences between the two Hfq proteins as reflected in the amino acid sequence, especially in the two Sm-domains (Fig. 2A).
Complete deletion of F. novicida hfq only resulted in a modest growth defect compared to wild-type U112 (Fig. 5). The transposon mutant analysis illustrates the importance of the F. novicida Hfq N-terminal domain during 42°C growth (Fig. 3B). The N-terminal insertion of strain Tnhfq2 would disrupt a region similar to the Sm1 domain of E. coli Hfq, while the C-terminal disruption of strain Tnhfq1 would leave both Sm domains intact (Fig. 2A) [41]. The Sm motifs found in Hfq proteins are vital for hexameric formation and riboregulation, while the exact function of the C-terminal half remains unclear [22], [42]. However, stability of the two transposon mutant variants is unknown since a Western blot signal could not be observed using the F. novicida Hfq specific antisera (data not shown). While neither insertion strain exhibited a more dramatic phenotype than the complete hfq deletion strain during growth at 37°C, the lag phase of Tnhfq2 during heat shock was more pronounced than the hfq knockout strain (Fig. 3B and Fig. 6D). The reason for this discrepancy is unclear, but further studies are currently underway to elucidate this phenotype.
The importance of Hfq for stress tolerance has been reported in a variety of other bacteria, including pathogens such as Pseudomonas aeruginosa and Neisseria gonorrhoeae [43]. As such, the importance of F. novicida Hfq in growth and survival during stress conditions was examined by comparing phenotypic differences between the hfq mutant strain and wild-type U112, as well as monitoring changes in hfq transcription. The slightly faster growth rate for both U112/pKK214 and Δhfq/pKK214 when exposed to increased salt concentration suggests that additional NaCl provides a growth advantage to both strains (Figs. 5, 6A, and 6B), while the increased lag phase of Δhfq/pKK214 when exposed to 2% additional NaCl and the subsequent reduced transcription of hfq during growth in 1% NaCl (Fig. 6B and 7) indicate a potential contribution for Hfq during osmotic stress. This finding is also supported by previous work in F. tularensis subspecies holarctica [13]. Similar phenotypes have been observed in Gram-negative pathogens such as Burkholderia cepacia, Moraxella catarrhalis, and Neisseria gonorrhoeae [43].
Tolerance to low pH is important in F. tularensis due to its ability to survive and escape the phagosome during intracellular infection of certain host cell types such as macrophages. Phagosomes have previously been shown to become acidic in response to F. tularensis infection [44]. While both U112 and Δhfq exhibited a reduced rate during growth in acidic media, the prolonged lag phase of Δhfq (Fig. 6C) as well as increased expression of hfq after one hour in acidic conditions (Fig. 7) suggest a role for Hfq in the initial response to low pH. Interestingly, growth of the complemented strain and strains harboring empty pKK214 was severely inhibited by the decreased pH suggesting difficulties in plasmid maintenance during growth at pH 5, and was therefore not included in this analysis. Additionally, we examined the growth rate of U112/pKK214 and Δhfq/pKK214 during heat shock. Compared to the salt and low pH exposure, heat shock had a more negative effect on the growth rate of Δhfq/pKK214 and resulted in entry into stationary phase at a lower optical density than all other conditions tested (Fig. 6). This phenotype suggests a role for Hfq in the heat shock response even though transcription of hfq was initially repressed upon introduction of the stressor before returning to normal levels (Fig. 7).
Reactive oxygen species are one defense mechanism used by macrophages, monocytes, and neutrophils to combat intracellular pathogens. F. tularensis has been reported to inhibit particular neutrophil killing mechanisms such as the respiratory burst as part of its infection lifecycle [45], [46]. Oxidative stress induced by addition of hydrogen peroxide had a severe effect on both U112/pKK214 and Δhfq/pKK214 as indicated by their pronounced lag phases (Fig. 6E). However, the additional six-hour lag phase exhibited by Δhfq/pKK214 indicates that Hfq is necessary for combating initial exposure to H2O2. Similar growth responses to oxidative stress have been reported for hfq mutants in an array of pathogens [43]. While expression of hfq was not assayed under this condition, expression of mglA was upregulated in the mutant (Fig. 9). Previous studies using a F. novicida mglA mutant have shown that lack of MglA results in increased resistance to H2O2 [39]. It is therefore tempting to speculate that the increased expression of mglA in Δhfq is directly linked to the increased lag phase of the strain during H2O2 exposure. However, further studies are necessary to determine the role Hfq plays in regulating mglA expression. As a whole, these experiments demonstrate a role for F. novicida Hfq during growth under a variety of environmental conditions.
Figure 9. Quantification of selected gene transcription using quantitative real-time PCR.
Changes in gene expression in the hfq mutant compared to wild-type U112 were examined during both exponential (exp) and stationary phase (stat) using RNA derived from growth in TS broth supplemented with cysteine at 37°C. Each gene was analyzed using RNA derived from three separate cultures and each reaction was performed in triplicate. Transcript levels were normalized to the level of DNA helicase (FTN_1594).
The highly pleiotropic phenotype of an E. coli hfq mutant is largely due to the importance of Hfq in sRNA-mediated regulation. A number of sRNA targets are global regulators such as the alternative sigma factors rpoH, encoding the heat-shock sigma factor σ32, and rpoS, which encodes the general stress signal factor σS [47], [48]. F. novicida lacks an annotated rpoS gene and to date no known master regulator controlling the transition from exponential to stationary phase has been characterized. Interestingly, we observed the hfq mutant reaching a higher cell density before entry into stationary phase compared to wild-type under most conditions tested (Figs. 5 and 6). The complemented strain (Δhfq/pKK214hfq) also surpassed the cell density of U112/pKK214 before entering stationary phase. Because the native promoter region directly upstream of hfq is solely responsible for expression of Hfq in the complemented strain, the presence of multiple promoter regions controlling transcription of hfq in U112 suggests that the partial-complementation observed in Δhfq/pKK214hfq is due to insufficient cellular concentration of Hfq, especially as cellular density increases. The importance of controlling intracellular Hfq concentration has been shown in several bacteria, including Acinetobacter baylyi, where overexpression led to an inability of the cells to assemble into chains [49]. Additionally, while investigating the role of Hfq in Yersinia pseudotuberculosis, Bai and colleagues found that hfq mRNA levels in a partially complemented strain were lower than those of the wild-type due to differential transcription from multiple promoters. A multicopy construct was required to express hfq to levels necessary for full complementation to be observed [50]. Guina and coworkers have also shown that MglA is important for F. novicida survival during stationary-phase growth [39]. Thus the increased expression of mglA in Δhfq reported here could also be directly related to the increased cell density observed during stationary-phase. Additionally, global analysis of gene expression in a F. tularensis LVS hfq mutant strain revealed upregulation of rpoD, the gene encoding the exponential phase σ70 [13]. Farewell et al. demonstrated that overexpression of σ70 in E. coli dramatically alters gene expression and inhibits development of σS-dependent phenotypes, mimicking the effect of an rpoS mutation [51]. Despite the lack of an annotated σS in F. novicida, we speculate that Hfq plays a role in the transition from exponential to stationary phase and that this role is dose-dependent.
Biofilm formation plays an important part in the transmission and environmental persistence of many bacterial pathogens. Because F. novicida has recently been shown to form biofilms on natural and chitin surfaces [33], we wanted to examine if Hfq contributes to biofilm formation in F. novicida. While no significant differences in crystal violet staining were observed between the two strains after growth in TS broth, the higher crystal violet staining exhibited by the hfq mutant strain when grown in the more restrictive MH broth is indicative of increased biofilm formation (Fig. 8). This level of biofilm formation was similar to that reported for F. novicida on chitin surfaces [33]. Biofilm formation in Campylobacter jejuni is significantly increased during growth in MH broth compared to more nutrient-rich media, so it is possible that F. novicida grown in rich TS broth is unable to form a substantial enough biofilm to observe a phenotypic difference between wild-type and the hfq mutant strain [52]. However, a ready explanation for increased biofilm formation in the hfq mutant during growth in MH broth is not apparent. Attia et al. demonstrated a growth advantage for a Moraxella catarrhalis hfq mutant over the wild-type in a continuous flow biofilm system. The authors suggest this phenotype results from altered gene expression in the Δhfq background leading to changes in the overall outer membrane architecture [53]. In accordance with this hypothesis, Meibom et al. found expression of an outer membrane protein (FTL_0535) and several Type IV pili-associated proteins (FTL_0797, FTL_0798, and FTL_0827) upregulated in a F. tularensis LVS hfq mutant strain [13]. While further expression analyses are needed to confirm this hypothesis, it seems possible that changes in outer membrane protein architecture could account for the increased biofilm formation observed in the F. novicida hfq mutant.
Hfq is an important regulator for a variety of genes in numerous bacteria, including F. tularensis LVS [13], [20], [40]. In order to evaluate its role in F. novicida, we monitored expression of nine genes identified by microarray analysis of a F. tularensis LVS hfq mutant [13]. Seven of these genes showed significant (greater than 2-fold) changes in expression. Two genes, pdpA and pdpB, encoded within the Francisella pathogenicity island were downregulated at different times in the hfq mutant. Previous studies have shown that inactivation of these two genes in F. novicida leads to growth defects within host cells and attenuation of virulence [6], [8]. Another downregulated gene, katG, encodes a protein annotated as possessing catalase/peroxidase activity, potentially a factor in the severe Δhfq growth defect observed during oxidative stress (Fig. 6E), as inactivation of katG increased susceptibility of F. novicida to hydrogen peroxide [39]. Three of the genes examined in this study, katG, dnaG, and pyrF, exhibited similar expression changes to those previously reported by Meibom and colleagues while three other genes, pdpA, pdpB, and bioD, exhibited different changes in expression [13]. While the reason behind these differences is unclear, it is interesting to note that two of the genes, pdpA and pdpB, are virulence factors encoded in the FPI, of which there is only one copy in the F. novicida genome compared to more virulent strains of Francisella which contain two copies [54]. Additionally, the transcriptional regulator MglA was upregulated during stationary phase growth in the hfq mutant. MglA has been implicated in coordinating both virulence gene expression and the stress response of F. tularensis by interacting with SspA (Stringent starvation protein A) and binding RNA polymerase [12], [39]. Absence of MglA has also been shown to result in increased katG expression in F. novicida [39], thus the downregulation of katG observed in this study could be a consequence of increased mglA expression in the Δhfq strain. While we are unable to determine if the expression changes observed for the profiled genes are directly or indirectly affected by Hfq, the protein still appears to play a role in their expression.
The ability of Hfq to affect such wide-ranging cellular functions is likely due to its role as an RNA chaperone for sRNAs. Hfq facilitates pairing interactions between small RNAs and their mRNA targets in several bacteria leading to post-transcriptional regulation of the target gene [55]. To date, no small RNAs have been identified in F. novicida so the exact regulatory function of Hfq remains to be elucidated, but it is likely that many of the effects observed here are due to interactions with as yet unidentified sRNAs. Our work demonstrates the importance of Hfq in cell growth, stress resistance, gene expression, and biofilm formation in F. novicida. Information from these studies is critical to understanding how this pathogen responds to and survives in the diverse environments it occupies.
Supporting Information
Expression of qRT-PCR reporter gene uvrD . Relative change in uvrD expression across multiple time points using RNA derived from U112 growth in TS broth supplemented with cysteine at 37°C.
(TIF)
Acknowledgments
We thank Tendai Mutangadura, University of Missouri, for creating the F. novicida hfq deletion cassette, Brian Beal, AgReliant Genetics LLC, for expression of the F. novicida Hfq for antisera generation, and Gisela Storz, National Institute of Health, for providing the E. coli GS081 (hfq mutant) and MC4100 strains.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) through an Academic Research Enhancement Award #RAI082502A to K. S. Bender. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McLendon MK, Apicella MA, Allen L-AH. Francisella tularensis: Taxonomy, genetics, and immunopathogenesis of a potential agent of biowarfare. Annual Reviews in Microbiology. 2006;60:167–185. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellis J, Oyston PCF, Green M, Titball RW. Tularemia. Clinical Microbiology Reviews. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mörner T. The ecology of tularemia. Revue scientifique et technique. 1992;11:1123–1130. [PubMed] [Google Scholar]
- 4.Parker RR, Steinhaus EA, Kohls GM, Jellison WL. Contamination of natural waters and mud with Pastuerella tularensis. Bulletin National Institute of Health. 1951;193:7–33. [PubMed] [Google Scholar]
- 5.Sjöstedt A. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes and Infection. 2006;8:561–567. doi: 10.1016/j.micinf.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KKM, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. Journal of Bacteriology. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmerk CL, Duplantis BN, Wang D, Burke RD, Chou AY, et al. Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology. 2009;155:1489–1497. doi: 10.1099/mic.0.025379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss DS, Brotcke A, Henry T, Margolis JJ, Chan K, et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proceedings of the National Academy of Sciences USA. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su J, Yang J, Zhao D, Kawula TH, Banas JA, et al. Genome-wide identification of Francisella tularensis virulence determinants. Infection and Immunity. 2007;75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean RE, Ireland PM, Jordan JE, Titball RW, Oyston PC. RelA regulates virulence and intracellular survival of Francisella novicida. Microbiology. 2009;155:4104–4113. doi: 10.1099/mic.0.031021-0. [DOI] [PubMed] [Google Scholar]
- 11.Brotcke A, Monack DM. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infection and Immunity. 2008;76:3473–3480. doi: 10.1128/IAI.00430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charity JC, Costante-Hamm MM, Balon EL, Boyd DH, Rubin EJ, et al. Twin RNA Polymerase-associated proteins control virulence gene expression in Francisella tularensis. PLoS Pathogens. 2007;3:e84. doi: 10.1371/journal.ppat.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meibom KL, Forslund A-L, Kuoppa K, Alkhuder K, Dubail I, et al. Hfq, a novel pleiotropic regulator of virulence-associated genes in Francisella tularensis. Infection and Immunity. 2009;77:1866–1880. doi: 10.1128/IAI.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Molecular Microbiology. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schiano CA, Bellows LE, Lathem WW. The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infection and Immunity. 2010;78:2034–2044. doi: 10.1128/IAI.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellin JR, McClure R, Lopez D, Green O, Reinhard B, et al. Role of Hfq in iron-dependent and -independent gene regulation in Neisseria meningitidis. Microbiology. 2010;156:2316–2326. doi: 10.1099/mic.0.039040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 18.Møller T, Franch T, Højrup P, Keene DR, Bächinger HP, et al. Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Molecular Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 19.Christiansen JK, Larsen MH, Ingmer H, Søgaard-Anderson L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: Role in stress tolerance and virulence. Journal of Bacteriology. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietrich M, Munke R, Gottschald M, Ziska E, Boettcher JP, et al. The effect of hfq on global gene expression and virulence in Neisseria gonorrhoeae. FEBS Journal. 2009;276:5507–5520. doi: 10.1111/j.1742-4658.2009.07234.x. [DOI] [PubMed] [Google Scholar]
- 21.Sonnleitner E, Schuster M, Sorger-Domenigg T, Greenberg EP, Bläsi U. Hfq-dependent alterations of the trancriptome profile and effects on quorum sensing in Pseudomonas aeruginosa. Molecular Microbiology. 2006;59:1542–1558. doi: 10.1111/j.1365-2958.2006.05032.x. [DOI] [PubMed] [Google Scholar]
- 22.Tsui H-CT, Leung H-CE, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Molecular Microbiology. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 23.Geng J, Song Y, Yang L, Feng Y, Qiu Y, et al. Involvement of the post-transcriptional regulator Hfq in Yersinia pestis virulence. PLoS ONE. 2009;4:e6213. doi: 10.1371/journal.pone.0006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Molecular Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 25.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003798. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meibom KL, Dubail I, Dupuis M, Barel M, Lenco J, et al. The heat-shock protein ClpB of Francisella tularensis is invoved in stress tolerance and is required for multiplication in target organs of infected mice. Molecular Microbiology. 2008;67:1384–1401. doi: 10.1111/j.1365-2958.2008.06139.x. [DOI] [PubMed] [Google Scholar]
- 27.Bell BL, Mohapatra NP, Gunn JS. Regulation of virulence gene transcripts by the Francisella novicida orphan response regulator PmrA: role of phosphorylation and evidence of MglA/SspA interaction. Infection and Immunity. 2010;78:2189–2198. doi: 10.1128/IAI.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, et al. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Research. 2004;32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton HL, Schwartz KJ, Dillard JP. Insertion-duplication mutagenesis of Neisseria: Use in characterization of DNA transfer genes in the Gonococcal genetic island. Journal of Bacteriology. 2001;183:4718–4726. doi: 10.1128/JB.183.16.4718-4726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abd H, Johansson T, Golovliov I, Sandström G, Forsman M. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Applied and Environmental Microbiology. 2003;69:600–606. doi: 10.1128/AEM.69.1.600-606.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LoVullo ED, Sherrill LA, Perez LL, Pavelka MS., Jr Genetic tools for highly pathogenic Francisella tularensis subsp. tularensis. Microbiology. 2006;152:3425–3435. doi: 10.1099/mic.0.29121-0. [DOI] [PubMed] [Google Scholar]
- 32.Djordjevic D, Wiedmann M, McLandsborough LA. Microtiter plate assay for assesment of Listeria monocytogenes biofilm formation. Applied and Environmental Microbiology. 2002;68:2950–2958. doi: 10.1128/AEM.68.6.2950-2958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margolis JJ, El-Etr S, Joubert L-M, Moore E, Robinson R, et al. Contributions of Francisella tularensis subsp. novicida chitinases and Sec secretion system to biofilm formation on chitin. Applied and Environmental Microbiology. 2010;76:596–608. doi: 10.1128/AEM.02037-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsui H-CT, Winkler ME. Transcriptional patterns of the mutL-miaA superoperon of Escherichia coli K-12 suggest a model for posttranscriptional regulation. Biochimie. 1994;76:1168–1177. doi: 10.1016/0300-9084(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 36.Tsui H-CT, Feng G, Winkler ME. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Eσ32-Specific promoters during heat shock. Journal of Bacteriology. 1996;178:5719–5731. doi: 10.1128/jb.178.19.5719-5731.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallagher LA, Ramage E, Jacobs MA, Kaul R, Brittnacher M, et al. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proceedings of the National Academy of Sciences USA. 2007;104:1009–1014. doi: 10.1073/pnas.0606713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Durham-Colleran MW, Verhoeven AB, van Hoek ML. Francisella novicida forms in vitro biofilms mediated by an orphan response regulator. Environmental Microbiology. 2009;59:457–465. doi: 10.1007/s00248-009-9586-9. [DOI] [PubMed] [Google Scholar]
- 39.Guina T, Radulovic D, Bahrami AJ, Bolton DL, Rohmer L, et al. MglA regulates Francisella tularensis subsp. novicida (Francisella novicida) response to starvation and oxidative stress. Journal of Bacteriology. 2007;189:6580–6586. doi: 10.1128/JB.00809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates σE expression. Molecular Microbiology. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 41.Olsen AS, Møller-Jensen J, Brennan RG, Valentin-Hansen P. C-Terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. Journal of Molecular Biology. 2010;404:173–182. doi: 10.1016/j.jmb.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 42.Večerek B, Rajkowitsh L, Sonnleitner E, Schroeder R, Bläsi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Research. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao Y, Vogel J. The role of Hfq in bacterial pathogens. Current Opinion in Microbiology. 2010;13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Chong A, Wehrly TD, Nair V, Fischer ER, Barker JR, et al. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infection and Immunity. 2008;76:5488–5499. doi: 10.1128/IAI.00682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCaffrey RL, Allen L-AH. Francisella tularensis LVS evades killing by human neutrophils via inhibition of the respiratory burst phagosome escape. Journal of Leukocyte Biology. 2006;80:1222–1223. doi: 10.1189/jlb.0406287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulert GS, McCaffrey RL, Buchan BW, Lindemann SR, Hollenback C, et al. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagensis of strain LVS. Infection and Immunity. 2009;77:1324–1336. doi: 10.1128/IAI.01318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA Polymerase. Microbiology and Molecular Biology Reviews. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. Journal of Bacteriology. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schilling D, Gerischer U. The Acinetobacter baylyi hfq gene encodes a large protein with an unusual C terminus. Journal of Bacteriology. 2009;191:5553–5562. doi: 10.1128/JB.00490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai G, Golubov A, Smith EA, McDonough KA. The importance of the small RNA chaperone Hfq for growth of epidemic Yersinia pestis, but not Yersinia pseudotuberculosis, with implications for plague biology. Journal of Bacteriology. 2010;192:4239–4245. doi: 10.1128/JB.00504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farewell A, Kvint K, Nyström T. Negative regulation by RpoS: a case of sigma factor competition. Molecular Microbiology. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 52.Reeser RJ, Medler RT, Billington SJ, Jost BH, Joens LA. Characterization of Campylobacter jejuni biofilms under defined growth conditions. Applied and Environmental Microbiology. 2007;73:1908–1913. doi: 10.1128/AEM.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Attia AS, Sedillo JL, Wang W, Liu W, Brautigam CA, et al. Moraxella catarrhalis expresses an unusual Hfq protein. Infection and Immunity. 2008;76:2520–2530. doi: 10.1128/IAI.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nano FE, Schmerk C. The Francisella pathogenicity island. Annals of the New York Academy of Sciences. 2007;1105:122–137. doi: 10.1196/annals.1409.000. [DOI] [PubMed] [Google Scholar]
- 55.Brennan RG, Link TM. Hfq structure, function, and ligand binding. Current Opinion in Microbiology. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 56.Larson CL, Wicht W, Jellison WL. A new organism resembling P. tularensis isolated from water. Public Health Reports. 1955;70:253–258. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of qRT-PCR reporter gene uvrD . Relative change in uvrD expression across multiple time points using RNA derived from U112 growth in TS broth supplemented with cysteine at 37°C.
(TIF)