Abstract
Poly(ADP-Ribose) (PAR) polymerase (PARP) inhibitors represent a promising class of novel anticancer agents. The present study explores the molecular rationale for combining veliparib (ABT-888) with camptothecin (CPT) and its clinical derivatives, topotecan and irinotecan. ABT-888 inhibited PAR induction by CPT and increased CPT-induced cell killing and histone γH2AX. Increased DNA breaks by ABT-888 were not associated with a corresponding increase of topoisomerase I cleavage complexes and were further increased by inactivation of tyrosyl-DNA phosphodiesterase 1. SiRNA knockdown for the endonuclease XPF–ERCC1 reduced the ABT-888-induced γH2AX response in non-replicating and replicating cells but enhanced the antiproliferative effect of ABT-888 in CPT-treated cells. Our findings indicate the involvement of XPF–ERCC1 in inducing γH2AX response and repairing topoisomerase I-induced DNA damage as an alternative pathway from PARP and tyrosyl-DNA phosphodiesterase 1.
INTRODUCTION
DNA topoisomerase I (Top1) is the target of clinically approved anticancer agents (topotecan, irinotecan and belotecan) derived from the plant alkaloid camptothecin (CPT) (1–4). It is essential in metazoans for the relaxation of DNA supercoiling generated during transcription and replication. Relaxation proceeds by formation of Top1 cleavage complexes (Top1cc), in which one DNA strand is cleaved by the covalent linkage of Top1 to the 3′-end of a DNA phosphodiester bond [reviewed in (3–6)]. Top1cc are normally very transient. Following DNA relaxation, Top1 is released by religation of the DNA. Top1cc can be stabilized (or `trapped’) under at least three conditions (2): (i) by drugs such as CPTs and non-CPT Top1 inhibitors (3,7); (ii) when the DNA template is altered (2); and (iii) during apoptosis (8). Abnormally stabilized Top1cc can be highly damaging when they interfere with the movement of replication and transcription complexes (9–12). Such collisions convert the Top1cc into DNA double-strand ends (DSE) with Top1 remaining covalently attached to the 3′-end of the broken DNA.
The repair of Top1-associated DNA damage in human cells is not fully understood (2). In budding yeast, two main pathways can remove Top1 adducts: hydrolysis of the Top1–DNA bond by tyrosyl–DNA phosphodiesterase 1 (TDP1) (13–15), and endonucleolytic excision of the Top1cc along with a section of the covalently attached DNA segment by different endonuclease complexes including Rad1–Rad10, Mre11–Rad50–Xrs2, Mus81–Mms4 and Slx1–Slx4 (2,16–18). The redundancy between the TDP1 and the endonuclease pathways has been demonstrated in yeast where inactivation of TDP1 has minimal impact on CPT action unless the Rad1–Rad10 endonuclease is simultaneously inactivated (16–18). Rad1–Rad10 is the heterodimeric ortholog of the human endonuclease XPF–ERCC1 (19). Those endonucleases cleave the duplex DNA segment immediately 5′ from the damaged region where the two DNA strands are separated (3′-flap, splayed arm or bubble) (19). XPF–ERCC1 is also a critical 5′-endonuclease in nucleotide excision repair (NER) both for global and transcription-coupled repair (TCR) (20).
A role for poly(ADP-ribose) polymerase (PARP) in the repair of Top1-associated DNA damage is well-established. Genetic inactivation of PARP sensitizes mammalian cells to CPT (21–23); PARP inhibitors enhance the effects of CPT and its clinical derivatives both in cell culture (22,24–30) and in xenograft systems (29,30). However, the molecular mechanisms by which PARP acts in the repair of Top1-induced DNA damage have not been elucidated and yeast cannot be used because the PARP pathway is not present in yeast cells. In mammalian cells, PARP inhibitors increase DNA breaks in response to Top1cc (22,24,27) but without concomitant increase in Top1–DNA complexes (27). PARP inactivation is associated with Tdp1 deficiency (2) and with toxic interference of Ku and DNA-PK in the homologous recombination (HR) pathway (23), which is critical for the repair of Top1cc (17,23,31–36).
The purpose of the present study is to elucidate the molecular mechanisms involved in the sensitization of cancer cells to CPT by PARP inhibitors. For this purpose, we used veliparib (ABT-888), one of the leading PARP inhibitors in clinical development (37,38). ABT-888 is a benzimidazole derivative with high potency against both PARP-1 and PARP-2 enzymes (Ki = 5 nM) (28). ABT-888 is orally bioavailable (39) and in clinical trials in combination with temozolomide, cyclophosphamide, platinum derivatives (cis-platin and oxaliplatin), mitomycin, radiation therapy and CPT derivatives (irinotecan and topotecan) (http://clinicaltrialsfeeds.org/clinical-trials/results/term=Drug:+veliparib+(ABT-888)?recr=Open). Our study examines the molecular effects of ABT-888 on CPT-induced cytotoxicity and γH2AX response, and the role of XPF–ERCC1 in the repair of Top1cc-induced DNA damage, which is relevant to the ongoing clinical trials combining ABT-888 with CPT derivatives.
MATERIALS AND METHODS
Cell lines and drugs
Human U2OS osteosarcoma cells and human HT29 colon carcinoma cells were obtained from the Developmental Therapeutics Program (DTP, National Cancer Institute, Bethesda, MD, USA) and maintained in RPMI 1640 medium complemented with 10% fetal bovine serum (FBS) at 37°C, 5% CO2. Human peripheral lymphocytes were obtained from the Blood Bank at the National Institutes of Health and maintained in RPMI 1640 supplemented with 10% FBS. CPT, veliparib (ABT-888, Abbott Laboratories, Abbott Park, IL, USA) and flavopiridol (FLV) were obtained from the Drug Synthesis and Chemistry Branch, Division of Cancer Treatment (National Cancer Institute, Bethesda, MD, USA). 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB) and the proteasome inhibitor, MG-132 were obtained from Sigma (St Louis, MO, USA). TDP1+/+ and TDP1−/− murine embryonic fibroblasts (MEFs) (40) were a kind gift from Dr. Cornelius F. Boerkoel (Center for Molecular Medicine and Therapeutics; University of British Columbia, Canada).
Cytotoxicity assay
The ATPlite assay (PerkinElmer Life Sciences, Waltham, MA, USA) was used to determine the cytotoxicity of CPT in the absence or presence of ABT-888. The ATP level in untreated cells was defined as 100%. Survival of treated cells was defined as ATP treated cells/ATP untreated cells × 100.
Western blotting
Proteins were detected by western blotting with corresponding specific primary antibodies. XPF monoclonal antibody (Ab-1) was purchased from Lab Vision, Thermo Fisher Scientific, Fremont, CA, USA, ERCC1 monoclonal antibody (D-10) from Santa Cruz Biotechnology, Santa Cruz, CA, β-actin monoclonal antibody (A5441) from Sigma, anti-phosphorylated-H2AX (γH2AX, clone JBW301) from Upstate Biotech, Millipore, Billerica, NY, USA and GAPDH monoclonal antibody (14C10) from Cell signaling Technology, Danvers, MA, USA. The figures show representative data that were reproducible in separate experiments.
COMET assays
DNA double-strand breaks (DSBs) were evaluated using the neutral single cell gel electrophoresis (neutral COMET assay). The COMET assays were performed according to the manufacturer's instructions (Trevigen, Gaithersburg, MD, USA). Data are expressed as mean ± SD.
Immunofluorescence assays
Cells plated in 4-well chamber slides (Nalgene Nunc International, Rochester, NY, USA) and cytospins of human lymphocytes were processed for immunofluorescence microscopy as described (41). For the simultaneous detection of γ-H2AX and replication foci, cells were labeled with 30 μmol/l 5-ethenyl-2′-deoxyuridine (EdU, Invitrogen, Carlsbad, CA, USA) for 90 min. During the last 30 min, cells were treated with CPT in the absence or presence of ABT-888. Following treatment, the medium was aspirated out and the cells were washed in phosphate buffer saline (PBS). Cells were immediately fixed and permeabilized by a 20-min incubation at room temperature with 4% paraformaldehyde and an overnight incubation in ice-cold 70% ethanol at 4°C. The staining for γH2AX was done first as described (41). EdU staining was then done with the Click-iT EdU Alexa Fluor® 647 flow cytometry assay kit from Invitrogen following the manufacturer’s instructions. The primary antibody for γH2AX (clone JBW301) was from Upstate (Millipore). The anti-PAR polymer rabbit polyclonal antibody was the product of Trevigen (Gaithersburg, MD, USA). The anti-53BP1 antibody (NB100-904) was from Novus Biologicals (Littleton, CO, USA).
γH2AX flow cytometry
Human lymphocytes were processed for flow cytometry as described (12) using the anti-γH2AX antibody (ab11174) from Abcam (Cambridge, MA, USA) and a Becton Dickinson FACScan flow cytometer (BD Biosciences, San Jose, CA, USA). Percentages of γH2AX positive cells were determined using CellQuest software (BD Biosciences).
Alkaline elution assay
DNA damage was detected using alkaline elution assays as described earlier (42,43). Briefly, cells were radiolabeled with [3H]-thymidine (1.0 μCi/ml) for 72 h and chased overnight (16 h) with radioisotope-free medium before receiving drug treatments. Cells were treated as indicated; after which, they were harvested by scraping into ice-cold Hank’s balanced salt solution (HBSS). Total DNA breaks [DNA strand breaks (SB)] were detected using DNA-denaturing conditions (pH 12.1) under deproteinizing conditions. To assess DNA–protein cross-links (DPC), cells were treated as indicated followed by irradiation with 30 Gy to break the DNA. Samples were lysed and evaluated for binding of the protein-crosslinked DNA by its retention on a 0.8 µm polyvinyl chloride/acrylic copolymer. Following alkaline elution, filters were incubated at 65°C with 1 M HCl for 45 min and 0.04 M NaCl was added for an additional 45 min of incubation. Radioactivity in each fraction was measured by liquid scintillation (Packard Instruments, Meridien, CT, USA).
Cellular Top1–DNA complexes detection (ICE Bioassay)
Top1–DNA complexes were detected as described earlier (43). Briefly, cells were pelleted and immediately lysed in 1% sarkosyl after drug treatment. Following homogenization with a Dounce homogenizer and pestle B, cell lysates were gently layered on cesium chloride step gradients and centrifuged at 165 000g for 20 h at 20°C. Half-milliliter fractions were collected and the fractions 6–10 were pooled together. The pooled fractions were then diluted with 25 mmol/l sodium phosphate buffer (pH 6.5) to make a 1×, 2×, 4×, or 8× scaled dilution for better resolution of differences and applied to Immobilon-P membranes (Millipore) in a slot-blot vacuum manifold. Top1–DNA complexes were detected using the C21 Top1 monoclonal antibody (gift from Yung-Chi Cheng, Yale University, New Haven, CT, USA) using standard western blotting procedures.
siRNA transfection
Gene-specific siRNAs for XPF (L-019946-00) or ERCC1 (L-006311-00) were products of Dharmacon (Lafayette, CO, USA). About 50 nM siRNAs were transfected to U2OS cells with Dharmafect transfection reagent (Dharmacon) for 48 h according to manufacturer’s instructions. Then culture medium was removed and cells were treated with CPT in the absence or presence of ABT-888. Cells transfected with negative control siRNA (D-001810-10, Dharmacon) were used as control.
Clonogenic assay
After drug treatment, cells were plated at a density of 100, 1000 and 10 000 per well in six-well plates and incubated for 10 days to allow formation of colonies. Cells were fixed with methanol, stained with 0.1% crystal violet (Sigma) for 30 min and washed with distilled water. Colonies were counted after air drying. Plating efficiency (PE) was defined as the number of colonies counted/the number of cells seeded. The survival fraction (SF) of untreated negative siRNA-transfected cells was defined as 100. SF were calculated as: PE treated/PE untreated × 100.
Statistical analyses
The data are represented as mean ± SD or mean ± SEM. The significance of differences between means was assessed by the Student’s t-test, with P < 0.05 being considered statistically significant. Three-way ANOVA tests were performed to compare the difference between γH2AX levels in individual CPT-treated cells in the presence or absence of ABT-888 (Figure 3 and Supplementary Figure S4A). Four-way ANOVA tests were used to compare the difference of γH2AX enhancement in si-XPF and si-Negative cells (Figure 7 and Supplementary Figure S4B).
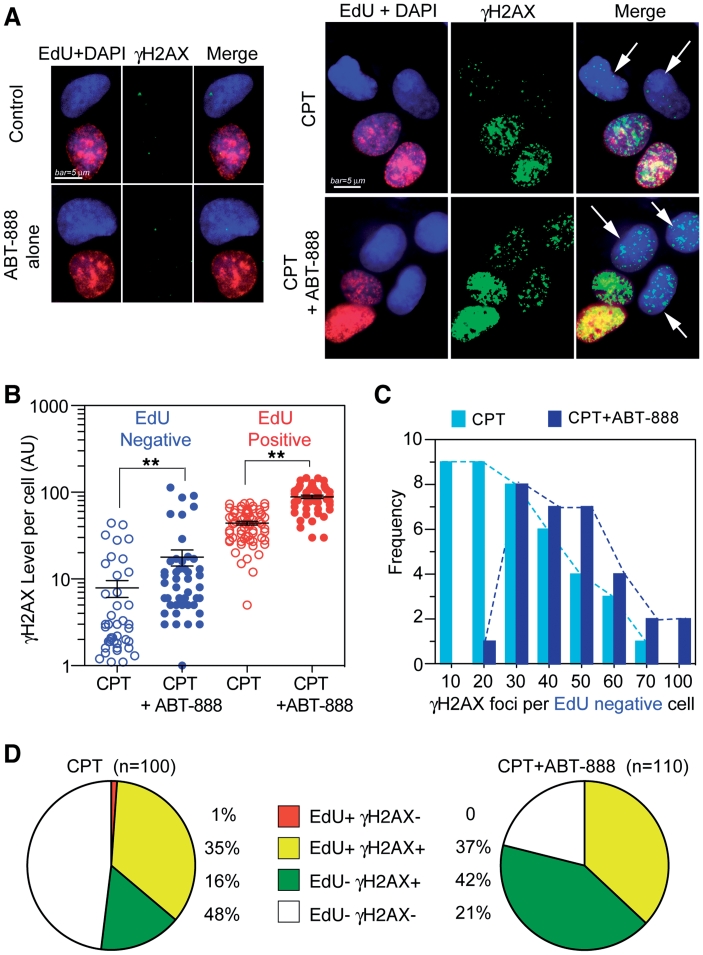
Figure 3.
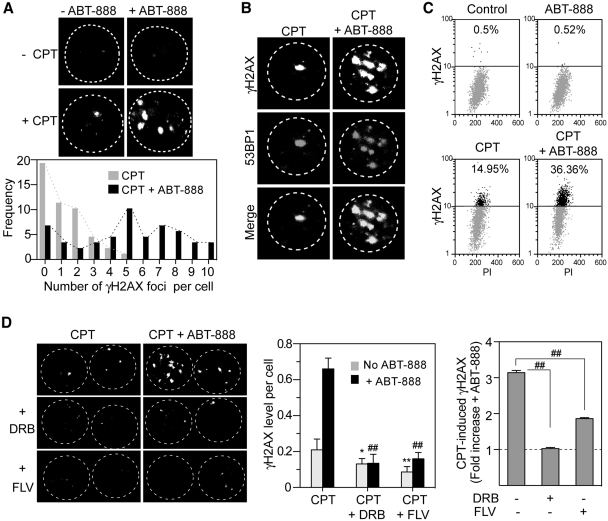
ABT-888 enhances both replication-dependent and independent γH2AX induced by CPT. Human osteosarcoma U2OS cells were treated with CPT (1 µM) for 30 min in the presence or absence of ABT-888 (0.5 µM). (A) The fluorescent thymidine analog EdU was used to identify S-phase cells by labeling their DNA (red signal). γH2AX is shown in green signal and nuclei labeled with DAPI are in blue. Representative immunofluorescence images (bar = 5 µm). White arrows indicate EdU negative but γH2AX positive cells. (B) Scattered-dot plot derived from the analyses of γH2AX level in individual cells in one representative experiment (see Supplementary Figure S4 for an independent experiment). Mean values ± SEM are shown as black bars. Number of cells analyzed: EdU-/CPT: n = 67; EdU + /CPT: n = 42; CPT + ABT: n = 46 (EdU+), n = 60 (EdU-). Standard t-tests were used for statistical analyses of the data from the representative experiment, **P < 0.01; (C) Distribution of γH2AX foci numbers per EdU negative cells in the presence and absence of ABT-888. Dotted curves link the edges of columns (Light blue, CPT alone; Dark blue, CPT + ABT-888). (D) Percentage distribution based on EdU and γH2AX showing γH2AX enhancement in EdU negative cells.
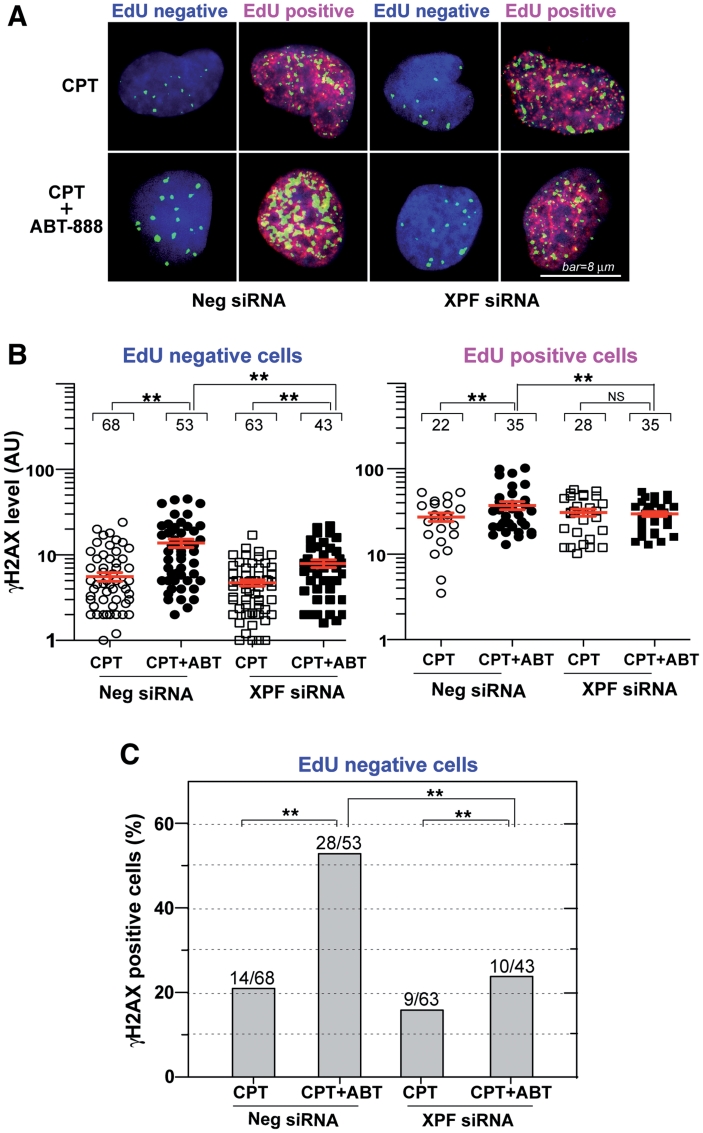
Figure 7.
XPF–ERCC1-dependent γH2AX enhancement by ABT-888 in non-replicating cells. U2OS cells were transfected with XPF siRNAs or control siRNAs (Neg siRNA). Two days later, the cells were labeled with EdU and treated with CPT (1 µM) for 30 min in the absence or presence of ABT-888 (0.5 µM). Cells were fixed and stained for immunofluorescence assays. (A) Representative confocal microscopy images (red signal: EdU; green signal: γH2AX; blue signal: DAPI to stain nuclei); bar = 8 µm. (B) Quantitation of γH2AX signals in individual cells (represented as scattered dots) from one representative experiment; Mean values ± SEM are shown as red lines. Numbers above each cluster indicate the number of cells counted. Standard t-tests were used for statistical analyses of the data from the representative experiment, **P < 0.01; NS, no significant difference. (C) XPF-dependent effect of ABT-888 in non-replicating (EdU negative) cells. Percentages of γH2AX-positive cells were scored based on drug treatment and XPF knockdown. The number of γH2AX positive cells and total number of cells scored in each group are indicated above each bar.
RESULTS
Induction of poly(ADP-ribosyl)ation by CPT
First, we tested PAR levels in CPT-treated human cancer cells and the inhibitory effect of ABT-888 on CPT-treated cells. In both human colon cancer HT-29 cells (Figure 1A) and human osteosarcoma U2OS cells (Supplementary Figure S1), PAR polymer levels increased after 30-min CPT treatments. This PAR response was inhibited by ABT-888, which demonstrates rapid PAR induction by Top1 inhibition with CPT and efficient inhibition by ABT-888.
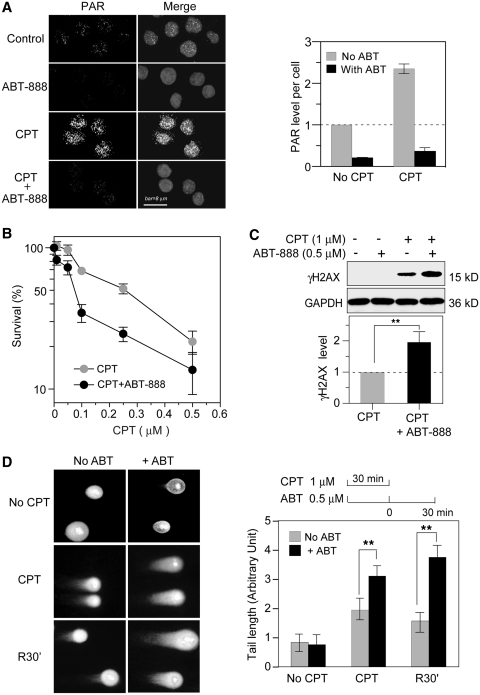
Figure 1.
Potentiation of CPT-induced DNA damage by the PARP inhibitor ABT-888 in human colon cancer HT-29 cells. (A) Fluorescence images of PAR. Cells were treated with 1 µM CPT for 30 min in the absence or presence of ABT-888 (0.5 µM). Nuclei were labeled with PI (gray signals in the Merge images) and PAR polymers are shown as white dots. Bar = 8 µm. Right, PAR quantitation (mean values ± SD) using untreated Control set as one. (B) Cytotoxicity assays. Cells were treated with CPT for 72 h in the absence or presence of ABT-888 (0.5 µM). The survival of untreated cells was defined as 100%. Data are shown as mean values ± SD (n = 3). (C) Enhancement of CPT-induced γH2AX by ABT-888. Cells were treated with CPT for 30 min in the presence or absence of ABT-888 (0.5 µM). γH2AX was determined by Western blotting: Top: representative experiment; Bottom: γH2AX levels were quantified from three independent experiments (mean values ± SD) using CPT-induced γH2AX levels set as one (**P < 0.01). (D) DSB measured by neutral COMET assays. Cells were co-treated with CPT and ABT-888 for 30 min; R30′: cells were examined 30 min after CPT removal in the absence or presence of ABT-888 (0.5 µM). Left, representative images; Right top: treatment schedule; Right bottom: quantitation of average tail lengths. At least 50 cells were quantified in each data set. Data are shown as mean values ± SD. Standard t-tests were used for statistical analyses; **P < 0.01.
The PARP inhibitor ABT-888 (Veliparib) increases CPT-induced cytotoxicity and DNA breaks without increasing CPT-induced Top1cc
Next, we determined the effect of ABT-888 on the cytotoxicity of CPT by treating HT-29 cells with CPT in the presence or absence of ABT-888. The results shown in Figure 1B demonstrate that ABT-888 potentiates the cytotoxicity of CPT under conditions where ABT-888 (0.5 µM) had no detectable cytotoxicity (data not shown). With 30-min exposure of CPT in the presence of ABT-888, the molecular marker of DNA DSB (44), γH2AX was increased by ABT-888 in CPT-treated cells (Fig.1C). Neutral COMET assays also showed increased CPT-induced DSBs in the presence of ABT-888 (Figure 1D). Upon CPT removal, those DSBs were more persistent than in the absence of ABT-888 (Figure 1D). Taken together, these experiments demonstrate increased DSBs induced by CPT in the presence of the PARP inhibitor ABT-888. They also show enhancement of the CPT-induced γH2AX response.
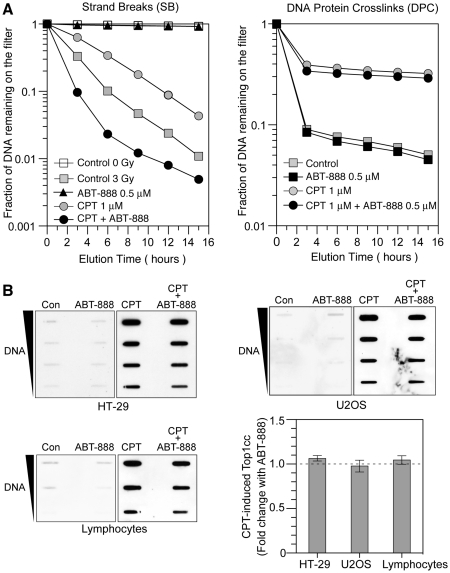
To investigate the mechanism(s) by which ABT-888 enhances CPT-induced DNA damage, we measured CPT-induced Top1cc as DNA–protein crosslinks (DPC) by alkaline elution (42,43) and by the ICE-bioassay (43). Figure 2A shows a representative experiment in which DPC and DNA strand breaks (SB) were measured in the same cells. Consistent with the COMET assays, ABT-888 increased the frequency of DNA SB (Figure 2A, left panel). Noticeably, under these conditions, the DPC remained at the same frequency in the presence of ABT-888 as in its absence (DPC; Figure 2A, right panel). Similarly, ICE-bioassays showed similar levels of Top1cc in the absence and presence of ABT-888 not only in HT29 cells, but also in U2OS osteosarcoma cells, and normal human peripheral lymphocyte cells (Figure 2B). Together these results demonstrate that PARP inhibition by ABT-888 induces the formation of additional DNA breaks in response to CPT without increasing Top1cc.
Figure 2.
ABT-888 induces DNA breaks without increasing CPT-induced Top1cc. (A) Representative alkaline elution assays in HT-29 cells; Left, CPT-induced SB under DNA denaturing conditions. Ionizing radiation (3 Gy) was used as positive control. Cells were exposed to either ionizing radiation or 1 µM CPT in the presence or absence of ABT-888 (0.5 µM), or were left untreated; Right, CPT-induced DPC. Cells were treated with 1 µM CPT for 30 min in the presence or absence of ABT-888 (0.5 µM) followed by irradiation with 30 Gy. DPC were measured by DNA retention on protein-adsorbing filters. (B) Top1cc measured by ICE-bioassay in three different cell lines. HT-29 cells (top left) or U2OS cells (top right) were treated with 1 µM CPT for 30 min in the presence or absence of ABT-888 (0.5 µM). Human peripheral lymphocytes require higher drug concentrations to elicit signals and were treated with 20 µM CPT for 2 h with or without ABT-888 (5 µM) (bottom left). Quantitation of the lack of significant effect of ABT-888 on CPT-induced Top1cc in three cell lines (bottom right). For each cell line, fold change was defined as Top1cc (CPT + ABT-888)/Top1cc (CPT alone). Data are mean values ± SD (n = 3).
Enhancement of both replication-dependent and independent γH2AX by ABT-888
Because CPT-induced DNA damage results from both replication (10,11,45,46) and transcription interference (2,8,12,47–49), we tested the relationship between the ABT-888-induced γH2AX and DNA replication. Immunofluorescence microscopy was used to quantitate the γH2AX fluorescent signals in individual cells (Supplementary Figure S2A) (45,50). The distribution of γH2AX levels showed two groups of cells with low and high γH2AX levels, respectively. In the CPT + ABT-888 treated-cells, both peaks were shifted to the right, demonstrating the levels of γH2AX were increased by PARP inhibition in both groups (Supplementary Figure S2B). To clarify whether those two groups were related to DNA replication, EdU incorporation (51) was used to detect the replicating cells and γH2AX co-staining was used to observe the relationship between DNA synthesis and γH2AX induction (52). Figure 3A demonstrates the induction of γH2AX foci both in replicating cells (EdU positive) and in non-replicating cells (EdU negative cells, indicated by arrows). EdU positive cells presented higher levels of γH2AX than EdU negative cells (Figure 3B), suggesting high γH2AX levels correspond to replicating cells. ABT-888 induced more and larger CPT-induced γH2AX foci in both EdU positive (replicating) and EdU negative (non-replicating) cells (Figure 3A and B). These results were reproducible in an independent experiment (Supplementary Figure S4A). ANOVA analysis of the data from two independent experiments indicated significant difference between CPT and CPT + ABT-888 in EdU negative cells (P < 0.001), and EdU positive cells (P < 0.001). Noticeably, CPT-induced γH2AX in the EdU negative cell population was augmented from 16% (16 out of 100 cells) for CPT alone to 42% (46 out of 110 cells) for the combination CPT + ABT-888 (Figure 3C). Moreover, ABT-888 almost doubled the average CPT-induced γH2AX foci per cell (26 ± 16 for CPT alone versus 45 ± 13 for CPT + ABT-888) (Figure 3D). Taken together, those data demonstrate that PARP inhibition enhances CPT-induced γH2AX both in replicating cells and non-replicating cells.
Enhancement of transcription-related γH2AX by ABT-888 in non-replicating cells
To further elucidate the replication-independent effects of ABT-888, we tested human peripheral lymphocytes treated with CPT, which we previously reported induce transcription-induced DSBs in response to CPT (12,49). Immunofluorescence imaging of γH2AX and analyses of the number of γH2AX foci, showed a significant increase in γH2AX foci in the presence of ABT-888 (5.1 ± 3.2 foci/cell versus 1.3 ± 1.2 foci/cell for CPT alone) (Figure 4A). Those γH2AX foci were co-localized with p53 binding protein 1 (53BP1), consistent with the formation of DSBs (Figure 4B). Flow cytometry also showed ∼2-fold increased γH2AX in the presence of ABT-888 (Figure 4C). These data, together with those obtained in Figure 3 indicate that PARP inhibition enhances both CPT-induced replication-dependent and independent DNA damage.
Figure 4.
Enhancement of γH2AX by ABT-888 in non-replicating human peripheral blood lymphocytes. Lymphocytes were treated with CPT (20 µM) for 2 h in the absence or presence of ABT-888 (5 µM). (A) Top: γH2AX foci induction observed by confocal microscope; Bottom: single cell analysis of the distribution of γH2AX foci numbers. (B) Co-localization of γH2AX and 53BP1. (C) γH2AX response determined by 2D flow-cytometry. (D) Inhibition of the γH2AX enhancement by transcription inhibitors. Lymphocytes were pretreated for 1 h with DRB (100 µM) or FLV (1 µM) before the addition of CPT and ABT-888. Left: representative images of γH2AX foci by confocal microscopy. Middle: average γH2AX level per nucleus. Mean and SD from three experiments are shown. Standard t-tests were used for statistical analyses; *P < 0.05 versus CPT, **P < 0.01 versus CPT, ##P < 0.01 versus CPT + ABT-888. Right: fold induction of CPT-induced γH2AX by ABT-888. Fold change was defined as γH2AX (CPT + ABT-888)/γH2AX (CPT). ##P < 0.01.
Next we tested whether the effect of ABT-888 in non-replicating cells was transcription-linked. Lymphocytes were pretreated with transcription inhibitors before the addition of CPT with or without ABT-888. The transcription inhibitors DRB and FLV (8,12,49) not only prevented CPT-induced γH2AX (12), but also reduced the enhancement of the CPT-induced γH2AX by ABT-888 (Figure 4D). These results demonstrate the involvement of PARP in the repair of transcription-linked DNA damage induced by CPT.
PARP is involved in a common repair pathway with TDP1
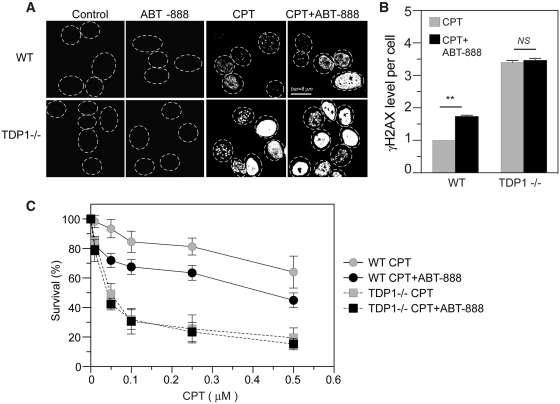
Because TDP1 represents a major pathway for the repair of Top1cc (2,15), the effects of ABT-888 were examined in TDP1 knockout cells (40). Figure 5 (panels A and B) shows a more intense CPT-induced γH2AX response in the TDP1 knockout cells than the corresponding wild-type cells (53), which is consistent with the defective repair of Top1cc and increased DNA breaks in TDP1-deficient cells (40,43,54,55). Notably, ABT-888 failed to further enhance the γH2AX response in the TDP1−/− cells, which indicates that PARP inhibition no longer enhances Top1-induced DNA damage once TDP1 is inactivated. The effects of ABT-888 on CPT-induced cytotoxicity were also measured in TDP1−/− cells. Figure 5C shows that ABT-888 was unable to potentiate the cytotoxicity of CPT in the TDP1−/− cells. These data suggest that PARP acts in the same pathway as TDP1 for the repair of Top1cc.
Figure 5.
TDP1-dependent enhancement of γH2AX by ABT-888. (A) Representative immunofluorescence images of γH2AX in wild-type (WT) MEF or TDP1−/− MEF cells treated with CPT (1 µM) for 30 min with or without ABT-888 (0.5 µM). (B) Quantitation of γH2AX levels per cell. The average γH2AX level in CPT-treated WT cells was defined as one. Data are mean values ± SD. Standard t-tests were used for statistical analyses; **P < 0.01; NS, no significant difference. (C) Cytotoxicity of CPT with or without ABT in WT and TDP1−/− cells. MEF cells were treated with CPT for 72 h in the absence or presence of ABT-888 (0.5 µM). The survival of untreated WT cells was defined as 100%. Data are mean values ± SD (n = 3 independent experiments).
Because the repair of Top1cc requires the proteasomal degradation of Top1 following its ubiquitination (46,48), we tested whether the enhancement of γH2AX by ABT-888 was dependent on the proteasome. Cells treated with CPT and ABT-888 in the presence of the proteasome inhibitor, MG-132, failed to induce γH2AX (Supplementary Figure S3) demonstrating that PARP acts downstream from the proteasome for the repair of Top1–DNA complexes.
The XPF–ERCC1 complex is involved in the repair of CPT-induced DNA damage
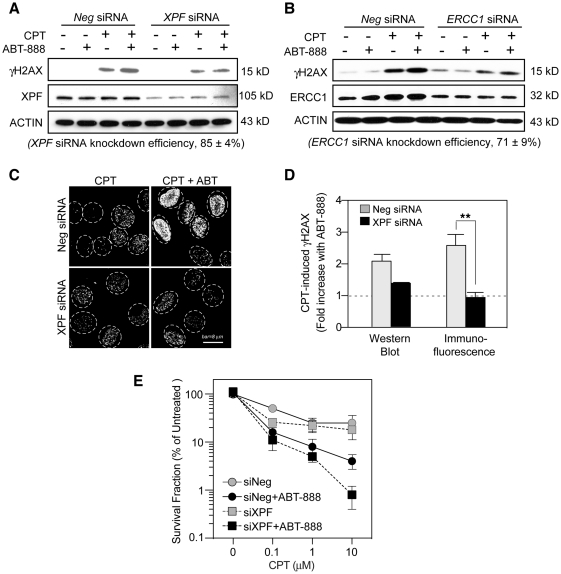
Based on our finding that ABT-888 induces the formation of DNA breaks in response to CPT (see above), and on the genetic data from yeast showing the importance of the endonuclease Rad1–Rad10 complex as an alternative pathway for the repair of Top1-induced DNA damage (‘Introduction’ section), we tested the possible implication of the mammalian Rad1–Rad10 ortholog XPF–ERCC1 complex in the repair of CPT-induced DNA damage. Knocking-down XPF by siRNA resulted in a marked attenuation of the γH2AX enhancement by ABT-888 (Figure 6A, C and D). Similar results were observed by knocking down the DNA binding partner of XPF, ERCC1 (Figure 6B). Notably, we found that knocking down XPF also reduced ERCC1 expression and vice versa (data not shown) suggesting the cross-stabilization of XPF and ERCC1. Immunofluorescence microscopy confirmed that XPF knockdown blocked the enhancing effect of ABT-888 on the CPT-induced γH2AX formation (Fig. 6C and D). XPF knockdown had no effect on the γH2AX level of untreated or ABT-alone-treated cells (data not shown). Together, these experiments demonstrate that the enhancement of CPT-induced γH2AX by PARP inhibition is dependent on XPF–ERCC1.
Figure 6.
Involvement of XPF–ERCC1 and PARP in CPT-induced γH2AX and cytotoxicity. XPF or ERCC1 siRNAs were transfected into U2OS cells for 48 h before CPT treatment (1 µM for 30 min) in the presence or absence of ABT-888 (0.5 µM). (A) Effect of XPF knockdown on the γH2AX enhancement by ABT-888. γH2AX levels were detected by western blotting. XPF siRNA knockdown efficiency was determined as mean values ± SD (n = 4). (B) Effect of ERCC1 knockdown on the γH2AX enhancement by ABT-888. γH2AX levels were detected by western blotting. ERCC1 siRNA knockdown efficiency was determined as mean values ± SD (n = 4). (C) Representative γH2AX immunofluorescence images in XPF siRNA-transfected cells. (D) Fold induction of CPT-induced γH2AX by ABT-888. Results were from western blotting and immunofluorescence assays, respectively. Data are mean values ± SD (n = 2 independent experiments for western blotting, n = 3 independent experiments for immunofluorescence assay). Standard t-tests were used for statistical analyses of immunofluorescence data; **P < 0.01). (E) Effect of XPF knockdown on the survival of cells treated with CPT in the presence or absence of ABT-888. U2OS cells transfected with XPF siRNA or negative siRNA for 48 h were treated with CPT for 30 min in the absence or presence of ABT-888 (0.5 µM). Cells were cultured for 10 days following drug removal to allow colony formation. The SF of untreated cells transfected with negative siRNA (Neg siRNA) was defined as 100%. Data are mean values ± SD (n = 3 independent experiments).
Clonogenic assays were performed to determine the functional implication of XPF–ERCC1 in the repair of CPT-induced DNA damage in the absence and presence of ABT-888. Figure 6E shows that XPF-inactivation sensitized cells to CPT and that ABT-888 increased the cytotoxicity of CPT both in the presence or absence of XPF (Figure 6E). These results demonstrate that ABT-888 further enhances the killing of XPF–ERCC1-deficient cells in response to CPT.
Involvement of XPF–ERCC1 in the replication-independent γH2AX induced by ABT-888 in CPT-treated cells
Next we wished to determine the γH2AX response as a function of DNA replication in XPF-knockdown cells in the absence or presence of ABT-888. As in Figure 3, EdU co-staining was used to differentiate the non-replicating and replicating cells. XPF siRNA reduced the γH2AX enhancement by ABT-888 in both cell populations (Figure 7A). Further analyses of γH2AX levels in individual cells also showed that XPF siRNA reduced γH2AX levels enhancement both in the EdU negative and EdU positive cells (Figure 7B). Four-way ANOVA analyses of γH2AX data from two independent experiments demonstrated significant difference of γH2AX enhancement between XPF siRNA- and Negative siRNA-transfected cells (P < 0.05). The dependence on XPF was further analyzed in the EdU negative cells (Fig. 7C). In those cells, XPF knockdown significantly attenuated the γH2AX response to ABT-888. Together these results demonstrate the involvement of XPF–ERCC1 both in replication-dependent and independent γH2AX activation by PARP inhibition in CPT-treated human cells.
DISCUSSION
The present study provides new insights into alternative pathways for the repair of Top1-induced DNA damage in human cells and on the rationale for combining Top1 and PARP inhibitors. Our experiments demonstrate that ABT-888 enhances the DNA damaging activities of CPT (cytotoxicity, DNA breaks, γH2AX induction) at concentrations where ABT-888 does not have detectable effects on its own. Our results are consistent with the reported effects of other PARP inhibitors (NU1025, AG14361) in combination with CPT derivatives (25,27) and provide mechanistic insights for the recently initiated clinical trials combining topotecan or irinotecan and veliparib (ABT-888). Our data also suggest the usefulness of γH2AX as a clinical pharmacodynamic biomarker in such combination therapies (44,56).
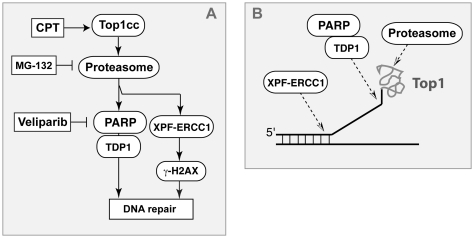
Although PARP has been reported to directly activate Top1 (57–59), our study shows that ABT-888 increases the cytotoxicity of CPT without affecting the levels of Top1cc (Figure 2). Similarly, another PARP inhibitor, AG14361 was reported to synergize with topotecan without affecting the Top1cc levels (27). Noticeably, our study also shows that PARP inhibition increases the levels of `frank breaks’ (i.e. non-Top1-associated) (Figures 1 and 2), which is in agreement with independent studies with different PARP inhibitors (24,27). Our interpretation is that these breaks correspond to new `repair lesions’ introduced by endonucleases such as XPF–ERCC1 in order to remove the Top1cc and process the damaged DNA into a suitable substrate for HR (Figure 8).
Figure 8.
Role of XPF–ERCC1 and PARP in parallel pathways for the repair of Top1cc. (A) Scheme illustrating the alternative repair pathways involving the proteasome, PARP, TDP1 and XPF–ERCC1 for the repair of Top1-induced DNA damage. Other alternative pathways involving Mus81–Eme1 and Mre11–Nbs1 and Rad50 (2) are not represented. (B) Hypothetical biochemical mechanisms for Top1cc excision by XPF–ERCC1 (endonuclease pathway) and TDP1 (PARP–TDP1 excision pathway).
Our data indicate that PARP acts downstream from the proteasome. Indeed, ABT-888 was unable to enhance the γH2AX response to CPT when cells were simultaneously treated with MG-132 (Supplementary Figure S3). Our results along with recent studies (46,48,60,61) place the proteasome as an early effector in the repair of Top1cc (Figure 8). It is plausible that Top1, which is a 100 kDa polypeptide encircling the DNA to which it is covalently bound (5) needs to be proteolyzed for the repair enzymes (including TDP1) to access the broken DNA ends (13,62–64). PARP is also a known cofactor of XRCC1, and XRCC1 is an established repair factor for Top1cc (54,63,65,66). XRCC1 forms repair complexes with TDP1 (53,63,67), and PARP1 knockout cells tend to be deficient in TDP1 activity [Figure 7 in (2)]. Together with our finding that ABT-888 failed to enhance the γH2AX and cytotoxic responses to CPT in TDP1−/− cells (53) (see Figure 5), these results suggest that PARP functions together with TDP1 in a common repair pathway (Figure 8A).
The present study provides the first evidence for a role of XPF–ERCC1 in the repair of Top1-induced DNA damage in human cells (Figure 8). This conclusion is consistent with genetic data in budding yeast where inactivation of Rad1–Rad10 (the yeast XPF–ERCC1 orthologs) sensitizes cells to CPT, especially when TDP1 is also inactivated (17,32) [reviewed in (2)]. Thus, we propose that XPF–ERCC1 functions as an alternative repair pathway besides the PARP-TDP1 pathway in mammalian cells (Figure 8). This can explain why inactivation of XPF–ERCC1 can further increase the cytotoxicity of ABT-888 in CPT-treated cells (Figure 6E). However, because we also found that inactivation of XPF–ERCC1 inhibits the γH2AX response (Figure 6A–D), it is plausible that XPF–ERCC1 repairs Top1cc by cleaving the DNA upstream from the Top1cc and generating `frank breaks’ (see beginning of ‘Discussion’ section). These breaks could then be responsible for the XPF–ERCC1-dependent activation of γH2AX (Figure 8B).
Our data suggest that XPF–ERCC1 is involved not only in the repair of replication-associated DNA damage but also in the repair of replication-independent DNA damage generated by Top1cc. This adds XPF–ERCC1 to the cellular responses to Top1cc-induced transcription-associated damage in addition to RNA polymerase II hyperphosphorylation, BRCA1-dependent proteolysis of Top1 (8), RNaseH1-dependent DSB induction with ATM activation (12) and altered RNA splicing (68). The increase in DSB and γH2AX by ABT-888 in normal peripheral lymphocytes raises the question as to whether the synergistic effect of PARP inhibitors is selective for cancer cells. The ongoing clinical trials with veliparib in association with topotecan or irinotecan should provide an answer to this question. Finally, our experiments suggest that maximum benefit for combining PARP and Top1 inhibitors might be achieved in tumors with XPF–ERCC1 deficiency.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Center for Cancer Research; National Cancer Institute Intramural Program; National Institutes of Health. Funding for open access charge: National Institutes of Health, US Government.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr HongFang Liu, Department of Biostatistics, Bioinformatics and Biomathematics, Georgetown Medical Center, for assistance with statistical analyses.
REFERENCES
- 1.Hsiang YH, Hertzberg R, Hecht S, Liu LF. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 2.Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, et al. Repair of topoisomerase I-mediated DNA damage. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pommier Y, Leo E, Zhang H, Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 6.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 7.Pommier Y, Cushman M. The indenoisoquinoline noncamptothecin topoisomerase I inhibitors: update and perspectives. Mol. Cancer Ther. 2009;8:1008–1014. doi: 10.1158/1535-7163.MCT-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sordet O, Larochelle S, Nicolas E, Stevens EV, Zhang C, Shokat KM, Fisher RP, Pommier Y. Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. J. Mol. Biol. 2008;381:540–549. doi: 10.1016/j.jmb.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holm C, Covey JM, Kerrigan D, Pommier Y. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- 10.Hsiang Y-H, Lihou MG, Liu LF. Arrest of DNA replication by drug-stabilized topoisomerase I–DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 11.Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sordet O, Redon CE, Guirouilh-Barbat J, Smith S, Solier S, Douarre C, Conti C, Nakamura AJ, Das BB, Nicolas E, et al. Ataxia telangiectasia mutated activation by transcription- and topoisomerase I-induced DNA double-strand breaks. EMBO Rep. 2009;10:887–893. doi: 10.1038/embor.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang S-W, Burgin AB, Huizenga BN, Robertson CA, Yao KC, Nash HA. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc. Natl Acad. Sci. USA. 1996;93:11534–11539. doi: 10.1073/pnas.93.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topo I covalent complexes. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 15.Dexheimer TS, Antony S, Marchand C, Pommier Y. Tyrosyl-DNA phosphodiesterase as a target for anticancer therapy. Anticancer Agents Med. Chem. 2008;8:381–389. doi: 10.2174/187152008784220357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng C, Brown JA, You D, Brown JM. Multiple endonucleases function to repair covalent topoisomerase i complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vance JR, Wilson TE. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl Acad. Sci. USA. 2002;99:13669–13674. doi: 10.1073/pnas.202242599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu C, Pouliot JJ, Nash HA. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl Acad. Sci. USA. 2002;99:14970–14975. doi: 10.1073/pnas.182557199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 20.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee S, Cheng M-F, Trivedi D, Petzold SJ, Berger NA. Camptothecin hypersensitivity in poly(adenosine diphosphate-ribose) polymerase-deficient cell lines. Cancer Commun. 1989;1:389–394. doi: 10.3727/095535489820875129. [DOI] [PubMed] [Google Scholar]
- 22.Bowman KJ, Newell DR, Calvert AH, Curtin NJ. Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br. J. Cancer. 2001;84:106–112. doi: 10.1054/bjoc.2000.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattern MR, Mong SM, Bartus HF, Mirabelli CK, Crooke ST, Johnson RK. Relationship between the intracellular effects of camptothecin and the inhibition of DNA topoisomerase I in cultured L1210 cells. Cancer Res. 1987;47:1793–1798. [PubMed] [Google Scholar]
- 25.Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 2008;183:1203–1212. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaney CA, Wang LZ, Kyle S, White AW, Calvert AH, Curtin NJ, Durkacz BW, Hostomsky Z, Newell DR. Potentiation of temozolomide and topotecan growth inhibition and cytotoxicity by novel poly(adenosine diphosphoribose) polymerase inhibitors in a panel of human tumor cell lines. Clin. Cancer Res. 2000;6:2860–2867. [PubMed] [Google Scholar]
- 27.Smith LM, Willmore E, Austin CA, Curtin NJ. The novel poly(ADP-Ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks. Clin. Cancer Res. 2005;11:8449–8457. doi: 10.1158/1078-0432.CCR-05-1224. [DOI] [PubMed] [Google Scholar]
- 28.Penning TD, Zhu GD, Gandhi VB, Gong J, Liu X, Shi Y, Klinghofer V, Johnson EF, Donawho CK, Frost DJ, et al. Discovery of the Poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1 H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J. Med. Chem. 2009;52:514–523. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- 29.Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, Ator M, Bihovsky R, Hudkins R, Chatterjee S, et al. Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol. Cancer Ther. 2003;2:371–382. [PubMed] [Google Scholar]
- 30.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, Durkacz BW, Hostomsky Z, Kumpf RA, Kyle S, et al. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J. Natl. Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 31.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 32.Pouliot JJ, Robertson CA, Nash HA. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells. 2001;6:677–687. doi: 10.1046/j.1365-2443.2001.00452.x. [DOI] [PubMed] [Google Scholar]
- 33.Malik M, Nitiss JL. DNA repair functions that control sensitivity to topoisomerase-targeting drugs. Eukaryot. Cell. 2004;3:82–90. doi: 10.1128/EC.3.1.82-90.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eng WK, Faucette L, Johnson RK, Sternglanz R. Evidence that DNA topoisomerase I is necessary for the cytotoxic effects of camptothecin. Mol. Pharmacol. 1988;34:755–760. [PubMed] [Google Scholar]
- 35.Knab AM, Fertala J, Bjornsti M-A. Mechanisms of camptothecin resistance in yeast DNA topoisomerase I mutants. J. Biol. Chem. 1993;268:22322–22330. [PubMed] [Google Scholar]
- 36.Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drew Y, Plummer R. PARP inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug Resist. Updat. 2009;12:153–156. doi: 10.1016/j.drup.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat. Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donawho CK, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, Ferguson DC, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin. Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 40.Hirano R, Interthal H, Huang C, Nakamura T, Deguchi K, Choi K, Bhattacharjee MB, Arimura K, Umehara F, Izumo S, et al. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007;26:4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang YW, Jones TL, Martin SE, Caplen NJ, Pommier Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J. Biol. Chem. 2009;284:18085–18095. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Covey JM, Jaxel C, Kohn KW, Pommier Y. Protein-linked DNA strand breaks induced in Mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 1989;49:5016–5022. [PubMed] [Google Scholar]
- 43.Miao ZH, Agama K, Sordet O, Povirk L, Kohn KW, Pommier Y. Hereditary ataxia SCAN1 cells are defective for the repair of transcription-dependent topoisomerase I cleavage complexes. DNA Repair. 2006;5:1489–1494. doi: 10.1016/j.dnarep.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 44.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. gammaH2AX and cancer. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell. Biol. 2007;27:5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin CP, Ban Y, Lyu YL, Liu LF. Proteasome-dependent processing of topoisomerase I-DNA adducts into DNA double-strand breaks at arrested replication forks. J. Biol. Chem. 2009;284:28084–28092. doi: 10.1074/jbc.M109.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A ubiquitin-proteasome pathway for the repair of topoisomerase I–DNA covalent complexes. J. Biol. Chem. 2008;283:21074–21083. doi: 10.1074/jbc.M803493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sordet O, Nakamura AJ, Redon CE, Pommier Y. DNA double-strand breaks and ATM activation by transcription-blocking DNA lesions. Cell Cycle. 2010;9:274–278. doi: 10.4161/cc.9.2.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guirouilh-Barbat J, Redon C, Pommier Y. Transcription-coupled DNA double-strand breaks are mediated via the nucleotide excision repair and the Mre11–Rad50–Nbs1 complex. Mol. Biol. Cell. 2008;19:3969–3981. doi: 10.1091/mbc.E08-02-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guirouilh-Barbat J, Zhang YW, Pommier Y. Induction of glutathione-dependent DNA double-strand breaks by the novel anticancer drug brostallicin. Mol. Cancer Ther. 2009;8:1985–1994. doi: 10.1158/1535-7163.MCT-09-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, Shiloh Y, Pommier Y. Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J. 2009;28:3667–3680. doi: 10.1038/emboj.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 55.Katyal S, el-Khamisy SF, Russell HR, Li Y, Ju L, Caldecott KW, McKinnon PJ. TDP1 facilitates chromosomal single-strand break repair in neurons and is neuroprotective in vivo. EMBO J. 2007;26:4720–4731. doi: 10.1038/sj.emboj.7601869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Redon CE, Nakamura AJ, Zhang YW, Ji JJ, Bonner WM, Kinders RJ, Parchment RE, Doroshow JH, Pommier Y. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin. Cancer Res. 2010;16:4532–4542. doi: 10.1158/1078-0432.CCR-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bauer PI, Buki KG, Comstock JA, Kun E. Activation of topoisomerase I by poly [ADP-ribose] polymerase. Int. J. Mol. Med. 2000;5:533–540. doi: 10.3892/ijmm.5.5.533. [DOI] [PubMed] [Google Scholar]
- 58.Malanga M, Althaus FR. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and induces DNA strand break resealing. J. Biol. Chem. 2004;279:5244–5248. doi: 10.1074/jbc.C300437200. [DOI] [PubMed] [Google Scholar]
- 59.Park SY, Cheng YC. Poly(ADP-ribose) polymerase-1 could facilitate the religation of topoisomerase I-linked DNA inhibited by camptothecin. Cancer Res. 2005;65:3894–3902. doi: 10.1158/0008-5472.CAN-04-4014. [DOI] [PubMed] [Google Scholar]
- 60.Cusack JC, Jr, Liu R, Houston M, Abendroth K, Elliott PJ, Adams J, Baldwin AS., Jr Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor-kappaB inhibition. Cancer Res. 2001;61:3535–3540. [PubMed] [Google Scholar]
- 61.Takeshita T, Wu W, Koike A, Fukuda M, Ohta T. Perturbation of DNA repair pathways by proteasome inhibitors corresponds to enhanced chemosensitivity of cells to DNA damage-inducing agents. Cancer Chemother. Pharmacol. 2009;64:1039–1046. doi: 10.1007/s00280-009-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Debethune L, Kohlhagen G, Grandas A, Pommier Y. Processing of nucleopeptides mimicking the topoisomerase I-DNA covalent complex by tyrosyl-DNA phosphodiesterase. Nucleic Acids Res. 2002;30:1198–1204. doi: 10.1093/nar/30.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plo I, Liao ZY, Barcelo JM, Kohlhagen G, Caldecott KW, Weinfeld M, Pommier Y. Association of XRCC1 and tyrosyl DNA phosphodiesterase (Tdp1) for the repair of topoisomerase I-mediated DNA lesions. DNA Repair. 2003;2:1087–1100. doi: 10.1016/s1568-7864(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 64.Interthal H, Chen HJ, Champoux JJ. Human Tdp1 cleaves a broad spectrum of substrates including phosphoamide linkages. J. Biol. Chem. 2005;280:36518–36528. doi: 10.1074/jbc.M508898200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barrows LR, Holden JA, Anderson M, D'Arpa P. The CHO XRCC1 mutant, EM9, deficient in DNA ligase III activity, exhibits hypersensitivity to camptothecin independent of DNA replication. Mutat. Res. 1998;408:103–110. doi: 10.1016/s0921-8777(98)00022-6. [DOI] [PubMed] [Google Scholar]
- 66.Park SY, Lam W, Cheng YC. X-ray repair cross-complementing gene I protein plays an important role in camptothecin resistance. Cancer Res. 2002;62:459–465. [PubMed] [Google Scholar]
- 67.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Solier S, Barb J, Zeeberg BR, Varma S, Ryan MC, Kohn KW, Weinstein JN, Munson PJ, Pommier Y. Genome-wide analysis of novel splice variants induced by topoisomerase I poisoning shows preferential occurrence in genes encoding splicing factors. Cancer Res. 2010;70:8055–8065. doi: 10.1158/0008-5472.CAN-10-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.