Abstract
Precise transcriptional control is dependent on specific interactions of a number of regulatory elements such as promoters, enhancers and silencers. Several studies indicate that the genome in higher eukaryotes is divided into chromatin domains with functional autonomy. Chromatin domain boundaries are a class of regulatory elements that restrict enhancers to interact with appropriate promoters and prevent misregulation of genes. While several boundary elements have been identified, a rational approach to search for such elements is lacking. With a view to identifying new chromatin domain boundary elements we analyzed genomic regions between closely spaced but differentially expressed genes of Drosophila melanogaster. We have identified a new boundary element between myoglianin and eyeless, ME boundary, that separates these two differentially expressed genes. ME boundary maps to a DNaseI hypersensitive site and acts as an enhancer blocker both in embryonic and adult stages in transgenic context. We also report that BEAF and GAF are the two major proteins responsible for the ME boundary function. Our studies demonstrate a rational approach to search for potential boundaries in genomic regions that are well annotated.
INTRODUCTION
Chromatin domain boundaries are elements that prevent cross talk between regulatory elements of neighboring genes and, thereby, facilitate proper transcriptional control. It has been proposed that boundary elements are present throughout the genome and functionally subdivide the genome into independent units called chromatin domains. Regulatory elements are restricted to the target genes within such domains as boundaries prevent inappropriate crosstalk among regulatory elements in different domains. However, only a handful of such boundary elements are known till date (1–13). Chromatin domain boundary elements prevent communication between enhancers and promoters when placed between the two but mechanisms involved in this process are poorly understood. Various assays have been developed to study boundaries that include insulator, enhancer blocker and barrier assays (14–19). Some of the boundary elements possess both enhancer blocker and barrier activities and these different functions are attributed to different proteins (20). These studies have opened exciting possibilities associated with boundaries in higher order chromatin organization but much remains to be understood about how boundaries function, primarily because very few boundaries have been identified and studied in detail. It is, therefore, highly desirable to identify new chromatin domain boundaries and their interacting partners. Identification of the proteins that associate with boundaries may also lead to understanding how these elements function. Few proteins responsible for boundary function of different known boundaries have been identified (21–26) but the molecular mechanism by which they contribute to the boundary function has remained largely unclear.
It is speculated that thousands of such elements must be present in every genome for proper gene regulation. Very few new boundaries have been identified of late and the reason for this slow progress is the lack of a rational and functional screening for new boundaries. One of the ways to identify new boundaries may be via genome-wide binding studies of boundary interacting proteins. Such studies have been carried out recently (27–30), but all the binding sites identified may not reflect boundaries as these proteins are also known to have additional functions. In the current study we present a boundary search approach based on the logic that closely spaced genes that are differentially expressed must be insulated from the regulatory elements of one another and one of the mechanisms to achieve this may be the presence of a boundary element separating the domains of the two neighboring genes. We analyzed the fourth chromosome of Drosophila with this rationale and mapped a chromatin domain boundary between myoglianin and eyeless genes. We refer to it as myoglianin–eyeless, ME boundary. Our studies show that ME boundary contains a DNaseI hypersensitive site and it functions as a boundary both in adult and embryonic stages of Drosophila. Sequence analysis of ME boundary revealed binding sites for known boundary interacting proteins BEAF (boundary element associated factor) (26,31,32) and GAF (GAGA factor, a.k.a. trithorax like, Trl) (33). Earlier studies have shown that binding sites of proteins on the boundary element and presence of normal levels of the proteins are needed for the activity of several boundary elements (25,32). Using genetic and molecular approaches, we further show that BEAF and GAF are positive regulators of the ME boundary function and they contribute to the boundary function of ME by directly binding to it.
MATERIALS AND METHODS
DNaseI hypersensitivity assay
Nuclei were prepared from 0–16 h old Canton-S embryos essentially as described earlier (34) and digested with DNase I in a controlled manner for 0.5 to 5 min. About 100 U/ml DNaseI concentration was used and reactions were terminated at every half minute interval by adding 20 mM EDTA and 1% SDS. DNA was isolated from chromatin using standard phenol/chloroform extraction. An aliquot from each digest was analyzed on agarose gel for appropriate DNaseI digestion pattern (smear pattern) and samples from 2 to 4 min digests were pooled and digested completely with EcoRI restriction endonuclease. After complete digestion with EcoRI, DNA was loaded on a 40 cm gel and electrophoresis was performed for 10–12 h at 40 V. The temperature of the buffer was kept constant by attaching the electrophoresis tank to a water bath. After electrophoresis the DNA was transferred from the gel to positively charged nylon membrane using vacuum transfer. Southern hybridization was performed according to the DIG protocol (Roche) using probe from the region mentioned in text.
Cloning of ME test fragments in P-element derived boundary assay vectors
Test fragments from ME region were PCR amplified using different set of primers and cloned into boundary assay vectors (Supplementary Figure S1). In brief, PCR product was first cloned in pTZ57R/T (a PCR cloning vector from Fermentas Life Sciences) and excised out as EcoRI-HindIII fragments and cloned into EcoRI/HindIII double digested pLML vector that contains loxP sites flanking the MCS (35,36). ME test DNA flanked by loxP sites was excised out from the LML construct and cloned into XhoI digested boundary assay vectors RW+, YW and pCfhL. As indicated in Supplementary Figures S1 and S3, 1583 bp ME fragment was used in RW+ and YW while 917 bp fragment was used in pCfhL vector.
P-element transformation and scoring of reporter gene expression
Transposon injections were done in early embryos of w1118 flies essentially as described (37) and transformants were selected by rescue of the eye color. Transgenic lines used in this study are listed in Supplementary Table S2. Levels of mini-white expression of the different transgenic lines were determined from 4-day-old flies of approximately the same size (38). Genetic interaction of transgenic lines with different mutations (listed in Supplementary Table S3) was checked by crossing virgin females from the mutant stocks with males from the transgenic ME lines. This was done to take advantage of possible maternal effect of the mutant loci.
Quantitative measurement of eye pigment
Adult heads were homogenized in 1 : 1 mixture of chloroform/ammonium hydroxide (0.1%) and this mixture was centrifuged. Supernatant was taken to read absorbance at 485 nm. Blank used was chloroform/ammonium hydroxide (0.1%).
Wing images
For taking wing images, flies were aged for 3–4 days and then kept in a solution of glycerol/ethanol (1 : 10) for 3–4 days. Finally the wings were cut in water, mounted on a slide in Hoyer's medium and images were taken.
β-Galactosidase assay
CS and transgenic embryos (0–6-h old) were collected, dechorionated in 50% bleach (NaOCl, sodium hypochlorite) and washed thoroughly with PBS. Then embryos were fixed in saturated heptane for 20 min at room temperature on rotating shaker. About 10 ml of heptane was saturated with 5 ml PBS and 5 ml gluteraldehyde (25%), phases were allowed to separate and the top phase of heptane was recovered. After fixation, fixing solution was removed and washed for additional 20–30 min with PBSTr (0.3% Triton X-100). Embryos were incubated in coloration solution for 30 min at 37°C and later replaced with coloration solution supplemented with 1.0% X-gal and incubated for additional 2–10 h at 37°C (covered with aluminum foil) until clear color pattern developed. After staining, coloring solution was removed, washed in PBST and finally, photographed. When testing effect of different mutations (BEAF and Dref), we used virgins from mutant stock balanced over CyO;wg-lacZ balancer chromosome to distinguish embryos carrying the mutant allele.
RNA in situ hybridization
Whole mount RNA in situ hybridizations were carried out using 0–6-h-old embryos of pCfhl-ME (917) transgenic and flipped out flies. DIG labeled RNA probes were made for lacZ and white mRNA and in situ hybridization was performed as described earlier (39,40) with a small change—instead of Proteinase K treatment 80% acetone was used as described in Nagaso et al. (41).
ImmunoFISH on polytene chromosomes
Flies were reared at 18°C under non-crowding conditions and third instar larvae were selected for polytene preparation. Polytene chromosomes were prepared as described earlier (42). The slides with good polytene preparations were rinsed in PBS for 15 min at room temperature, kept in 2× SSC at room temperature and then into a 65°C water bath for 45 min. They were then dehydrated by passing through 70% ethanol and 96% ethanol (twice for 5 min each). The chromosomal DNA was denatured by incubating in 0.07 M NaOH for 10 min. After denaturation the slides were washed in 2× SSC twice for 1 min each and finally for 5 min. The slides were dehydrated by passing through 70% ethanol and 96% ethanol as above and dried mildly after dehydration. Hybridization mixture was then added to the slide and covered by a cover slip and incubated at 80°C for 5 min (formamide 50%, SSC 2X, dextransulfate 10%, salmon sperm DNA 0.8 mg/ml). Dig labeled DNA (1 μg of ME DNA or lacZ DNA) was prepared using the Nick translation kit from Roche. The probe, purified by Qiagen reaction clean up kit, was mixed with hybridization solution and incubated at 95°C for 5 min. The denatured probe was added to the slide and covered with a cover slip. The slide was incubated in a humid chamber at 37°C for 12–16 h. After hybridization the cover slip was removed and the slide was washed in 2× SSC, three times for 5 min each at 42°C and once for 5 min at RT. Thereafter the slide was washed in PBS for 15 min. The slides were then incubated in blocking solution for 1 h at room temperature and then rinsed in PBS for a minute to remove the milk powder. Primary antibody [anti BEAF (1 : 10 dilution) or anti GAF (1 : 100 dilution)] diluted in PBTx was added and the slide was incubated in a humid chamber for 1 h at 37°C. After incubation the slides were rinsed with PBS twice, 5 min each at room temperature. The slides were then washed for 15 min in wash solution A (1× PBS, 300 mM NaCl, 0.2% Tween-20, NP40 0.2%) and for 15 min in wash solution B (1× PBS, 400 mM NaCl, 0.2% Tween-20, 0.2% NP40) while shaking the rack thoroughly. The slides were then rinsed in PBS for 5 min. Secondary antibody (anti-DIG Cy3 1 : 300 dilution) diluted in PBTx was added and the slide was incubated in a humid chamber for 1 h at RT. The slides were rinsed in PBS for 5 min. Second secondary antibody [anti-mouse FITC (1 : 200 dilution) or anti-rabbit FITC (1 : 200 dilution)] diluted in PBTx was added and the slide was incubated in a humid chamber for 1 h at RT. After incubation the slides were rinsed with PBS for 5 min. The slides were then washed for 15 min each in wash solution A and B while shaking the rack thoroughly. Finally, the slides were rinsed with PBS for 5 min. Mounting media with DAPI was added to the slide and covered with a cover slip. The slides were examined under confocal microscope.
Chromatin preparation
Chromatin was prepared from FLAG-BEAF expressing larvae (wild-type and different mutant background as indicated in the text), essentially as described earlier (43). In experiments where embryos were used for chromatin preparation (for GAF pull down shown in Figure 7b), one gram of freshly dechorionated Canton-S embryos (0–16 h) were washed several times in PBST, centrifuged at 500g for 1 min and resuspended in 10 ml crosslinking solution (Crosslinking solution: 50 mM Hepes pH 7.6, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 100 mM NaCl, 1.8% Formaldehyde) and 30 ml heptane. The embryos were cross-linked for 15 min at room temperature with vigorous shaking and then centrifuged for 1 min at 500g, resuspended in PBST-glycine (125 mM) and allowed to sediment. The embryos were then washed with ice cold PBST, and resuspended in ice-cold PBST containing protease inhibitors. They were then homogenized using Wheaton Dounce Tissue Grinder (10 times). Homogenate was centrifuged at 400g at 4°C for 1 min. The supernatant was then centrifuged at 1100g for 10 min at 4°C. Purified cells were resuspended in cell lysis buffer (5 mM Hepes pH 8.0, 85 mM KCl, NP-40 0.5%) containing DTT, PMSF and protease inhibitor mixture. The extract was centrifuged at 1300g for 4 min at 4°C and the pellet was resuspended in cell lysis buffer containing DTT, PMSF and protease inhibitor mixture. The extract was centrifuged at 2000g for 4 min at 4°C. The nuclei were washed once with cell lysis buffer and incubated in nuclei lysis buffer (Nuclei lysis buffer: 50 mM Tris.HCl pH 8.0, 10 mM EDTA, 1% SDS) containing DTT, PMSF and protease inhibitor mixture for 20 min on ice with gentle shaking. SDS concentration (0.1%) was maintained by adding immunopercipitation buffer to nuclei. The chromatin was sheared to an average of 300–600 bp by sonication (Biorupter™).
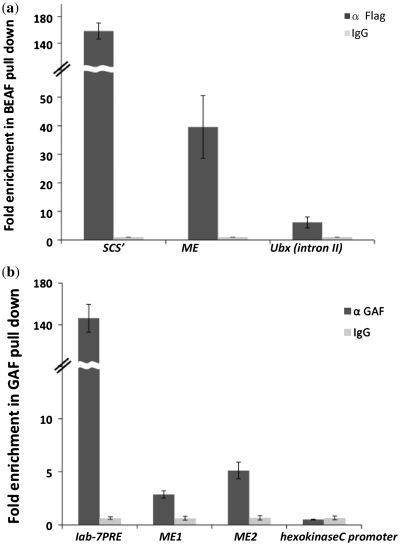
Figure 7.
ChIP analysis of BEAF and GAF in ME region. (a) Chromatin with Flag-tagged BEAF was immunoprecipitated after sonication using anti FLAG-M2 agarose and analyzed by real-time PCR. Fold enrichment at the endogenous ME, Ubx intron II region (negative control) and SCS’ (positive control) loci were calculated as relative enrichment over the mock control, and the averages from three independent experiments, with error bars, are shown. (b) Enrichment of GAF at the ME boundary, iab-7PRE (positive control) and hexo kinaseC promoter (negative control) regions. Chromatin was immunoprecipitated after sonication using anti GAF antibody and analyzed by real-time PCR. Fold enrichment were calculated as relative enrichment over the mock control, and averages from three independent experiments, with error bars, are shown.
Immunoprecipitation was performed using α-FLAG M2 agarose (Sigma) and α-GAF antibody generated in the lab (43,44) following the ChIP protocol provided with Upstate Biotechnology ChIP Assay Kit. In brief, ∼40 μg of the chromatin was incubated with 75 μl Protein A sepharose resin slurry (Upstate Biotechnology) for an hour at 4°C. Pre-cleared chromatin was incubated without antibody or with α-FLAG M2 agarose (Sigma) or α-GAF antibody overnight at 4°C. Chromatin from chromatin–antibody–resin complex was recovered by centrifugation followed by several washing steps. Chromatin was eluted in elution buffer at room temperature (Upstate kit). DNA was isolated by phenol/chloroform extraction and ethanol precipitation in the presence of 20 μg glycogen. Precipitated DNA was resuspended in equal volume and analyzed by real-time PCR.
Real-time PCR and quantification
Each PCR reaction contained 2 pmol of each primer, 1× SYBR Green master mix with HotStart Taq polymerase (Applied Biosystems). PCR was performed and monitored in 7900HT Fast Real-Time PCR System: 4 min activation of Taq at 95°C, followed by 40 cycles of 94°C 10 s, 56°C 20 s, 68°C 20 s. Product formation was detected at 60°C in the fluorescein isothiocyanate channel. Enrichment was calculated based on Δct values, difference of Ct between specific antibody and mock pull down. Primers used for PCR analysis is listed in Supplementary Table S4.
Mutagenesis of BEAF sites
Five BEAF binding sites in the ME (917) including two palindromic site in the core region (cloned in pLML vector) were mutated by site directed mutagenesis using QuickChangeTM Site directed mutagenesis Kit from Stratagene. BEAF motifs, CGATA, were changed to CTCGA sequentially, using overlapping primers having mutation at the corresponding sites, PCR amplified and cloned in CfhL.
RESULTS
Sequence analysis of fourth chromosome for potential boundary elements
The rationale used for a region to have boundary potential was that closely spaced but differentially expressed genes must have a boundary in between the two genes to prevent regulatory crosstalk. We selected the fourth chromosome of Drosophila melanogaster to look for new boundaries by this approach. A whole chromosome analysis of the fourth chromosome is easy as it is the smallest chromosome in fly. Three fourth of this chromosome is a heterochromatin block and the remaining ∼1.2 Mb of euchromatin region is also interspersed with heterochromatic regions (45). Another interesting feature that distinguishes the fourth chromosome from other chromosomes is the presence of large number of repeats (46,47) although the gene density on this chromosome is the same as on the other chromosomes.
We analyzed the intergenic regions and looked at various features associated with them, for example, presence of MARs, patterns of repeats, etc. The fourth chromosome sequence taken from NCBI (NC_004353.2) consists of 81 predicted genes (61.5% of the entire chromosome) of which 62 were found to be transcribed. There are 80 intergenic regions, the smallest intergene, 154 bp, is located between CG11076 and ATPsyn-beta and the longest intergene, 45434 bp, is located between CG2052 and lgs. Twenty-three intergenic regions were flanked by convergent transcribed genes whereas 24 intergenic regions were flanked by divergent transcribed genes; the rest of the intergenes were flanked by genes transcribed in the same direction.
The approach we took to identify chromatin domain boundaries in intergenic regions was to focus on small (few kb) intergenic regions that separate differentially expressed genes, as these are likely to possess insulator functions needed to prevent cross talk between regulatory elements of the flanking genes. Among various potential boundary containing regions, we selected few intergenic regions for further analysis based on features likely to be associated with boundary elements such as differential expression of genes flanking the intergene, presence of motifs for boundary binding proteins, MAR potential (48), etc. (Supplementary Table S1). Finally, we carried out detailed functional analysis of a 1.6 kb intergenic region (49ig50) which separates two differentially expressed genes myoglianin and eyeless (Supplementary Figure S1 and Supplementary Table S1).
The Drosophila, eyeless (ey), Pax-6 gene homolog, is expressed in the eye imaginal disc primordia and central nervous system. In third instar larvae, ey expression is visible in the optic lobes of the brain, in several spots of the ventral ganglion and in the eye imaginal discs, where it is restricted to the undifferentiated cells anterior to the morphogenetic furrow (49). The second intron of ey gene contains an enhancer that regulates the eye specific expression of the gene in the eye disc primordia of embryos and in the eye imaginal disc of the third instar larvae (50). myoglianin, on the other hand, belongs to the TGF-β superfamily and is closely related to the vertebrate muscle differentiation factor Myostatin and BMP-11 (51). myoglianin is expressed throughout in Drosophila life cycle. In situ data showed that in preblastoderm embryos, a high level of maternally deposited myoglianin transcript is uniformly distributed throughout the embryo. By stage 14, strong expression is evident in glial cells. This glial expression is completely lost in stage 15 embryos and instead expression is detected in the somatic, visceral, and heart musculature, which persists through late embryogenesis. In third instar larvae the expression pattern is observed in the brain and ventral nerve cord where it is expressed in the glial cells (51). Due to this, clearly distinct spatial expression profile and dynamics, we reasoned that the 1.6 kb region separating myoglianin and eyeless is likely to contain a boundary that prevents enhancer of eyeless present in the second intron from acting on the myoglianin promoter.
ME intergenic region contains DNaseI hypersensitive site
We used DNaseI hypersensitivity analysis to narrow down our search for the potential boundary elements because hypersensitive sites are found to be associated with regulatory elements and also because all known boundary sequences have DNaseI hypersensitive sites (4,52–56). We used nuclei from 16-h-old CS embryos to perform DNaseI hypersensitivity assay. In case of control where DNaseI digestion step was omitted, a 9.7 kb band was detected. Five extra bands appeared in the case when chromatin was subjected to DNaseI digestion prior to digestion with EcoRI revealing the presence of hypersensitive sites within 9.7 kb region (Figure 1a). Of the five hypersensitive sites, III and I map to the promoters of myoglianin and eyeless genes, respectively (Figure 1b). Two minor hypersensitive sites IV and V located in the region beyond the promoter of the myoglianin gene (Figure 1b) may reflect regulatory regions specific to this gene. Hypersensitive site II located in the 1.6 kb intergenic region was taken as the putative ME boundary for subsequent functional analysis (Supplementary Figure S1).
Figure 1.
Mapping of the DNaseI hypersensitive sites in the intergenic region ME separating myoglianin and eyeless genes. (a) Lane 1 is a 1 kb DNA ladder. Lane 2 is genomic DNA digested with EcoRI, a 9.7 kb band is generated by cleavage in the myoglianin and eyeless genes. Lane 3 is nuclei from 16-h-old embryos digested with DNaseI followed by purification and complete digestion with EcoRI. Samples were subjected to electrophoresis on a 1% agarose gel, transferred and covalently bound to positively charged nylon membrane and hybridized to probe shown in Figure 1b. DNaseI hypersensitive sites are marked with arrows. (b) A schematic presentation of the 9.7-kb region showing different DNaseI hypersensitive sites. Two hypersensitive sites III and I are near the promoter regions of myoglianin and eyeless genes, whereas two smaller hypersensitive sites IV and V are within the myoglianin gene. The hypersensitive site II located in the intergenic region is designated as ME boundary. The region used as a probe is shown as double headed arrow.
ME acts as an enhancer blocker
ME acts as an enhancer blocker in adult tissues
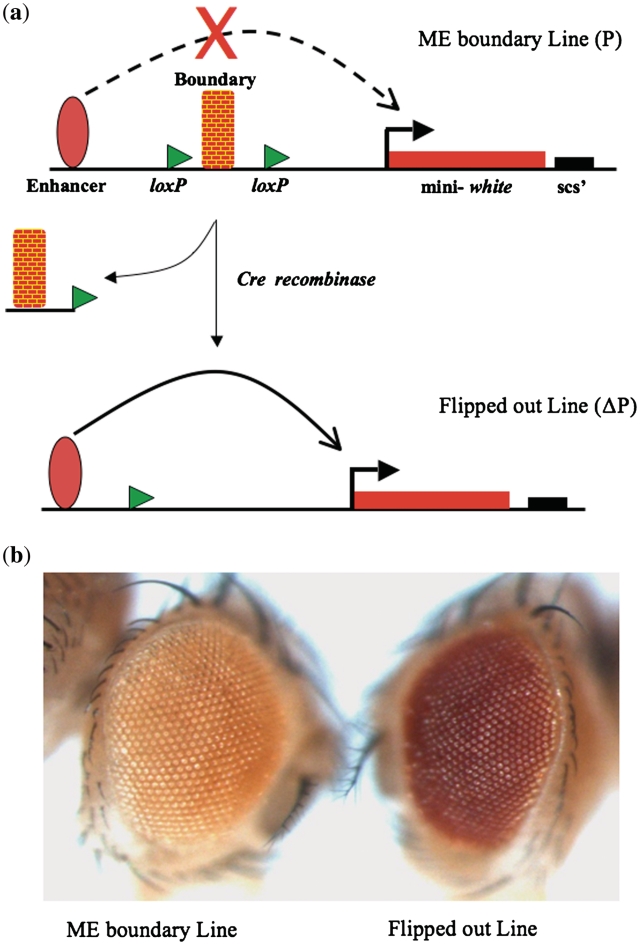
Enhancer blocker activity of boundary elements have been assayed earlier using a P-element vector, RW+ (57). In this assay vector, enhancer driving mini-white gene leads to a high level of white expression seen as red eye color. However, when a boundary is inserted between enhancer and promoter, the transgenic flies have a light eye color due to the enhancer blocking activity of the boundary element. We inserted ME region between the enhancer and promoter of the mini-white gene in the RW+ vector and generated transgenic flies (Figure 2a and Supplementary Table S2). As 8 lines out of 10 lines showed lighter eye color in heterozygous state, we reasoned that the ME region might be acting as an enhancer blocker in the RW+ construct. When these transgenic lines were crossed to flies carrying Cre recombinase to flip out the ME boundary DNA from the transgenic location, the eye color turned darker in all the cases (Figure 2b). This result showed that ME region acts as an enhancer blocker and that the light eye color is due to the boundary function of ME and not a position effect.
Figure 2.
Enhancer blocker assay in eye. (a) Schematic representation of the RW+ vector. The test fragment is inserted between the enhancer and promoter of mini-white. The test fragment is flanked on both sides with loxP sites. On crossing the fly with Cre expressing line the test fragment gets flipped out and the enhancer can now act on the promoter. (b) ME acts as an enhancer blocker in the eye. The eye on the left contains the test fragment in RW+ vector, whereas the eye on the right is the same line after the test fragment is flipped out.
Next, we tested enhancer blocker activity of ME in wings. We developed a P-element YW vector, which has two reporters, mini-white for screening the transgenes and yellow to score for the enhancer blocker activity (S. Krishnan and R. K. Mishra, personal communication). In this construct two enhancers are used, one wing enhancer and one body enhancer driving high expression of yellow gene in wing and body, respectively. When ME test fragment having enhancer blocker activity was inserted between the two enhancers, the body enhancer could still drive expression of the yellow gene and give dark pigmentation to the body (Supplementary Figure S2a). Wing enhancer, on the other hand, was inhibited by the presence of the ME boundary and yellow expression was weaker, seen as light wing pigmentation, supporting our earlier results that ME region indeed acts as an enhancer blocker. When we flipped out the ME part from the transgenic line, there was an increase in the wing pigmentation (Supplementary Figure S2b and c) whereas there was no change in eye color and body pigmentation (Supplementary Figure S2d and e). These results reinforce our observation that ME functions as enhancer blocker and that the reduction in reporter gene expression is not due to position effect. This also rules out any repressive activity in the test DNA.
ME acts as an enhancer blocker in embryos
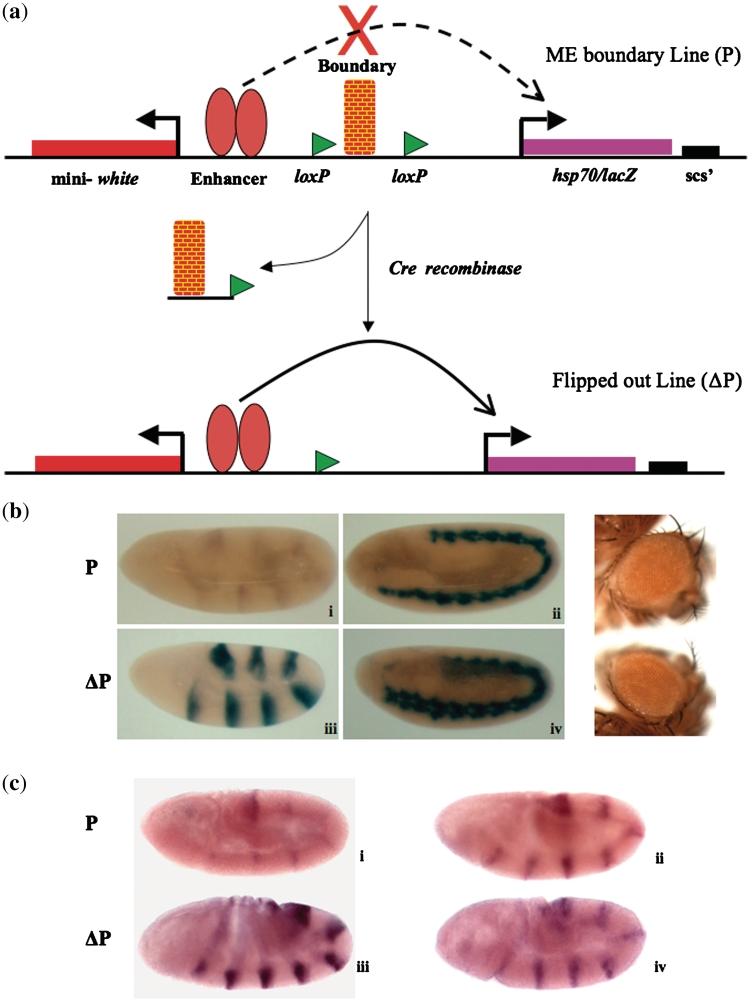
Having established that ME region acts as a boundary in eye and wing, we were interested in finding out if it could act as a boundary in embryonic stages as well. We inserted 917 bp DNA from the ME sequence (Supplementary Figure S3) between the ftz enhancers and the lacZ gene of the CfhL vector (Figure 3a) (58). ME sequence caused drastic reduction in the X-gal staining in embryos suggesting that ME was not allowing both the enhancers to act on lacZ (Figure 3b). Though blocking was seen both in case of young and old embryos, it was more prominent in case of young embryos and the boundary strength was similar to known gypsy derived boundary element (58), Supplementary Figure S4. Of the seven transgenic lines that we established, two were strong blockers, two were moderate blockers and three were weak blockers (Supplementary Table S2). This data suggested that ME region acts as enhancer blocker in the embryonic stages and that a shorter version of ME region was sufficient to act as a boundary. In the flipped out version of these lines the X-gal staining came back strongly, whereas the eye color remained the same before and after flipping out (Figure 3b). To finally confirm that ME functions as neutral boundary and not a repressor, we carried out RNA in situ hybridization in embryos to look for expression levels of both lacZ and white gene from the transegenic locus. As shown in Figure 3c, while lacZ expression was enhanced in the flipped out line as compared to the initial one, expression of white did not show any increase in the flipped out line. On the contrary RNA level of white gene shows a decrease in the flipped out line, presumably because after the removal of the boundary, ftz enhancer is competed by the lacZ gene. This conclusively establishes that ME region acts as a chromatin domain boundary and not a repressor.
Figure 3.
Enhancer blocker assay in embryo. (a) Schematic representation of the CfhL vector. The test fragment is inserted between the ftz enhancers and hsp70/lacZ gene. The test fragment is flanked on both sides with loxP sites. On crossing the fly with Cre expressing fly the test fragment gets flipped out and the enhancer can now act on the promoter. (b) Boundary function of ME is position independent. The upper panel (i and ii) shows early and late embryos from a line carrying ME. The lower panel (iii and iv) shows the early and late embryo from the same line after ME is flipped out. All the embryos shown are homozygous. As shown in the right panel the eye color of the initial and flipped out line does not vary indicating the boundary and not a repressor action of ME. (c) ME functions as boundary and does not affect ftz-driven expression of white in CfhL lines. RNA in situ hybridization shows lacZ expression (i and iii) and ftz-driven expression of white (ii and iv) in embryos. P and ΔP indicate initial and flipped out ME transgenic lines, respectively.
BEAF is required for the ME boundary function
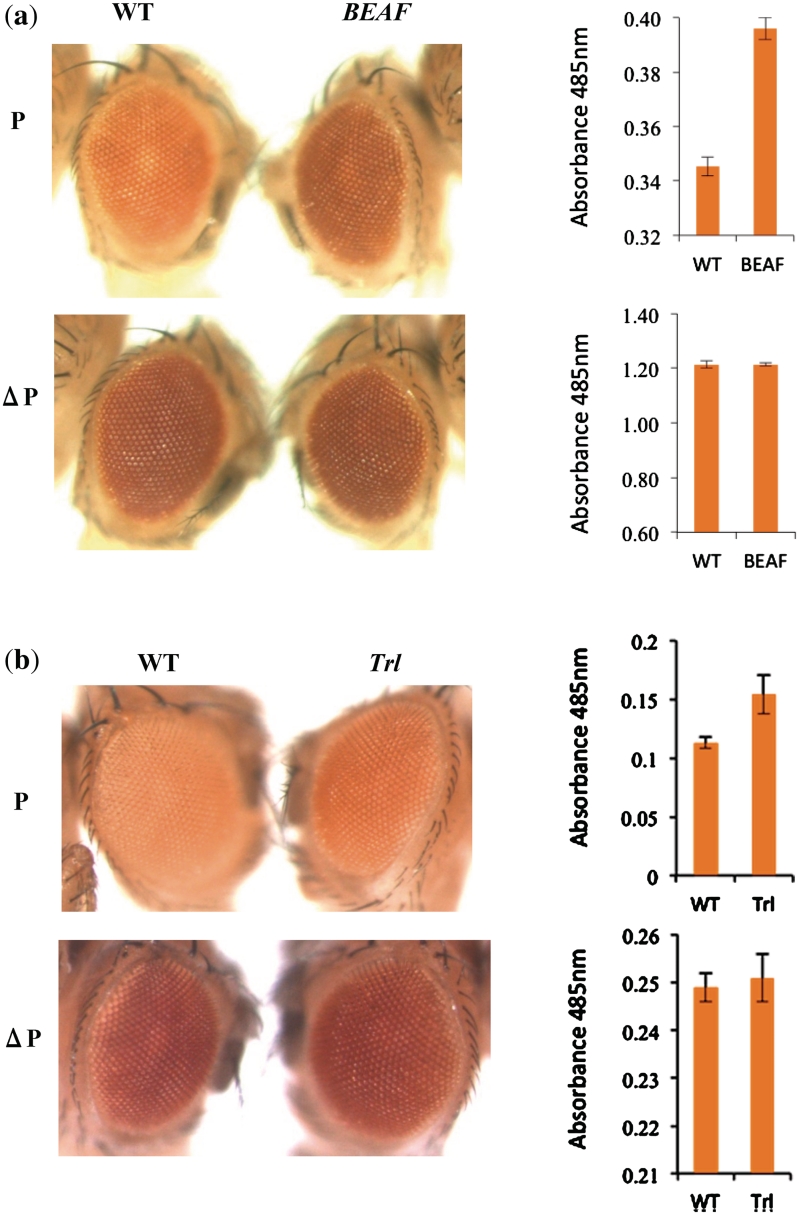
A close look at the sequence of ME boundary revealed that it contains binding sites for BEAF (Supplementary Figures S1 and S3). To test whether boundary activity of ME is dependent on BEAF, RW+ transgenic lines were placed in BEAF mutant background. We used BEAFAB-KO fly (59) to test the contribution of BEAF in enhancer blocking activity of ME in transgenic lines. In heterozygous BEAFAB-KO context, only a mild reduction in the insulator function of ME boundary was observed indicated by a slight increase in the eye color (data not shown). In the homozygous null mutant context of BEAF, a clear and significant increase in the eye color was observed (Figure 4a). Effect in the case of male flies was more pronounced as compared to female flies. On the contrary, we did not observe any change in eye color or the pigment level in case of the flipped out lines in the wild-type and homozygous BEAF mutant backgrounds (Figure 4a). Three independent initial lines and their flipped out versions were tested and all the lines showed comparable result, that BEAF mutation consistently weakened the wild-type ME boundary activity. These results indicate that BEAF contributes to the ME boundary function though it may not be the only factor involved.
Figure 4.
(a) BEAF is required for the boundary function of ME. In the first panel the left eye is from a line carrying ME on the third chromosome. The right eye is from the same line in the background of BEAFABKO/BEAFABKO. The lower panel shows the flipped out version of the same line in native and mutant background, respectively. The graph on the right shows pigment values. All the eyes shown are from males heterozygous for ME transgene. Error bars represent standard deviation form three independent experiments. (b) GAF is required for the boundary function of ME. In the first panel the left eye is from a line carrying ME on the third chromosome. The right eye is from the same line in the mutant background of TrlR85. The lower panel shows the flipped out version of the same line in native and mutant background, respectively. The graph on the right shows pigment values for these eyes. All the flies shown are females heterozygous for ME transgene.
It has been shown earlier that palindrome of BEAF recognition sequence also consists of DNA replication-related element (DRE), TATCGATA, which can be recognized by transcription factor DREF (29,60). Since DRE is present in ME boundary we wanted to test if Dref has any contribution to this boundary. For this purpose, we carried out lacZ staining of both initial and flipped out lines of ME boundary in the Dref mutant background. We did not find any effect of DrefKG09294 on ME boundary although this allele is known to affect the function of this protein (61), (Supplementary Figure S6). We, therefore, conclude that ME boundary function is not affected by Dref and that the BEAF recognition motif present in ME boundary responds only to BEAF.
GAF is required for the ME boundary function
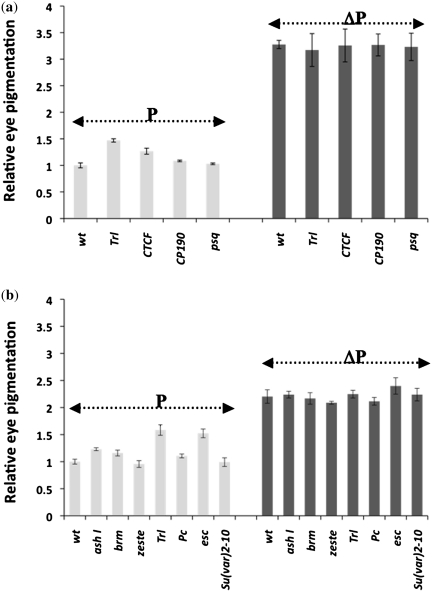
ME region also contains scattered GAF binding sites (Supplementary Figures S1 and S3). This prompted us to analyze the requirement of this protein to the enhancer blocker activity of ME in transgenic assay. In heterozygous GAF mutant background of TrlR85 (33), a mild relief from enhancer blocking by ME boundary in form of a slight increase in eye color was seen, which was also reflected in the pigment assay. In the case of GAF mutant background we observed a more pronounced effect in female flies (Figure 4b). When flipped out line of the same transgene was brought in the mutant background, no change in eye color or pigment level was seen. We also tested Trl13C (Figure 5a, Supplementary Figure S7) and, unlike in the case of TrlR85, observed similar effect in both male and female flies. Since GAGA sites are also known to be recognized by pipsqueak (62), we further tested if mutation in this gene (psqD91) could affect ME boundary function. As shown in Figure 5a and Supplementary Figure S7, psqD91 had no effect on ME boundary. This data suggested that GAF also is a positive regulator of ME boundary function although the effect of BEAF appears to be more pronounced indicating that the later is the major factor responsible for this boundary function.
Figure 5.
Effect of boundary interacting factors and PcG/trxG of genes on ME boundary. (a) Mutations of known boundary interacting factors (CTCF and CP190) and GAGA site interacting factors (GAF/Trl and pipsqueak) were tested. Relative pigmentation from eyes of male flies heterozygous for transgene as well as the mutation were estimated. P indicates transgenic initial ME line and ΔP indicates the flipped out version of the ME line. Virgin females from Trl13C, CTCFY+6, CP190H4-1, psqD91 crossed with males carrying the transgene. Male progeny carrying different mutations in combination with the ME transgene were used for eye color comparison (Supplementary Figure S7) and quantitative pigmentation assay. Error bars represent standard deviation form three independent experiments. (b) Mutations in selected PcG and trxG members were tested for their effect on ME boundary. Virgins from mutant stock were crossed with ME males and female progeny carrying both the mutation and the ME transgene were used for eye color comparison (Supplementary Figure S8) and quantitative pigmentation assay. Varying degree of effect is seen in case of ash1B1, brm2, Trl13C, Pc1 and esc2 while no noticeable effect is seen in case of z1 and Su(var)2-102. Error bars represent standard deviation form three independent experiments.
Effect of other boundary interacting factors on ME boundary
Several factors are known to contribute to the boundary function in Drosophila. These include CTCF and CP190 that are involved in several boundary elements especially gypsy insulator which also depends on PcG and trxG members (63–65). We tested mutations in CTCF and CP190 to see if ME boundary also depends on these factors. We saw mild effect of CTCF but no effect of CP190 on ME boundary function, Figure 5a and Supplementary Figure S7. When PcG and trxG mutations were tested, as reported earlier in the case of gypsy insulator (65), we observed noticeable but varying degree of effect of several of these mutations on ME boundary function, particularly in ash1, Trl and esc mutant context (Figure 5b and Supplementary Figure S8). Mutations in heterochromatin component Su(var)2-10 and PcG/trxG members, Pc, brm and Zeste, did not show any effect. ME boundary, therefore, appears to depend on a large number of factors that influence nuclear organization and chromatin structure (65). BEAF and GAF, however, may be the direct factors responsible for its boundary activity as ME boundary region contains binding sites for these proteins.
BEAF and GAF contribute to the ME boundary function by direct binding to it
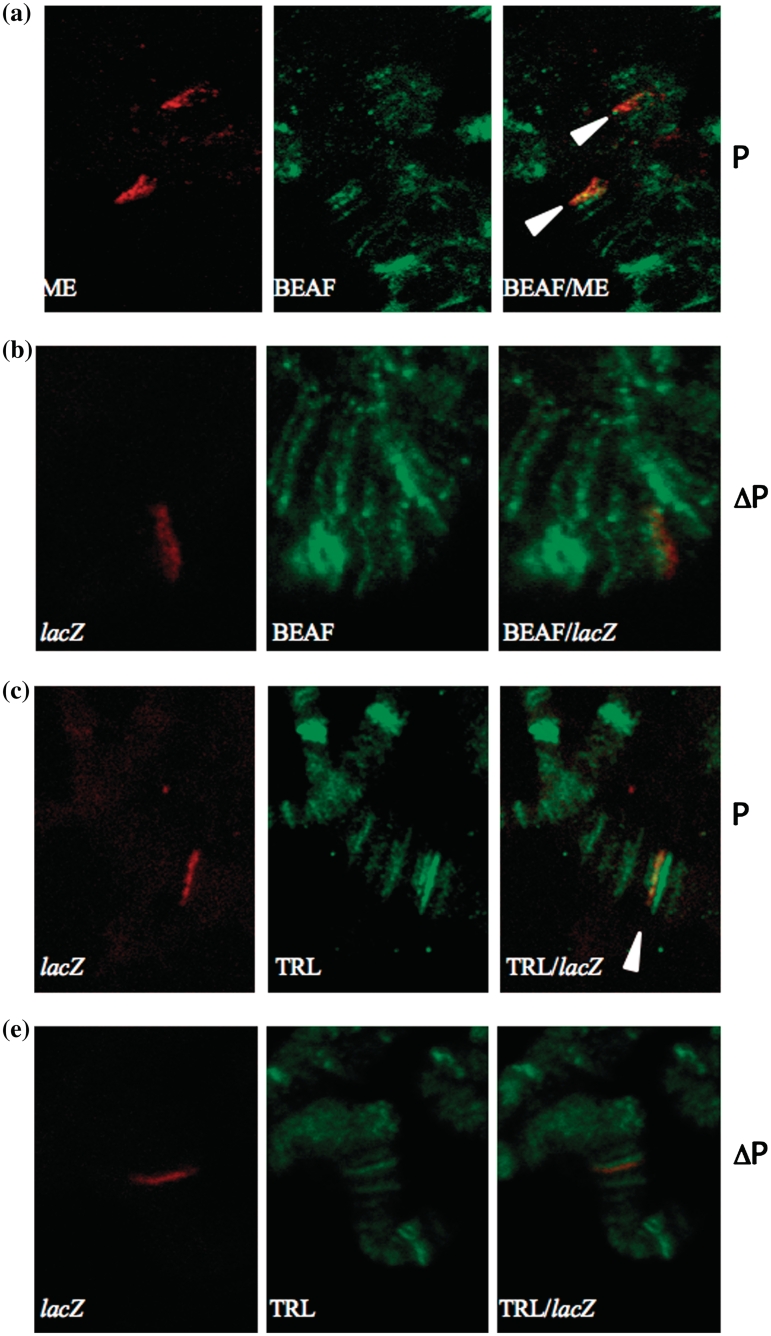
While the above experiments show the role of BEAF and GAF in ME boundary function, it does not rule out an indirect effect of the mutatnts used. To check whether BEAF binds to ME, immunoFISH was performed on polytene chromosomes using anti-BEAF antibody (34). Clear localization of BEAF at the ME region was observed both on the transgene insertion site and the native location of ME boundary on the fourth chromosome (Figure 6a). This data indicated that BEAF indeed binds to ME region. To cross check our result, we performed immunoFISH on the flipped out line as a control. No colocalization was observed in case of flipped out lines (Figure 6b). We also used anti-GAF antibody (35,43,44), to test if GAF binds to ME region. Clear colocalization of GAF and lacZ was observed, suggesting that GAF also binds to ME region (Figure 6c). In the flipped out version of the same transgenic line, GAF did not colocalize with lacZ (Figure 6d). These results, Figure 6 and Supplementary Figure S5 (full image of polytene spread), clearly showed that BEAF and GAF directly bind to the ME boundary and, thereby, contribute to its boundary function.
Figure 6.
Colocalization of BEAF and ME on polytene chromosomes. Transgenic larvae carrying ME on the third chromosome were dissected and used for immunoFISH. (a) Probe ME is in red, anti BEAF antibody is in green. Clear colocalization can be seen both on third and fourth chromosome (white arrowheads). (b) BEAF does not colocalize to the transgene in case of flipped out line. Probe lacZ is in red, anti BEAF antibody is in green. (c) Colocalization of GAF and ME at the site of transgene insertion probed by lacZ on polytene chromosomes. Probe lacZ is in red, GAF antibody signal is in green. Clear colocalization can be seen (white arrowhead). (d) GAF does not colocalize to the transgene in case of flipped out line.
To further confirm our findings that BEAF and GAF are involved in boundary function of ME by direct binding, we performed ChIP experiments. Since BEAF antibody did not work for the ChIP experiments, we used FLAG tagged BEAF expressing flies, generated in the lab from FLAG-BEAF fusion construct under a constitutive Pc promoter, to prepare chromatin (34). Anti FLAG M2 agarose was used to pull down the chromatin. More than 10-fold enrichment of ME is seen in BEAF pull down as compared to the negative control of Ubx intron II, which does not contain binding sites for BEAF, Figure 7a. As a positive control scs’ boundary was used on which BEAF is known to bind (26) that showed ∼40-fold enrichment with respect to the negative control. This ChIP data confirmed that BEAF directly binds to the ME boundary.
ChIP was also performed for calculating the occupancy of GAF at ME using anti GAF antibody. We used hexo-kinaseC promoter region, which does not contain binding sites for GAF as a negative control and iab-7PRE, which contains GAF binding sites (43) as a positive control. We tested two regions of ME for GAF, ME1 which is in the middle region of ME adjacent to the BEAF sites and ME2 which is towards the end of the ME region. We observed ∼3-fold enrichment in case of ME1 and ∼5-fold enrichment in case of ME2 as compared to the negative control, Figure 7b. Although the enrichment is relatively less compared to the positive control, these results clearly suggest that GAF interacts with the ME boundary.
BEAF binding sites are essential for the ME boundary function
Once we established that BEAF is the major factor that contributes to the ME boundary function (Figure 4) by directly binding to it, we wanted to analyze whether BEAF binding sites in the ME boundary are essential for its boundary function. To this end, we mutated all the five BEAF binding sites, including two palindromic sites in the 917 bp core region of ME boundary (Supplementary Figure S1). BEAF binding CGATA motifs were changed to CTCGA, which is reported to abolish BEAF binding (66). The mutated fragment was cloned in the CfhL vector and transgenic lines were generated (Supplementary Table S2). On staining the embryos for the lacZ activity dark staining was observed suggesting that mutated ME does not function as a boundary and as a result the ftz enhancers are able to drive the expression of lacZ gene, Figure 8. On flipping out the mutated ME region from these transgenic lines, no difference in the lacZ activity was observed (Figure 8). Also, there was no significant change in eye color and eye pigment observed in transgenes before and after flipping out the mutated ME (data not shown). To further confirm that loss of boundary function in mutated ME lines is due to absence of BEAF from the mutated region, we carried out ChIP experiments on wild-type and mutated ME lines. We observed complete loss of BEAF in mutated ME region as opposed to clear occupancy in the wild-type ME (Supplementary Figure S9). This suggests that BEAF binding sites are essential for the ME boundary function.
Figure 8.
BEAF binding sites are essential for the ME boundary function. Embryos from different transgenic lines were stained for lacZ activity. In the upper panel left embryo (i) is from a control line carrying only the vector, right embryo (ii) is from a control line carrying 1 kb λ DNA in place of test fragment. In the middle panel left embryo (iii) is from a transgenic line carrying ME boundary, right embryo (iv) is from the flipped out version of same line. In the lower panel left embryo (v) is from a line carrying ME fragment with mutated BEAF sites, right embryo (vi) is from the flipped out version of same line.
DISCUSSION
Chromatin domain boundaries are essential for proper transcriptional control of the genome. Although their importance in regulation of higher order chromatin organization is well established, very few boundary elements have been identified and studied till date. Sequence comparison of the known boundaries does not reveal any sequence similarity, which makes identification of new boundaries difficult. We looked for the pairs of genes which are closely spaced and differentially expressed and applied the rationale that such genes must be separated by chromatin domain boundary elements for their proper regulation. ME is one such intergenic region which separates the two differentially expressed genes. We show that ME boundary maps to a hypersensitive site and has binding sites for BEAF which is a well studied boundary associated protein (26). Finally, by using three independent transgenic assays, we show the functionality of the ME boundary at different stages of development and in different tissues.
BEAF is the major player in the boundary function of ME boundary as evident from genetic data and the effect that BEAF has on the ME boundary is by direct binding to the ME region as is evident from the ImmunoFISH and ChIP data. It is already known that BEAF binds to the scs’ boundary as a heterotrimer at the CGATA sites (66) and ME boundary has similar arrangement of the CGATA sites (Supplementary Figure S1). Some scattered CGATA motifs are also present in the ME boundary. ChIP data shows that BEAF binds to the core region of ME where two palindromic CGATA sites and one additional CGATA site are present. The importance of these BEAF binding sites is also evident from the fact that when we mutate these sites, boundary activity of ME is lost.
The ME boundary also contains binding sites for GAF. The pattern of GAF binding sites in ME boundary is similar to that seen in the case of Fab-7 boundary present in the bithorax complex of D. melanogaster. This prompted us to examine whether GAF has any effect on the boundary activity of ME (Supplementary Figure S1) (25). Our results show that GAF is also a positive regulator of the ME boundary function as loss of single copy of GAF results in partial loss of the boundary function of ME. This effect is by direct binding of GAF to the ME sequence as seen in the ImmunoFISH and ChIP experiments. In case of GAF, we observed that the effect of loss of GAF was more dramatic in female flies, which was opposite to what we see in the case of BEAF. Since both these proteins, specially GAF, regulate a large number of loci and GAF has also been implicated in dosage compensation (67), it is likely that the sex specific effect seen here in the case of ME boundary may be a result of complex and indirect interaction of multiple factors.
We show that both BEAF and GAF are needed for ME boundary activity. However, either BEAF or GAF (Trl) mutations alone were not sufficient for the complete loss of the boundary function. Since flies with BEAFAB-KO/BEAFAB-KO;P/TrlR85 genotype were lethal in our hands, it remains an open question whether BEAF and GAF can account for the complete boundary function of ME. Synthetic lethality in the double mutant BEAFAB-KO/BEAFAB-KO;P/TrlR85 does, however, suggest that these two proteins act in combination at the key loci and that this combination is essential for viability. There might be several such loci working as boundary elements and the double mutant combination, by abolishing or weakening a number of such boundaries, would cause misregulation of associated genes and lead to lethality.
ME boundary function is by recruitment of BEAF and GAF along with, perhaps, several other proteins although BEAF appears to be the major player as mutation in BEAF binding sites abolishes boundary function. Relatively lower level of GAF enrichment at ME, as seen in our ChIP experiments, may also indicate an indirect role of this protein at this locus. We also noticed minor but distinct effect of Polycomb and trithorax group mutations on ME boundary function. Our data, although suggestive and preliminary, indicate that ME boundary functions by recruiting multiple proteins, mutants of which lead to a partial loss of the boundary function. This mode of boundary function is similar to the other well studied gypsy boundary which depends on large number of factors including Su(Hw) (68), Mod(mdg4) (69), CP190 (64) and dTopors that associate with lamina (70). Boundary function of gypsy was also shown to depend on Polycomb and trithorax group of proteins (65). While we did not see any prominent effect of CTCF or CP190 on ME boundary activity, which is expected as ME region does not contain binding sites for these proteins, genome wide ChIP studies do detect association of these factors with ME (27,28,30). It is possible that ME may be part of nuclear structures where multiple boundaries cluster and number of factor participate even if not by direct binding to each boundary (71).
In conclusion, a rationale to look for boundary elements in short intergenic regions that separate differentially expressed genes can be applied successfully. Although expression pattern of a number of genes has not been analyzed in many organisms, analysis in other model organisms and human can be used and by homology criteria, large part of a genome can be mapped for potential boundary elements. Once a boundary region has been identified, the precise mapping of the functional boundary element can be accomplished by DNaseI hypersensitivity and transgene based assays available in model systems. Such studies will help us in understanding the genomic organization and regulatory environment of genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Council of Scientific and Industrial Research, India, fellowships (to H.S. and S.V.); Human Frontier Science Program young investigator grant (to R.K.M. Laboratory); Indo-French Centre for the Promotion of Advanced Research (IFCPAR) research grant; Department of Biotechnology (Government of India) and Department of Science & Technology (Government of India) research grants. Funding for open access charge: Centre for Cellular and Molecular Biology, Hyderabad, India.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Craig Hart for providing BEAFAB-KO flies and Paul Schedl for BEAF antibody. We also thank Pamela Geyer for DNA construct used to develop the YW vector. We also thank D. Vasanthi for help with fly work and A.G. Arimbasseri for help with the ChIP experiments. We thank Navneet K. Matharu and Surabhi Srivastava for critical reading of the article.
REFERENCES
- 1.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 2.Chung JH, Whiteley M, Felsenfeld G. A 5' element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 3.Di Simone P, Di Leonardo A, Costanzo G, Melfi R, Spinelli G. The sea urchin sns insulator blocks CMV enhancer following integration in human cells. Biochem. Biophys. Res. Commun. 2001;284:987–992. doi: 10.1006/bbrc.2001.5082. [DOI] [PubMed] [Google Scholar]
- 4.Galloni M, Gyurkovics H, Schedl P, Karch F. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 1993;12:1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gdula DA, Gerasimova TI, Corces VG. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl Acad. Sci. USA. 1996;93:9378–9383. doi: 10.1073/pnas.93.18.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerasimova TI, Corces VG. Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu. Rev. Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- 7.Geyer PK, Spana C, Corces VG. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 1986;5:2657–2662. doi: 10.1002/j.1460-2075.1986.tb04548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 10.Iqbal H, Mishra RK. Chromatin Domain Boundaries: defining the functional domains in genome. Proc. Indian Natl Sci. Acad. 2007;73:239–253. [Google Scholar]
- 11.Kanduri C, Holmgren C, Pilartz M, Franklin G, Kanduri M, Liu L, Ginjala V, Ulleras E, Mattsson R, Ohlsson R. The 5' flank of mouse H19 in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr. Biol. 2000;10:449–457. doi: 10.1016/s0960-9822(00)00442-5. [DOI] [PubMed] [Google Scholar]
- 12.Mishra RK, Karch F. Boundaries that demarcate structural and functional domains of chromatin. J. Biosci. 1999;24:377–399. [Google Scholar]
- 13.Robinett CC, O'Connor A, Dunaway M. The repeat organizer, a specialized insulator element within the intergenic spacer of the Xenopus rRNA genes. Mol. Cell. Biol. 1997;17:2866–2875. doi: 10.1128/mcb.17.5.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bi X, Broach JR. UASrpg can function as a heterochromatin boundary element in yeast. Genes Dev. 1999;13:1089–1101. doi: 10.1101/gad.13.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 17.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 18.Kellum R, Schedl P. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl Acad. Sci. USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 22.Gaszner M, Vazquez J, Schedl P. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 1999;13:2098–2107. doi: 10.1101/gad.13.16.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith ST, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkhurst SM, Harrison DA, Remington MP, Spana C, Kelley RL, Coyne RS, Corces VG. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 1988;2:1205–1215. doi: 10.1101/gad.2.10.1205. [DOI] [PubMed] [Google Scholar]
- 25.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 27.Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang N, Emberly E, Cuvier O, Hart CM. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 2009;29:3556–3568. doi: 10.1128/MCB.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith ST, Wickramasinghe P, Olson A, Loukinov D, Lin L, Deng J, Xiong Y, Rux J, Sachidanandam R, Sun H, et al. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev. Biol. 2009;328:518–528. doi: 10.1016/j.ydbio.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuvier O, Hart CM, Kas E, Laemmli UK. Identification of a multicopy chromatin boundary element at the borders of silenced chromosomal domains. Chromosoma. 2002;110:519–531. doi: 10.1007/s00412-001-0181-1. [DOI] [PubMed] [Google Scholar]
- 32.Cuvier O, Hart CM, Laemmli UK. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 1998;18:7478–7486. doi: 10.1128/mcb.18.12.7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371:806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 34.Pathak RU, Rangaraj N, Kallappagoudar S, Mishra K, Mishra RK. Boundary element-associated factor 32B connects chromatin domains to the nuclear matrix. Mol. Cell. Biol. 2007;27:4796–4806. doi: 10.1128/MCB.00305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vasanthi D, Anant M, Srivastava S, Mishra RK. A functionally conserved boundary element from the mouse HoxD locus requires GAGA factor in Drosophila. Development. 2010;137:4239–4247. doi: 10.1242/dev.058701. [DOI] [PubMed] [Google Scholar]
- 36.Sowpati DT, Thiagarajan D, Sharma S, Sultana H, John R, Surani A, Mishra RK, Khosla S. An intronic DNA sequence within the mouse Neuronatin gene exhibits biochemical characteristics of an ICR and acts as a transcriptional activator in Drosophila. Mech. Dev. 2008;125:963–973. doi: 10.1016/j.mod.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Pirrotta V, Steller H, Bozzetti MP. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 1985;4:3501–3508. doi: 10.1002/j.1460-2075.1985.tb04109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vazquez J, Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tautz D. In: Nonradioactive Analysis of Biomolecules. 2nd edn. Kessler C, editor. Berlin: Springer; 2000. pp. 573–580. [Google Scholar]
- 40.Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- 41.Nagaso H, Murata T, Day N, Yokoyama KK. Simultaneous detection of RNA and protein by in situ hybridization and immunological staining. J. Histochem. Cytochem. 2001;49:1177–1182. doi: 10.1177/002215540104900911. [DOI] [PubMed] [Google Scholar]
- 42.Zink D, Paro R. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 1995;14:5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chopra VS, Srinivasan A, Kumar RP, Mishra K, Basquin D, Docquier M, Seum C, Pauli D, Mishra RK. Transcriptional activation by GAGA factor is through its direct interaction with dmTAF3. Dev. Biol. 2008;317:660–670. doi: 10.1016/j.ydbio.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK. Vertebrate Homologue of Drosophila GAGA Factor. J. Mol. Biol. 2010;400:434–447. doi: 10.1016/j.jmb.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Sun FL, Cuaycong MH, Craig CA, Wallrath LL, Locke J, Elgin SC. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl Acad. Sci. USA. 2000;97:5340–5345. doi: 10.1073/pnas.090530797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartolome C, Maside X, Charlesworth B. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 2002;19:926–937. doi: 10.1093/oxfordjournals.molbev.a004150. [DOI] [PubMed] [Google Scholar]
- 47.Locke J, Podemski L, Roy K, Pilgrim D, Hodgetts R. Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res. 1999;9:137–149. [PMC free article] [PubMed] [Google Scholar]
- 48.Namciu SJ, Blochlinger KB, Fournier RE. Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell. Biol. 1998;18:2382–2391. doi: 10.1128/mcb.18.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- 50.Hauck B, Gehring WJ, Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc. Natl Acad. Sci. USA. 1999;96:564–569. doi: 10.1073/pnas.96.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lo PC, Frasch M. Sequence and expression of myoglianin, a novel Drosophila gene of the TGF-beta superfamily. Mech. Dev. 1999;86:171–175. doi: 10.1016/s0925-4773(99)00108-2. [DOI] [PubMed] [Google Scholar]
- 52.Emerson BM, Lewis CD, Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985;41:21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- 53.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 54.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGhee JD, Wood WI, Dolan M, Engel JD, Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981;27:45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- 56.Emerson BM, Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc. Natl Acad. Sci. USA. 1984;81:95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 59.Roy S, Gilbert MK, Hart CM. Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics. 2007;176:801–813. doi: 10.1534/genetics.106.068056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hart CM, Cuvier O, Laemmli UK. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–383. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- 61.Kim YS, Shin MJ, Yang DJ, Yamaguchi M, Park SY, Yoo MA. Transcriptional regulation of the Drosophila ANT gene by the DRE/DREF system. Genes Cells. 2007;12:569–579. doi: 10.1111/j.1365-2443.2007.01075.x. [DOI] [PubMed] [Google Scholar]
- 62.Huang DH, Chang YL, Yang CC, Pan IC, King B. pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol. Cell. Biol. 2002;22:6261–6271. doi: 10.1128/MCB.22.17.6261-6271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Gerasimova TI, Corces VG. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92:511–521. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 66.Hart CM, Zhao K, Laemmli UK. The scs' boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Greenberg AJ, Yanowitz JL, Schedl P. The Drosophila GAGA factor is required for dosage compensation in males and for the formation of the male-specific-lethal complex chromatin entry site at 12DE. Genetics. 2004;166:279–289. doi: 10.1534/genetics.166.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parkhurst SM, Corces VG. Interactions among the gypsy transposable element and the yellow and the suppressor of hairy-wing loci in Drosophila melanogaster. Mol. Cell. Biol. 1986;6:47–53. doi: 10.1128/mcb.6.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 70.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 71.Kumar RP, Senthilkumar R, Singh V, Mishra RK. Repeat performance: how do genome packaging and regulation depend on simple sequence repeats? Bioessays. 2010;32:165–174. doi: 10.1002/bies.200900111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.