SUMMARY
Cytosine methylation is the major covalent modification of mammalian genomic DNA and plays important roles in transcriptional regulation. The molecular mechanism underlying the enzymatic removal of this epigenetic mark, however, remains elusive. Here we show that 5-methylcytosine (5mC) hydroxylase TET1, by converting 5mCs to 5-hydroxymethylcytosines (5hmCs), promotes DNA demethylation in mammalian cells through a process that requires the base-excision repair pathway. While expression of the 12 known human DNA glycosylases individually did not enhance removal of 5hmCs in mammal cells, demethylation of both exogenously introduced and endogenous 5hmCs is promoted by the AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA-editing enzyme complex) family of cytidine deaminases. Furthermore, Tet1 and Apobec1 are involved in neuronal activity-induced, region-specific, active DNA demethylation and subsequent gene expression in the dentate gyrus of the adult mouse brain in vivo. Our study suggests a TET1-induced oxidation-deamination mechanism for active DNA demethylation in mammals.
INTRODUCTION
The mammalian genome is methylated predominantly at the C5-position of cytosine bases within CpG dinucleotides (Suzuki and Bird, 2008; Zhu, 2009), whereas non-CpG methylation has been observed at relatively low levels in pluripotent stem cells (Lister et al., 2009; Ramsahoye et al., 2000). 5mCs, especially when clustered, are important transcriptional silencers at gene promoters and endogenous retrotransposons in the genome (Jaenisch and Bird, 2003). DNA methylation also plays critical roles in tissue-specific gene expression, X chromosome inactivation, gene imprinting and nuclear reprogramming (Bonasio et al., 2010; Feng et al., 2010b). Cytosine methylation is catalyzed by a family of DNA methyltransferases (DNMTs) and deficiencies in these enzymes result in profound developmental defects (Goll and Bestor, 2005; Reik, 2007).
During early development, the mammalian DNA methylome is dramatically reprogrammed at two stages (Feng et al., 2010b; Gehring et al., 2009). In the zygote, DNA methylation of the paternal, but not maternal, pronucleus is rapidly lost at the genome-wide scale (Mayer et al., 2000; Oswald et al., 2000; Surani et al., 2007). In primordial germ cells, DNA methylomes undergo a second wave of reprogramming, resulting in a large decrease in the global 5mC level (Hajkova et al., 2010; Surani et al., 2007). In contrast, post-developmental DNA demethylation occurs in a highly locus-specific fashion and has been shown to regulate gene expression in various tissues (Ma et al., 2009a; Wu and Zhang, 2010; Zhu, 2009). In addition, DNA demethylation is required for epigenetic resetting during somatic reprogramming by nuclear transfer (Simonsson and Gurdon, 2004), cell fusion (Bhutani et al., 2010), or transcription factor-based derivation of induced pluripotent stem cells (Mikkelsen et al., 2008). In the adult mammalian brain, while the DNA epigenome is stable at the genome-wide level (Ma et al., 2009b), emerging evidence suggests the presence of active DNA modifications at specific genomic loci and these modifications are critical for certain types of brain plasticity (Day and Sweatt, 2010; Ma et al., 2010). For example, deletion of both Dnmt1 and Dnmt3a leads to decreased CpG methylation at several genomic loci and impacts synaptic functions of adult forebrain neurons (Feng et al., 2010a). Deletion of Gadd45b abolishes neuronal activity-induced DNA demethylation in the adult mouse dentate gyrus at specific genomic loci, including brain-derived neurotrophic factor (Bdnf) and fibroblast growth factor 1 (Fgf1) promoters, and attenuates activity-induced adult hippocampal neurogenesis (Ma et al., 2009b). Pharmacological inhibition of DNA methylation changes also affects synaptic plasticity, learning and memory (Day and Sweatt, 2010), further suggesting the importance of epigenetic DNA modification mechanisms in the adult nervous system.
Although DNA demethylation in mammals has been observed both at the genome-wide and locus-specific levels, mechanisms underlying the removal of methyl groups from the genomic DNA have been under intensive debate (Ooi and Bestor, 2008). Several mammalian proteins have been reported to exhibit DNA demethylase activity, however, such activities remain controversial (Wu and Zhang, 2010). Emerging evidence has implicated DNA repair as a potential mechanism for active DNA demethylation (Gehring et al., 2009; Ma et al., 2009a; Zhu, 2009). In plants, a group of 5mC-specific DNA glycosylases, including ROS1, DME and DML2/3, can specifically excise 5mC to initiate the base-excision repair (BER) to achieve active DNA demethylation (Zhu, 2009). However, no orthologue of these 5mC-specific DNA glycosylases has been identified in mammalian genomes. Alternatively, it has been proposed that glycosylation of 5mC may be preceded by deamination of 5mC and generation of a T:G mismatch, which can be efficiently repaired by mammalian glycosylases TDG and MBD4 (Gehring et al., 2009; Morgan et al., 2004; Rai et al., 2008). One cytidine deaminase, activation-induced deaminase (AID), is required for OCT4 and NANOG promoter demethylation during in vitro nuclear reprogramming (Bhutani et al., 2010). Aid-null mice also exhibit significant defect in genome-wide DNA demethylation in primordial germ cells (Popp et al., 2010). However, AID is unable to act on double-stranded DNA (dsDNA) and the 5mC reactivity for AID deamination in vitro is much lower than for unmethylated cytosines (Bransteitter et al., 2003; Conticello et al., 2007; Di Noia and Neuberger, 2007; Larijani et al., 2005), raising the question of whether AID, if it does initiate demethylation, directly deaminates 5mCs in vivo.
TET proteins, a group of Fe(II)/2-oxoglutarate-dependent dioxygenases, have recently been identified as 5mC hydroxylases that oxidize 5mCs to produce 5hmCs (Ito et al., 2010; Tahiliani et al., 2009). 5hmC may achieve passive DNA demethylation by excluding DNMT1, which maintains symmetric CpG methylation during DNA replication (Valinluck and Sowers, 2007). Interestingly, Tet1 knockdown in mouse embryonic stem cells (ESCs) results in Nanog promoter hypermethylation and defects in ESC self-renewal (Ito et al., 2010). 5hmC has also been hypothesized as a potential intermediate for active DNA demethylation (Bonasio et al., 2010; Feng et al., 2010b; Hajkova et al., 2010; Ito et al., 2010; Wu and Zhang, 2010; Xu et al., 2011). However, direct evidence for active demethylation of 5hmC-containing DNA is lacking. In two recent studies, loss-of-function mutations of TET2 appeared to display opposite effects on DNA methylation status (Figueroa et al., 2010; Ko et al., 2010), further raising the question of a general role of TET proteins and 5hmC in DNA demethylation. Notably, 5hmCs exist in relative abundance in various brain regions (Kriaucionis and Heintz, 2009). The physiological function of 5hmC and TET proteins in the brain remains to be determined.
Here we show that overexpression of TET1 in human cells reactivates a methylation-silenced plasmid reporter and promotes DNA demethylation of both exogenous non-replicable methylated reporter plasmids and multiple endogenous genomic loci. We provide direct evidence that human cells possess a robust demethylating activity towards 5hmC-containing DNA, which is DNA replication-independent and requires an intact BER pathway. Furthermore, AID/APOBEC cytidine deaminases promote 5hmC demethylation both in cultured human cells and in the adult mouse brain. Similar to deamination, 5hmC demethylation is processive, transcription-dependent, and strand-biased. Finally, TET1 is both sufficient and required for neuronal activity-induced, region-specific, and active DNA demethylation in the adult mouse brain in vivo. Our study identified TET1 as a critical factor to initiate an oxidation-deamination mechanism underlying active DNA demethylation in mammals.
RESULTS
TET1 promotes DNA demethylation in human cells
We first confirmed the 5mC hydroxylase activity of human TET1 by analysis of purified genomic DNA from HEK293 cells overexpressing the HA-tagged human TET1 catalytic domain (aa1418–2136, referred to as TET1) using immunoblotting and immunocytochemistry (Figures 1A and S1A). The specificity of 5hmC antibodies was confirmed by immunoblotting of linear DNA that contained almost exclusively unmethylated cytosines, 5mCs, or 5hmCs (Figure S1B). To examine whether hydroxylation of 5mC by TET1 promotes active DNA demethylation, we utilized a reporter assay using an in vitro methylated GFP expressing plasmid (Figure S1C) (Ma et al., 2009b). Complete methylation of all CpGs on the plasmid by the bacterial methyltransferase SssI resulted in strong silencing of GFP expression after transfection into HEK293 cells (Figure 1A). Overexpression of TET1, but not a catalytically inactive TET1 mutant (H1671Y/D1673A; TET1m), promoted reactivation of GFP expression, resulting in a significant increase in the number of GFP+ cells as quantified by FACS (Figure 1A). Thus, TET1 hydroxylase activity promotes the reversal of the silencing effect of cytosine methylation.
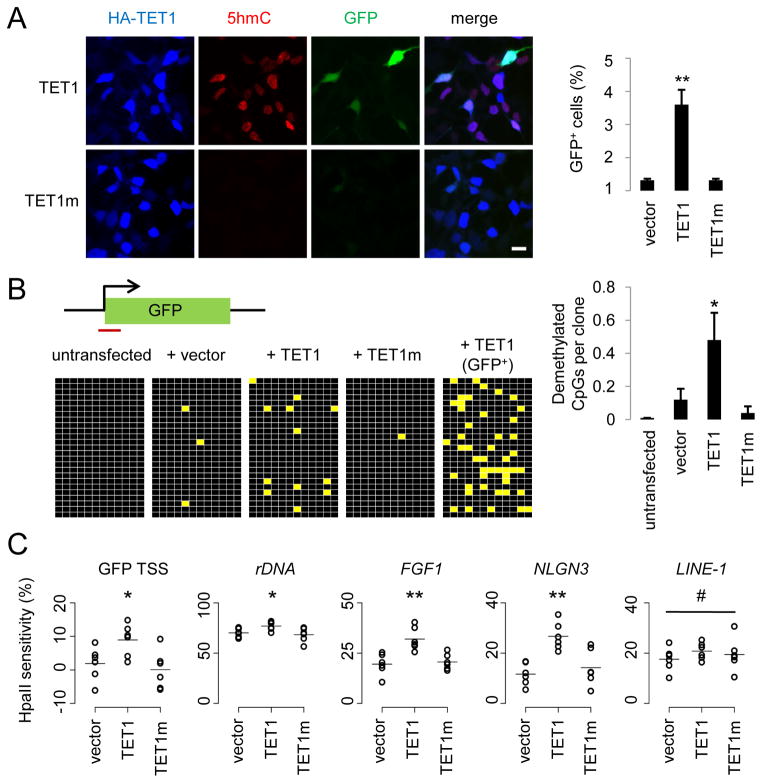
Figure 1. TET1 Catalyses 5mC Hydroxylation and Promotes Demethylation of Exogenous SssI-Methylated Reporter Plasmids and Endogenous Genomic Loci.
(A) Reactivation of methylation-silenced reporter plasmids by TET1. Shown on the left are sample images of immunostaining of HEK293 cells co-transfected with SssI-methylated GFP expression plasmids and expression constructs for TET1 or TET1m. Scale bar: 10 μm. Shown on the right is a summary of quantification of GFP+ cells as measured by FACS. Values represent mean ± SEM (n = 3; **: P < 0.01; Student’s t-test).
(B) Bisulfite sequencing analysis of reporter plasmids. Shown on the top is a schematic diagram of the region around GFP transcription start site (TSS) for bisulfite sequencing (indicated by the red line). Shown on the left are illustrations of methylation status of CpGs within the sequenced region. Black: methylated; yellow: unmethylated. Note the much higher level of demethylation in GFP+ sorted cells. Shown on the right is a summary of mean numbers of CpGs that are demethylated within each clone. Values represent mean ± SEM (n = 3; *: P < 0.05; Student’s t-test).
(C) Effects of TET1 and TET1m overexpression on the HpaII sensitivity of the GFP TSS region and several endogenous genomic loci in HEK293 cells. Open circles represent data from individual experiments and lines represent mean values (**: P < 0.01; *: P < 0.05; #: P > 0.1; Student’s t-test).
Cytosine methylation near transcription start sites (TSSs) of genes is known to be critical for transcriptional silencing (Suzuki and Bird, 2008). Bisulfite sequencing around the GFP TSS showed very sparse CpG demethylation 48 hours after transfection of SssI-methylated plasmids into HEK293 cells (Figure 1B). Expression of TET1, but not TET1m, significantly enhanced the demethylation of these CpGs (Figure 1B). As expected, sorted GFP+ cells with TET1 expression displayed much increased CpG demethylation (Figure 1B). We also used an independent approach to measure CpG methylation levels based on quantitative PCR (qPCR) after HpaII digestion, which is blocked by both CpG methylation and hydroxymethylation (Jin et al., 2010). Quantitative analysis showed a significant increase of HpaII sensitivity around GFP TSS in the presence of TET1, but not TET1m (Figure 1C and Table S1). Because the reporter plasmid is incapable of replication in HEK293 cells, the DNA demethylation observed around the GFP TSS is active in nature.
We next examined whether 5mC hydroxylation also promotes demethylation of the endogenous genomic DNA. Indeed, overexpression of TET1, but not TET1m, significantly increased HpaII sensitivity at multiple loci from distinct genomic subregions (Figure 1C), including promoters of ribosomal DNA (rDNA), FGF1 and NLGN3 (Neuroligin-3), but not at LINE-1s (long interspersed non-repetitive elements). Thus, TET1 overexpression also promotes region-specific DNA demethylation at endogenous genomic loci in human cells.
5hmC in both CpG and CpH contexts can be demethylated in human cells
To ascertain TET1-induced DNA demethylation is mediated by 5hmC generation, we examined whether pre-modified 5hmC-containing DNA is demethylated in human cells. By replacing deoxycytidine triphosphates (dCTPs) with either 5-methyl-deoxycytidine triphosphates (5mdCTPs) or 5-hydroxymethyl-deoxycytidine triphosphates (5hmdCTPs) in PCR reactions, we generated linear GFP expression cassettes in which almost all cytosines, except for primer regions, are either 5mCs or 5hmCs (~99.3% of all cytosines; Figures 2A and S2A; Table S1). As expected, HpaII cannot digest either fully methylated or fully-hydroxymethylated DNA fragments (Figure S2B). Overexpression of TET1, but not TET1m, led to significant HpaII sensitivity near GFP TSS in 5mC-GFP DNA after transfection into HEK293 cells (Figure 2B), which required demethylation of all four cytosines in the CCGG motif. Importantly, HpaII sensitivity at the same position arose on 5hmC-GFP DNA after transfection, and was not further increased by TET1 overexpression (Figure 2B). These results suggest that TET1-induced DNA demethylation is a direct result of 5hmC generation in human cells.
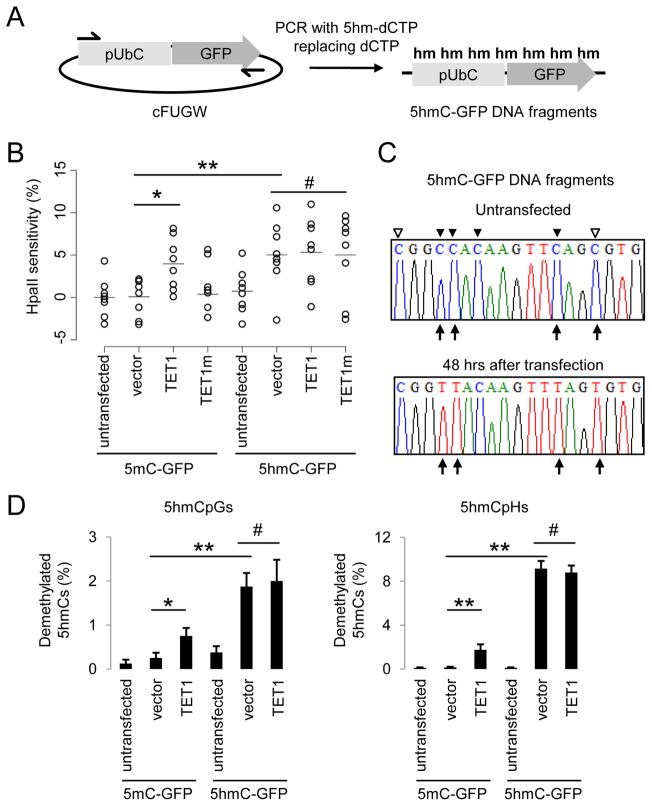
Figure 2. 5hmC-containing DNA Are Demethylated in HEK293 Cells.
(A) A schematic diagram of a PCR-based approach to generate 5mC-, or 5hmC-containing DNA fragments, that contain an Ubiquitin promoter (pUbC) followed by the GFP open reading frame.
(B) Summary of CpG methylation levels of GFP TSS regions under different conditions as quantified by the HpaII sensitivity. Open circles represent data from individual experiments and lines represent mean values (**: P < 0.01; *: P < 0.05; #: P > 0.1; Student’s t-test).
(C and D) Bisulfite sequencing analysis of 5mC-/5hmC-GFP DNA fragments under different conditions. Shown in (C) are sample traces from bisulfite sequencing of 5hmC-GFP DNA before or after transfection. Open and filled triangles indicate cytosines in CpG and CpH contexts, respectively. Arrows indicate demethylated 5hmCs. Shown in (D) are quantifications of demethylated CpGs (left) and CpHs (right) on 5mC-/5hmC-GFP DNA. Values represent mean ± SEM (n = 3; **: P < 0.01; *: P < 0.05; #: P > 0.1; Student’s t-test).
To directly quantify cytosine demethylation, we carried out bisulfite sequencing analysis of 5mC-GFP and 5hmC-GFP DNA using a modified protocol (Table S1). Consistent with HpaII-digestion results, bisulfite sequencing of 5mC-GFP DNA showed negligible demethylation after transfection into HEK293 cells (Figures 2C and 2D). Expression of TET1 led to ~0.8% of 5mCpGs demethylated in each 500bp clone (Figure 2D). Surprisingly, 5mCs in non-CpG contexts (5mCpH, H = A/C/T) showed an even higher frequency of demethylation in the presence of TET1 (Figure 2D). Importantly, robust demethylation occurred on transfected 5hmC-GFP DNA, with ~2% of 5hmCpGs and ~9% of 5hmCpHs demethylated in each clone, which accounts for ~8.5% of all modified cytosines in this region (Figures 2D and S2C). Collectively, these results demonstrate the presence of a DNA replication-independent 5hmC-demethylating activity and an unexpected capacity for active demethylation of 5hmCs in non-CpG contexts in mammalian cells.
5hmC demethylation requires the BER pathway
To characterize the mechanism underlying 5hmC demethylation, we used pharmacological inhibitors to examine the potential involvement of the BER pathway, which has been implicated for DNA demethylation in vertebrates (Gehring et al., 2009; Ma et al., 2009a). ABT-888 (ABT) inhibits poly(ADP-ribose) polymerases (PARPs), which signals single-strand breaks (Donawho et al., 2007; Purnell and Whish, 1980), while CRT0044876 (CRT) inhibits apurinic/apurimidinic endonuclease APE1 (Madhusudan et al., 2005). Both enzymes are critical components in the BER pathway and these two inhibitors block DNA demethylation in mouse zygotes (Hajkova et al., 2010). Interestingly, HpaII sensitivity around GFP TSS on 5hmC-GFP DNA was abolished in the presence of ABT and CRT (Figure 3A). Bisulfite sequencing analysis further confirmed the reduced DNA demethylation of 5hmC-GFP DNA after treatment of these two inhibitors (Figure 3B). In addition, TET1-induced demethylation of endogenous genomic loci, including FGF1 and rDNA promoters, was abolished by ABT and CRT (Figure 3C). Neither of the inhibitors interfered with the TET1 hydroxylase activity, as indicated by largely unchanged overall 5hmC levels in HEK293 cells with TET1 overexpression (Figure S3A). Taken together, these results suggest that the BER pathway is required for demethylation of both exogenous 5hmC and endogenous loci after TET1 overexpression.
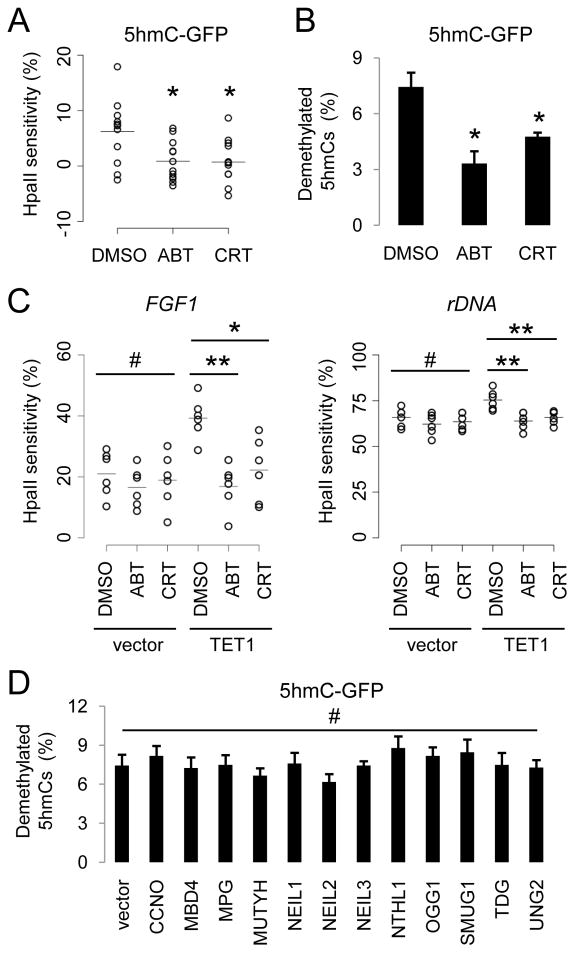
Figure 3. BER Enzyme Activities Are Required for 5hmC Demethylation.
(A and B) Effects of PARP1 inhibitor ABT-888 (50 μM) and APE inhibitor CRT0044876 (50 μM) on demethylation of transfected 5hmC-GFP DNA fragments. Shown in (A) is a summary of HpaII sensitivity assay. Open circles represent data from individual experiments and lines represent mean values (*: P < 0.05; Student’s t-test). Shown in (B) is a summary of bisulfite sequencing analysis. Values represent mean ± SEM (n = 3; *: P < 0.05; Student’s t-test).
(C) Effects of ABT and CRT on TET1-induced increases of HpaII sensitivity of FGF1 (left) and rDNA (right) promoters. Open circles represent data from individual experiments and lines represent mean values (**: P < 0.01; *: P < 0.05; #: P > 0.1; Student’s t-test).
(D) Effects of overexpression of human DNA glycosylases on demethylation of 5hmC-GFP DNA fragments. Shown is a summary of bisulfite sequencing analysis of 5hmC-GFP DNA after co-transfection with one of the 12 human DNA glycosylase cDNAs. Values represent mean ± SEM (n = 3; #: P > 0.1; one-way ANOVA).
The central components of the BER pathway include DNA glycosylases that recognize and remove specific types of damaged or mismatched DNA bases (Wood et al., 2005). Overexpression of each of the 12 known human DNA glycosylases in HEK293 cells (Figure S3B) did not lead to any noticeable decrease of 5hmC levels in the genomic DNA, as measured by immunoblotting and immunocytochemistry (Figures S3C and S3D). Similarly, no significant enhancement of demethylation of transfected 5hmC-GFP DNA was detected after overexpression of these glycosylases (Figure 3D). Thus, 5hmC does not appear to be directly recognized and removed by any of these glycosylases under our experimental conditions.
AID/APOBEC deaminases facilitate 5hmC demethylation
Which pathway could process 5hmC in mammalian cells? Previous studies have suggested that coupling between the AID/APOBEC family of cytidine deaminases and T:G mismatch glycosylases may mediate DNA demethylation in vertebrate cells (Morgan et al., 2004; Rai et al., 2008). However, 5mCs appear to be less favorable substrates than unmethylated cytosines for AID in vitro (Bransteitter et al., 2003; Larijani et al., 2005), suggesting that AID might not directly deaminate 5mCs during the process of active DNA demethylation. Therefore, we examined whether the AID/APOBEC family of cytidine deaminases might play a role in 5hmC demethylation. Interestingly, AID overexpression led to a significant decrease of 5hmC levels induced by TET1 in HEK293 cells (Figures 4A, 4B and S4A). Bisulfite sequencing analysis of transfected 5hmC-GFP DNA showed that AID significantly increased 5hmC demethylation (Figure 4C). Importantly, these demethylated cytosines were not unrepaired deamination products, because sequences of non-bisulfite converted 5hmC-GFP DNA co-transfected with AID showed no significant level of C-to-T transitions (Figure S4C).
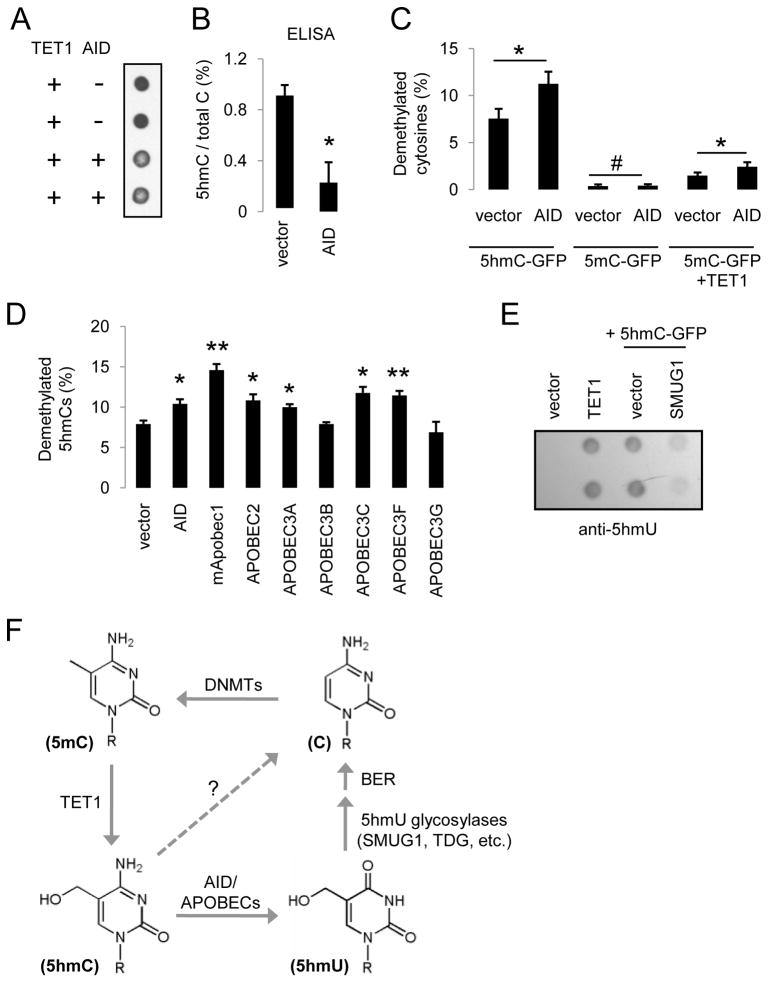
Figure 4. AID/APOBEC Deaminases Promote 5hmC Demethylation.
(A and B) Overexpression of AID results in a decrease in the abundance of 5hmCs in genomic DNA samples from HEK 293 cells as monitored by immunoblotting (A) and ELISA (B) using anti-5hmC antibodies.
(C) Effects of AID on demethylation of 5hmC-GFP and 5mC-GFP DNA. Values represent mean ± SEM (n = 3; *: P < 0.05; #: P > 0.1; Student’s t-test)
(D) Effects of overexpression of AID/APOBEC deaminases on 5hmC demethylation as measured by bisulfite sequencing. Values represent mean ± SEM (n = 3; **: P < 0.01; *: P < 0.05; Student’s t-test).
(E) Immunoblotting analysis of 5hmU levels in HEK293 cells after TET1 expression or transfection of 5hmC-GFP DNA, with or without the expression of 5hmU glycosylase SMUG1.
(F) An oxidation-deamination-BER model of TET1-induced active DNA demethylation in mammalian cells.
Does the demethylation-promoting effect of AID specifically target 5hmCs, or both 5hmCs and 5mCs? Overexpression of AID showed no significant effect on the demethylation of 5mC-GFP DNA (Figure 4C). Immunostaining also did not show any noticeable change in genomic 5mC levels in AID-overexpressing cells (Figure S4B). As expected, co-expression of AID and TET1 led to a significant increase in demethylation of 5mC-GFP DNA (Figure 4C). Thus, AID appears to specifically promote 5hmC demethylation, but does not directly deaminate 5mCs to induce demethylation.
We next examined whether other cytidine deaminases of the AID/APOBEC family also promote demethylation of 5hmCs. Overexpression of mouse Apobec1 (mApobec1), human APOBEC2, APOBEC3A, 3C, and 3E, but not APOBEC3B or 3G, significantly increased demethylation of 5hmC-GFP DNA after transfection into HEK293 cells (Figures 4D and S4D), suggesting a general property of the majority of AID/APOBEC deaminases in promoting 5hmC demethylation. Notably, APOBEC1 and APOBEC2 have both been implicated in active DNA demethylation in vertebrates (Morgan et al., 2004; Rai et al., 2008).
Deamination of 5hmC would produce 5-hydroxyluracil (5hmU). While there is no detectable endogenous 5hmU in HEK293 cells, expression of TET1 resulted in a significant amount of 5hmU (Figures 4E and S4E). Direct transfection of 5hmC-GFP DNA into HEK293 cells also led to a significant amount of 5hmU, which was decreased by co-expression of 5hmU glycosylase SMUG1 (Boorstein et al., 2001)(Figure 4E). In addition, overexpression of strong enhancers of 5hmC demethylation, including mApobec1, APOBEC3A and 3C, appeared to produce higher levels of 5hmU (Figure S4F). Expression analysis of HEK293 cells revealed the presence of many AID/APOBEC deaminases and DNA glycosylases (Figure S4G; Table S1), including known 5hmU glycosylases SMUG1, TDG (Baker et al., 2002), NEIL1 and NTHL1 (Zhang et al., 2005).
Collectively, these results suggest the following model for active DNA demethylation in mammalian cells (Figure 4F): 5mCs are first oxidized to 5hmCs by TET proteins. 5hmCs are then deaminated by AID/APOBEC deaminases into 5hmU. Finally, 5hmU can be excised by 5hmU glycosylases and repaired by the BER pathway with unmethylated cytosines.
5hmC demethylation recapitulates properties of AID-mediated deamination
Much effort has been made to characterize catalytic properties of AID, with a focus on the involvement of AID-mediated deamination in somatic hypermutation in B cells (Di Noia and Neuberger, 2007). If active 5hmC demethylation entails deamination, does it exhibit known catalytic features of AID-mediated deamination?
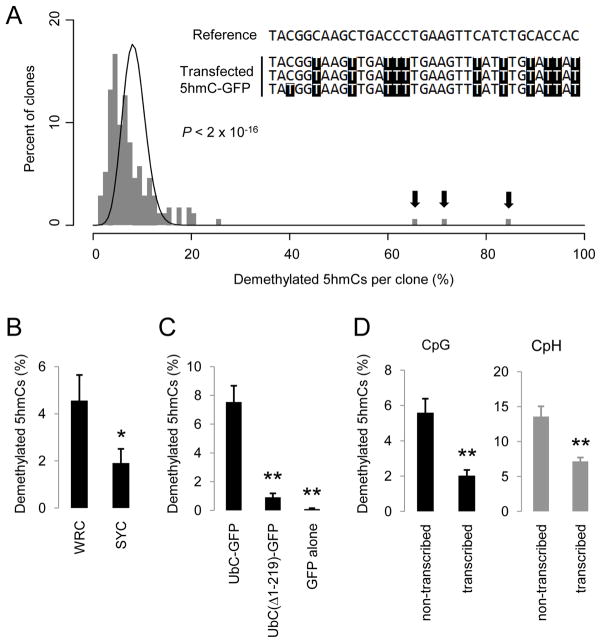
First, AID-mediated deamination is a processive reaction (Pham et al., 2003). Extensive bisulfite sequencing of 174 clones of 5hmC-GFP DNA after transfection showed a broad distribution of numbers of demethylated 5hmCs in individual clones, which significantly deviated from a distributive Poisson model (P < 2 × 10−26; Figure 5A). In 3 clones, 67%, 73% and 86% of all 5hmCs on each clone were demethylated, indicating high processsivity of 5hmC demethylation.
Figure 5. 5hmC Demethylation Recapitulates Known Properties of AID-Catalysed Deamination.
(A) A histogram of clone distribution of the percentage of demethylated 5hmCs. A Poisson distribution with λ = 8.4% is shown as a black line. Arrows indicate 3 highly processively demethylated 5hmC-GFP clones. A small portion of the reference sequence and the bisulfite sequencing results for these 3 clones are shown in the inset.
(B) Occurrence of single demethylated 5hmCs at WRC and SYC motifs. WRC motif: W = A/T; R = A/G; SYC motif: S = C/G; Y =C/T. Only demethylated 5hmCs that are at least 5 nucleotides away from any other demethylated 5hmCs are analyzed. Values represent mean ± SEM (n = 3; *: P < 0.05; Student t-test).
(C) Demethylation of promoter-truncated 5hmC-GFP variants. The same region in the GFP open-reading frame is bisulfite sequenced for all variants. Values represent mean ± SEM (n = 3; **: P < 0.01; Student t-test).
(D) Demethylation of 5hmCs in the CpG (left) and CpH (right) contexts on two opposite strands. Demethylated 5hmCs of the forward (untranscribed) strand and the reverse (transcribed) strand are represented by C-to-T and G-to-A transitions in bisulfite sequencing (Figure S5C), respectively. Values represent mean ± SEM (n = 3; **: P < 0.01; Student’s t-test).
Second, AID exhibits DNA sequence selectivity for deamination with hot spots (WRC motifs; W = A/T; R = A/G) and cold spots (SYC motifs; S = C/G; Y = C/T) (Pham et al., 2003). To minimize the non-specific targeting effect of the processive activity, only single demethylated 5hmCs at least 5 nucleotides away from any other demethylated 5hmCs were analyzed. The observed frequency of 5hmC demethylation at WRC motifs was 2.4 fold higher than that at SYC motifs (Figure 5B), indicating a similar sequence preference between 5hmC demethylation and AID-mediated deamination. Notably, the observed sequence preference of 5hmC demethylation is less robust than that of AID deamination of unmethylated cytosines in vitro or in somatic hypermutation, consistent with the notion that other AID/APOBEC deaminases with different sequence selectivity also contribute to 5hmC demethylation.
Third, transcription has been shown to target AID to specific genomic loci for deamination (Chaudhuri et al., 2003). We generated two variants of 5hmC-GFP DNA fragments, one with a truncated UbC-promoter and one with no promoter (Figure S5A), which exhibited either much reduced or no detectable GFP expression after transfection (Figure S5B). Concomitantly, demethylation of these truncated 5hmC-GFP DNA fragments was almost completely abolished (Figure 5C), suggesting that sufficient transcriptional activity is a prerequisite for 5hmC demethylation.
Finally, it has been shown that on a transcribed dsDNA substrate, AID preferentially deaminates the untranscribed strand (Chaudhuri et al., 2003). This prompted us to discriminate demethylation of 5hmC-GFP DNA on two opposite strands of DNA with bisulfite sequencing analysis (Figure S5C). Indeed, the average frequency of demethylation on the untranscribed strand of 5hmC-GFP DNA is 2.6-fold higher than that on the transcribed strand at 48 hrs after transfection (Figure 5D). This strand preference was not due to an asymmetric distribution of cytosines because it was observed in both CpHs and symmetrically distributed CpGs (Figure 5D).
Taken together, these results reveal an unexpected similarity between two seemingly unrelated processes - 5hmC demethylation and AID-mediated somatic hypermutation, and further support the role of deamination in active 5hmC demethylation.
TET1 and AID/APOBEC deaminases promote region-specific DNA demethylation in the adult mouse brain
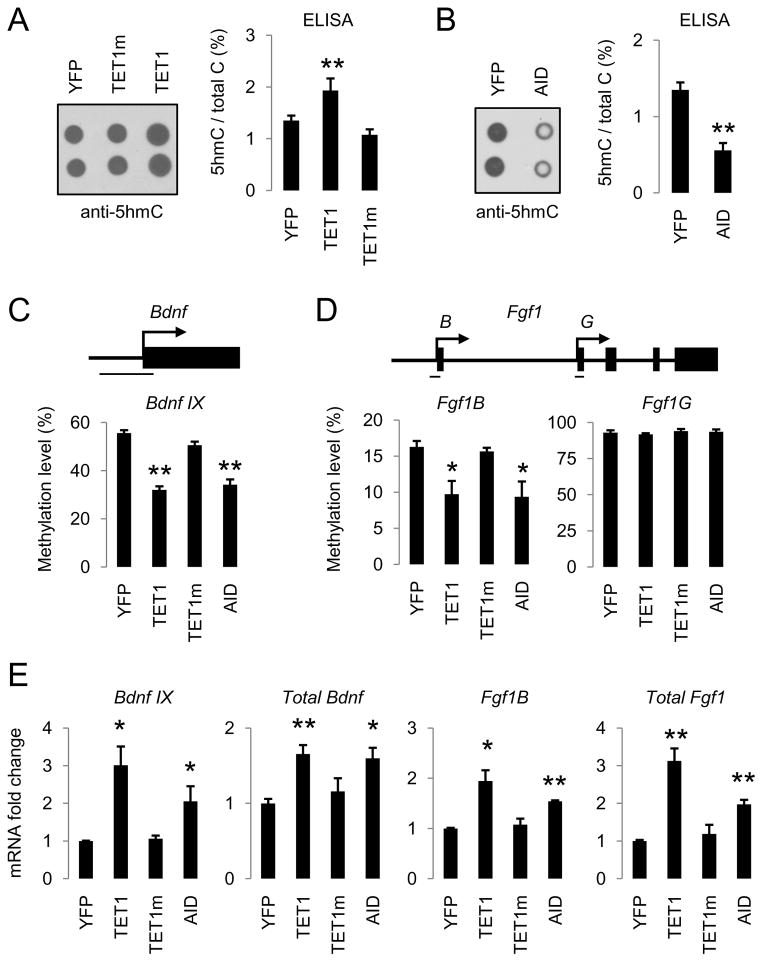
We next examined whether the TET1-induced oxidation-deamination mechanism regulates active DNA demethylation in mature neurons of the adult mouse brain in vivo. Notably, 5hmC has been shown to be abundant in various adult brain regions, including the hippocampal dentate gyrus (Munzel et al., 2010). We stereotaxically injected adeno-associated viruses (AAVs) overexpressing TET1, or TET1m, into the adult mouse dentate gyrus and measured 5hmC levels in micro-dissected dentate gyrus tissue 1 week after viral injection (Figure S6A). Despite the presence of a high basal level of 5hmCs (1.35% of all cytosines), overexpression of TET1, but not TET1m, further increased the 5hmC level by 43% (Figure 6A). On the other hand, AAV-mediated overexpression of AID significantly decreased the endogenous level of 5hmCs by 59% in the adult dentate gyrus (Figure 6B), supporting its role in 5hmC removal in vivo.
Figure 6. TET1 and AID Regulate Endogenous 5hmC Levels and Promote DNA Demethylation in the Adult Mouse Brain.
(A and B) Effects of AAV-mediated overexpression of control YFP, TET1, TET1m (A), and AID (B) on the endogenous 5hmC levels as measured by immunoblotting (left) and ELISA (right). Engineered AAV viruses were stereotaxically injected into the dentate gyrus of adult mice and dentate gyrus tissue was micro-dissected one week later for analysis. Values represent mean ± SEM (n = 3 animals for each condition; **: P < 0.01; Student’s t-test).
(C and D) Effects of YFP, TET1, TET1m and AID overexpression on methylation levels of Bdnf IX (C), Fgf1B and Fgf1G (D) alternative promoters in the adult dentate gyrus. Shown on top are diagrams of promoters and bars indicate regions for bisulfite sequencing analysis. Values represent mean ± SEM (n = 3 animals for each condition; **: P < 0.01; *: P < 0.05; Student’s t-test).
(E) Effects of YFP, TET1, TET1m and AID overexpression on specific isoform and total expression levels of Fgf1 and Bdnf in the adult dentate gyrus. Same groups of animals as in (C and D) were examined. Values represent mean ± SEM (**: P < 0.01; *: P < 0.05; Student’s t-test).
To determine whether TET1 and AID promote DNA demethylation in the adult brain in vivo, we examined the CpG methylation status of promoter IX of Bdnf (Bdnf IX) and a brain-specific promoter of Fgf1 (Fgf1B), two endogenous loci that have been shown to exhibit neuronal activity-induced active DNA demethylation in the adult dentate granule cells (Ma et al., 2009b). AAV-mediated overexpression of TET1 or AID, but not TET1m, led to significant decreases in the CpG methylation levels at these two genomic loci in dentate neurons from multiple animals (Figures 6C, 6D, S6B and S6C). In contrast, the CpG methylation status of a non-neuronal promoter of Fgf1 (Fgf1G) was not altered by these manipulations (Figures 6D and S6D). Consistent with our previous finding that the methylation status of Bdnf IX and Fgf1B promoters regulates their gene expression (Ma et al., 2009b), both TET1- and AID-induced demethylation was accompanied by a significant up-regulation in transcript levels of these two gene isoforms in vivo (Figure 6E). Taken together, these results suggest that TET1 and AID are sufficient to promote CpG demethylation of specific endogenous genomic loci and to increase expression of associated genes in the adult mouse brain.
Tet1 and Apobec1 are involved in neuronal activity-induced DNA demethylation in the adult mouse brain
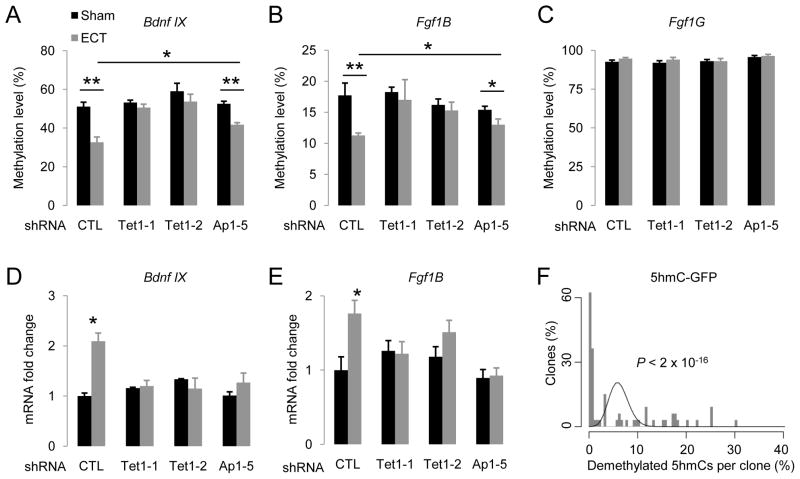
Our previous studies have shown that synchronous activation of adult dentate granule neurons in vivo by electroconvulsive stimulation (ECS) leads to CpG demethylation of Bdnf IX and Fgf1B promoters in these neurons within 4 hrs, but has no effect on the Fgf1G promoter (Ma et al., 2009b). To assess whether Tet1 is required for neuronal activity-induced DNA demethylation in vivo, we injected engineered AAVs that express short-hairpin RNAs (shRNAs) to knock down the endogenous Tet1 expression in the adult dentate gyrus (Figure S7A). The two shRNAs against mouse Tet1 have been previously characterized (Table S1) (Ito et al., 2010). We further confirmed the effectiveness of these two shRNAs in knocking down endogenous mouse Tet1 both in vitro and in vivo (Figure S7B). Interestingly, expression of either shRNA against mouse Tet1, but not the control shRNA, completely abolished ECS-induced demethylation of both Bdnf IX and Fgf1B, but not Fgf1G, promoters in dentate neurons from multiple animals (Figures 7A to 7C; S7E to S7G). ECS-induced expression of these two gene isoforms was also abolished (Figures 7D and 7E). Thus, endogenous Tet1 is required for neuronal activity-induced, region-specific, active DNA demethylation and gene expression in the adult brain.
Figure 7. Tet1 and Apobec1 are Involved in Neuronal Activity-Induced Region-Specific Demethylation in the Adult Mouse Brain.
(A to C) Effects of AAV-mediated shRNA knock-down of endogenous Tet1 and Apobec1 on ECS-induced CpG demethylation of Bdnf IX (A), Fgf1B (B) and Fgf1G (C) promoters. Engineered AAV viruses expressing control shRNA (CTL), two shRNAs against mouse Tet1 (Tet1-1 and Tet1-2), and one shRNA against mouse Apobec1 (Ap1-5), were stereotaxically injected into the dentate gyrus of adult mice. One week after viral injection, animals were subjected to ECS or sham treatment. Dentate gyrus tissue was micro-dissected 4 hrs later for analysis. Values represent mean ± SEM (n = 3–6 animals for each condition; **: P < 0.01; *: P < 0.05; Student’s t-test).
(D and E) Effects of Tet1 and Apobec1 knock-down on ECS-induced isoform-specific expression of Bdnf and Fgf1 in the adult dentate gyrus. Same groups of animals as in (A to C) were examined. Values represent mean ± SEM (*: P < 0.05; Student’s t-test).
(F) Demethylation of 5hmC-GFP DNA fragments after transfection into primary hippocampal neurons in culture. Shown is a histogram of clone distribution of the percentage of demethylated 5hmCs in 5hmC-GFP DNA fragments retrieved 7 days after transfection. A distributive Poisson model (λ = 6.2%) is shown by a black curve.
To directly examine whether 5hmCs can be demethylated in neurons, we transfected 5hmC-GFP DNA fragments into primary neurons in culture. Bisulfite sequencing analysis showed significant demethylation of 5hmC DNA in both CpG (~5%) and CpH (~6%) contexts. Interestingly, the numbers of demethylated 5hmCs in individual clones also exhibited a broad distribution, suggesting high processivity (Figure 7F).
To identify the endogenous Aid/Apobec that may contribute to neuronal activity-induced DNA demethylation in vivo, we examined the expression of these genes under normal conditions and 4 hrs after ECS in the adult dentate gyrus. While Aid, Apobec2 and Apobec4 appeared to be absent, Apobec1 and Apobec3 were expressed in the adult dentate gyrus at similar levels with or without ECS (Figure S7C). We obtained one shRNA (Ap1-5) that achieved effective knockdown of endogenous Apobec1 expression in culture and in the adult dentate gyrus after AAV-mediated expression (Figure S7D; Table S1). Interestingly, knockdown of Apobec1 significantly reduced ECS-induced demethylation of both Bdnf IX and Fgf1B promoters in dentate neurons from multiple animals in comparison to those expressing the control shRNA (Figures 7A to 7C; S7E to S7G). Furthermore, ECS-induced expression of these two gene isoforms was completely abolished (Figures 7D and 7E). It remains to be determined whether Apobec3 also plays a role in neuronal activity-induced DNA demethylation and expression of associated genes in the adult brain.
Taken together, these results demonstrated that neurons possess a significant capacity to demethylate 5hmCs and suggest that Tet1/Apobec1-mediated oxidation-deamination mechanism plays a critical role in neuronal activity-induced, region-specific, active DNA demethylation in the adult mouse brain.
DISCUSSION
Accumulating evidence supports the existence of active DNA demethylation in vertebrate cells, yet the underlying molecular mechanisms remain unclear (Wu and Zhang, 2010). The discovery of the TET family of 5mC hydroxylases has raised the question of its potential role in active DNA demethylation. Here we identify TET1-catalyzed 5mC hydroxylation as a key initiating step in DNA excision-repair based active DNA demethylation in mammalian cells both in vitro and in vivo. By introducing fully modified linear dsDNA substrates with 5hmCs into HEK293 cells and primary mouse neurons, we demonstrate that mammalian cells possess a robust, active demethylating machinery targeting 5hmC-containing DNA in both CpG and non-CpG contexts. We further provide mechanistic insights into this novel activity by pharmacological inhibition of key DNA repair enzymes and genetic manipulations of core enzymes in the BER pathway, including DNA glycosylases and AID/APOBEC deaminases. In cultured human cells, AID/APOBEC deaminases specifically promote 5hmC demethylation, but have no apparent effect on 5mCs. In the adult mouse brain, AID facilitates the removal of endogenous 5hmCs and, furthermore, both TET1 and AID promote region-specific DNA demethylation in dentate neurons. Finally, loss-of-function experiments establish an important role of endogenous Tet1 and Apobec1 in neuronal activity-induced DNA demethylation and expression of associated genes in dentate neurons in the adult mouse brain. Together, our results delineate a TET1/APOBEC-mediated oxidation-deamination mechanism underlying active, region-specific DNA demethylation in mammals. Previous studies have implicated either 5hmC or deaminated 5mC as the potential key intermediate for DNA demethylation. Our work thus reconciles those previous studies and suggests a convergence of two seemingly separate mechanisms into one single cooperative pathway for BER-mediated completion of active DNA demethylation.
Oxidation of 5mCs had been previously proposed to cause passive DNA demethylation by preventing DNMT1 recognition in vitro, which recognizes hemi-methylated CpGs after DNA replication to maintain a symmetric methylation pattern (Valinluck and Sowers, 2007). We present evidence for an unequivocal role of 5mC hydroxylation in promoting DNA demethylation in a DNA replication-independent manner by using non-replicating SssI-methylated plasmids and linear pre-modified dsDNA substrates. Consistent with a role in active DNA demethylation, TET proteins are expressed in mouse primordial germ cells (Hajkova et al., 2010) and in the dentate gyrus of the adult mouse brain (Szwagierczak et al., 2010), where genome-wide and locus-specific DNA demethylation occur (Hajkova et al., 2010; Ma et al., 2009b), respectively. The adult dentate gyrus provides a relative homogenous population of post-mitotic neurons in large numbers (Ma et al., 2009b), which is particularly important for epigenetic analysis because each diploid cell displays only two locus-specific binary modifications. Although mRNAs of all three Tet genes are detected in the mouse hippocampus (Szwagierczak et al., 2010), specific knock-down of Tet1 abolished neuronal activity-induced promoter demethylation of Bdnf IX and Fgf1B, suggesting non-redundant roles of different TET proteins in DNA demethylation. All three TET proteins exhibit 5 mC hydrolysis activity (Ito et al., 2010) and it will be interesting to explore their roles in genome-wide and region-specific DNA demethylation in different cell types.
Previous studies (Hajkova et al., 2010; Rai et al., 2008) and our current result support a role of BER in active DNA demethylation, yet none of the known mammalian DNA glycosylases has been shown to possess 5hmC glycosylase activity. Instead, our result suggests that 5mCs, once converted to 5hmCs by TET proteins, are processed by AID/APOBEC deaminases, a family of Zn2+-dependent cytidine deaminases that have been implicated in active DNA demethylation in vertebrates (Bhutani et al., 2010; Conticello et al., 2007). AID appears to specifically promote demethylation of 5hmCs, but not 5mCs in mammalian cells. In a recent study of reprogramming-associated demethylation of OCT4 promoter, AID was shown to occupy its target loci even before demethylation was initiated (Bhutani et al., 2010), suggesting a preceding event to AID catalysis. In support of a role of AID/APOBEC deaminases in TET1-induced DNA demethylation, 5hmC demethylation shares many known characteristics of AID-induced deamination, including high processivity, sequence selectivity, transcription-dependency, and DNA strand preference. Interestingly, the 5hmC demethylation enhancing ability is not limited to AID, but is also present in multiple APOBEC deaminases. Our results do not rule out the possibility that AID/APOBEC deaminases may act on tertiary intermediates rather than on 5hmCs directly, nor can we eliminate the possibility of other pathways that process 5hmCs independently of AID/APOBEC deaminases in mammalian cells (Figure 4F). AID/APOBEC deaminases are usually expressed in a highly restricted and tissue-specific manner and exhibit distinct sequence selectivity for deamination (Conticello et al., 2007), therefore different deaminases may mediate 5hmC demethylation cooperatively with their individual sequence preferences. Previous in vitro analysis has revealed different capacities of AID/APOBEC family members in deaminating unmethylated cytosines and 5mCs (Conticello et al., 2007). Future studies of in vitro 5hmC deamination will provide critical insight into the role of AID/APOBEC family members in DNA demethylation.
In summary, our results uncover a novel role of TET1, through conversion of 5mC into a critical intermediate product 5hmC, in promoting active DNA demethylation in mammalian cells both in vitro and in vivo. Besides a well-established role in cancer (Jones and Baylin, 2002), accumulating evidence implicates epigenetic aberrations in DNA methylation in neurodegenerative diseases (Urdinguio et al., 2009) and psychiatric disorders (Feng and Fan, 2009). Identification of novel roles for TET1 and 5hmC in neuronal activity-induced DNA demethylation will enhance our understanding of molecular mechanisms underlying dynamic changes of DNA methylation in the adult nervous system and provide new therapeutic targets for novel treatments.
EXPERIMENTAL PROCEDURES
DNA constructs
Human open reading frame entry clones were either obtained from Johns Hopkins HIT Center, or cloned from a human cDNA library. Fully modified linear UbC-GFP fragments and promoter-truncated variants were generated using dCTP, 5mdCTP (Amersham), or 5hmdCTP (Bioline) in PCR amplifications (Table S1C). AAV gene delivery vectors were constructed by cloning the EF1a-Gene-WPRE and U6-shRNA-EF1a-EYFP-WPRE cassette (Ge et al., 2006) into an AAV backbone. The efficacy of shRNAs against mouse Tet1 and Apobec1 (Table S1E) were tested in cell culture after electroporation and confirmed in adult mouse dentate gyri after AAV-mediated expression.
Methylated reporter assay and quantitative analysis of DNA methylation
NIT-GFP plasmid was used for methylation reporter assay as previously described (Ma et al., 2009b). The methylation-sensitive restriction assay was carried out using a StepOnePlus Real-Time PCR system (Applied Biosystems; Table S1A). HpaII sensitivity of a CCGG site was calculated by [1–2Ct(mock)-Ct(HpaII)] × 100%. Bisulfite (Zymo)-treated DNA was used as a template for PCR amplification of the region of interest as previously described (Table S1B) (Ma et al., 2008). Primers targeting non-bisulfite converted sequences were used in bisulfite sequencing analysis of 5hmC-GFP DNA fragments. Although these primers do not selectively amplify either strand, demethylation of the forward (non-transcribed) strand would appear as C-to-T transitions in Sanger sequencing results, whereas G-to-A transitions represent demethylation of the reverse (transcribed) strand (Figure S5C).
Detection and quantification of 5hmC and 5hmU by immunoblotting, ELISA and immunocytochemistry
DNA samples were applied onto Hybond-N+ membrane (Amersham), cross-linked by a UV stratalinker 1800 (Stratagene), and subjected to immunoblotting using antibodies against 5hmC (Active motif; 1:10000) or against 5hmU (Abcam; 1:2000). For ELISA quantification of 5hmC, DNA standards and samples were immobilized on a 96-well plate using Reacti-Bind DNA coating solution (Thermo Scientific). Chemiluminescent signals were developed using a TMB substrate (Thermo Scientific) and monitored by SpectraMax Plus 384 plate reader (Molecular Devices). Immunocytochemistry used following primary antibodies: anti-HA (rat; 1:500; Roche); anti-5hmC (rabbit; 1:500; ActiveMotif); anti-5mC (mouse; 1:250; Eurogentec); anti-V5 (goat; 1:500; Abcam).
Methylation and gene expression analysis of the adult mouse dentate gyrus
Adult mice (8–10 weeks old, male, C57BL/6 background) were used for analysis in accordance with protocols approved by the Institutional Animal Care and Use Committee. High titers of engineered AAV2/9 were stereotaxically injected into the dentate gyrus of adult mice as previously described (Ge et al., 2006). Mice were used one week after viral injection for analysis. ECS was administered as previously described (Ma et al., 2009b). Sham animals were similarly handled in parallel without the current delivery. Animals were analyzed at 4 hrs after ECS. DNA methylation and gene expression were carried out as previously described (Table S1B and S1D) (Ma et al., 2009b).
Supplementary Material
Acknowledgments
We thank D.K. Ma and K. Christian for comments, members of Song and Ming laboratories for discussion, Y. Gao for RNA-seq, H.C. Kang, J.S. Jeong, N.Y. Jung and J. Shin for help with molecular cloning, Y. Cai and L. Liu for technical support. This work was supported by AMRF and NIH (HD069184, NS048271) to G.L.M., by NIH (NS047344, AG024984, MH087874) and NARSAD to H.J.S., and by fellowships from FARMS to J.U.G, AMRF to Y.S., and MSCRF to C.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker D, Liu P, Burdzy A, Sowers LC. Characterization of the substrate specificity of a human 5-hydroxymethyluracil glycosylase activity. Chem Res Toxicol. 2002;15:33–39. doi: 10.1021/tx010113b. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein RJ, Cummings A, Jr, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J Biol Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1299–1440. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010a;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogamming in plant and animla development. Science. 2010b;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Goh EL, Sailor KA, Kitabatake Y, Ming GL, Song H. GABA regulates synaptic integration of newly generated neurons in the adult brain. Nature. 2006;439:589–593. doi: 10.1038/nature04404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Jeffries SJ, Lee C, Miller N, Jackson SP, Surani MA. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larijani M, Frieder D, Sonbuchner TM, Bransteitter R, Goodman MF, Bouhassira EE, Scharff MD, Martin A. Methylation protects cytidines from AID-mediated deamination. Mol Immunol. 2005;42:599–604. doi: 10.1016/j.molimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Chiang CH, Ponnusamy K, Ming GL, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming GL, Song H. DNA excision repair proteins and Gadd45 as molecular players for active DNA demethylation. Cell Cycle. 2009a;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009b;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Marchetto MC, Guo JU, Ming G, Gage FH, Song H. Epigenetic choreographers of neurogenesis in the adult mammalian brain. Nat Neurosci. 2010;13:1338–1344. doi: 10.1038/nn.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005;33:4711–4724. doi: 10.1093/nar/gki781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Munzel M, Globisch D, Bruckl T, Wagner M, Welzmiller V, Michalakis S, Muller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell MR, Whish WJ. Novel inhibitors of poly(ADP-ribose) synthetase. Biochem J. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Simonsson S, Gurdon J. DNA demethylation is necessary for the epigenetic reprogramming of somatic cell nuclei. Nat Cell Biol. 2004;6:984–990. doi: 10.1038/ncb1176. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8:1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577:275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of alpha-Ketoglutarate-Dependent Dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QM, Yonekura S, Takao M, Yasui A, Sugiyama H, Yonei S. DNA glycosylase activities for thymine residues oxidized in the methyl group are functions of the hNEIL1 and hNTH1 enzymes in human cells. DNA Repair (Amst) 2005;4:71–79. doi: 10.1016/j.dnarep.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.