Abstract
There are two main classes of small nucleolar RNAs (snoRNAs): the box C/D snoRNAs and the box H/ACA snoRNAs that function as guide RNAs to direct sequence-specific modification of rRNA precursors and other nucleolar RNA targets. A previous computational and biochemical analysis revealed a possible evolutionary relationship between miRNA precursors and some box H/ACA snoRNAs. Here, we investigate a similar evolutionary relationship between a subset of miRNA precursors and box C/D snoRNAs. Computational analyses identified 84 intronic miRNAs that are encoded within either box C/D snoRNAs, or in precursors showing similarity to box C/D snoRNAs. Predictions of the folded structures of these box C/D snoRNA-like miRNA precursors resemble the structures of known box C/D snoRNAs, with the boxes C and D often in close proximity in the folded molecule. All five box C/D snoRNA-like miRNA precursors tested (miR-27b, miR-16-1, mir-28, miR-31 and let-7g) bind to fibrillarin, a specific protein component of functional box C/D snoRNP complexes. The data suggest that a subset of small regulatory RNAs may have evolved from box C/D snoRNAs.
INTRODUCTION
Micro RNAs (miRNAs) are a family of short regulatory RNAs that post-transcriptionally regulate gene expression. In mammals, miRNAs have been found to perform their regulatory function mainly by translation inhibition of protein coding transcripts through base pairing to specific target sequences in the 3′-untranslated regions (UTRs) (1). While a subset of miRNAs are encoded in independent transcription units, many miRNAs are encoded in introns of protein-coding genes and are co-expressed with these host genes (2–4). Mature miRNAs are small RNAs of ∼22 nt in length that are processed out of ∼70 nt-long hairpin structures (called pre-miRNAs) (5). The canonical miRNA biogenesis pathway involves either excision of the miRNA precursors from the introns of their host gene transcripts or transcription from independent units, both followed by processing by the microprocessor complex in the nucleus, export to the cytoplasm and further processing by a dicer-containing complex (2–4). However, recent reports have identified several different non-canonical miRNA processing pathways (6–9). In particular, several groups have recently reported miRNAs and miRNA-like molecules derived from small nucleolar RNAs (snoRNAs), some of which have been immunoprecipitated with Ago proteins, functional protein interactors of mature miRNAs (7,8,10,11).
snoRNAs are a family of conserved nuclear RNAs concentrated in nucleoli where they either function in the modification of ribosomal RNA (rRNA) or participate in the processing of rRNA during ribosome subunit synthesis (12–15). Most snoRNAs have been found to be encoded in the introns of protein-coding genes (16). snoRNAs are processed out of these introns and carry out their function in complex with specific protein interactors, forming ribonucleoprotein complexes referred to as snoRNPs. Two main classes of snoRNAs have been identified: the box C/D snoRNAs and the box H/ACA snoRNAs, both of which serve as guide RNAs complementary to specific target sequences mainly in rRNA precursors. Box C/D snoRNAs catalyse 2′-O-ribose methylation and box H/ACA snoRNAs guide psuedouridine modifications.
Box C/D snoRNAs are characterized by the presence of conserved C and D sequence boxes that typically come into contact in the folded molecule and serve as a binding site for box C/D snoRNP proteins, including NOP56, NOP58, 15.5K and the highly conserved protein fibrillarin, which carries out the specific 2′-O-methylation. The guide sequence with complementarity to the target is located immediately 5′ to the box D or D′ region (e.g. see Figure 1). Several box C/D snoRNAs and a subset of their target sites in rRNA are conserved from yeast through to mammalian cells. However, numerous orphan box C/D snoRNAs have been identified that do not encode a region of complementarity to rRNA (17).
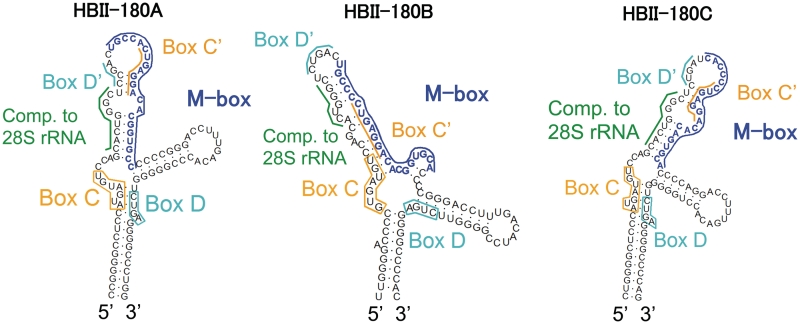
Figure 1.
Predicted secondary structure of the human HBII-180 box C/D snoRNAs. The characteristic features of members of the box C/D HBII-180 family are shown. Conserved C and D motifs are indicated by orange and cyan boxes/lines, respectively. The positions of rRNA complementary sequence are indicated by a green bar. The positions of M-box regions which can be altered to modulate target gene expression (snoMEN vector) (22) are indicated by a blue bar.
Through systematic investigation of human miRNAs, we have recently reported that a subset of miRNA precursors have box H/ACA snoRNA features, both in their primary and secondary structure as well as in the genomic region in which they are encoded. Five of these miRNA precursors show functional box H/ACA snoRNA characteristics by binding to dyskerin, a conserved protein component of the box H/ACA snoRNPs. This led us to propose an evolutionary relationship between miRNAs and snoRNAs, in which a subset of snoRNAs would have evolved to serve as precursors of miRNAs (10). snoRNAs have been characterized as mobile genetic elements with the capacity to copy themselves to other genomic locations (18,19), thus providing large numbers of potential miRNA-like precursors. Such copies of snoRNAs have been coined snoRTs [snoRNA retroposons, by Weber (18)]. Recent reports provide further complementary evidence supporting this hypothesis, including the discovery of nucleolar miRNAs (20) and the description of small miRNA size fragments derived from snoRNAs (11,21). In addition, we have recently described a family of closely related snoRNAs, the HBII-180s, which show typical features of box C/D snoRNAs but also contain a region of ∼20 nt with almost perfect complementarity to endogenous pre-mRNA sequences, termed the M-box (22). Although the detailed mechanism and relationship between the M-box sequences and the endogenous target RNAs to which they are complementary has not yet been characterized, we demonstrated that by altering the M-box region to make it complementary to selected target genes it is possible to suppress target gene mRNA and protein levels (22). This provides a knockdown vector system called snoMEN (snoRNA modulator of gene expression). Here, we show that RNA fragments corresponding to the M-box regions of HBII-180 snoRNAs can be generated from the precursor box C/D snoRNAs. Using an in-house algorithm, whose design is based on sequences of snoMEN molecules, as well as sequences of box C/D snoRNAs known to produce miRNA-like fragments, we addressed the broader relationship between box C/D snoRNAs and known miRNA precursors.
MATERIALS AND METHODS
RNase protection assay
RNase protection assay was performed by using mirVanaTM miRNA Detection Kit (Ambion). Full-length HBII-180C snoRNA was 32P labelled according to the manufacturer’s protocol. Labelled probes were mixed with HeLa cell total RNA and RNase treatment was performed according to the manufacturer’s protocol.
Northern and high sensitivity RNA blot analysis
HeLa cell extracts were fractionated using sucrose gradients, as previously described (23–25). Total HeLa cell RNA and RNA from separate cytoplasmic, nucleoplasmic and nucleolar fractions was isolated using the TRIzol method, with DNase I treatment, according to the manufacturer’s protocol (Invitrogen). Equal amounts of RNA from each sample were separated by 8 M urea polyacrylamide denaturing gel electrophoresis in 1× TBE buffer and the RNA transferred onto nylon membrane (Hybond-N; Amersham) by electro blotting. After UV or chemical cross-linking, the membrane was hybridized with 32P 5′-end labelled oligoribonucleotide probes specific for the following RNA species:
(HBII-239_mir-768: 5′-AGCAGUUUGAGUGUCAGCAUUG-3′,
hsa-mir-16-1: 5′-CGCCAAUAUUUACGUGCUGCUA-3′,
hsa-mir-27b: 5′-GCAGAACUUAGCCACUGUGAA-3′,
hsa-mir-31: 5′-CAGCUAUGCCAGCAUCUUGCCU-3′,
HBII-180A: 5′-GGGCACCGUGUCCUCAGUGGCA-3′,
HBII-180B: 5′-GUGCACCGUGUCCUCAGGGGCA-3′,
HBII-180C: 5′-GUGCACUGUGUCCUCAGGGGUG-3′,
tRNA-Ile 5′-UGGUGGCCCGUACGGGGAUCGA-3′,
HBII-180C-boxD region: 5′-CCCUCAGACCCCCAGGUGUCA-3′)
and for DNA species:
(U3: 5′-CCGTATTGGGGAGTGAGAGGGAGAGAACGCGGTCTGAGTG-3′,
U2: 5′-GGAGCAGGGAGATGGAATAGGAGCTTGCTCCGTCCACTCC-3′,
18S rRNA: 5′-ATCTTTGAGACAAGCATATGCTACTGGCAGGATCAACCAGGTA-3′). High sensitivity RNA blots were prepared as previously described (26).
Immunoprecipitation and quantitative RT-PCR
Immunoprecipitations were prepared as previously described (27,28). Nuclear lysates were prepared from HeLaYFP-Fibrillarin (29) and HeLaGFP (27) stable cell lines. Purified nuclei were resuspended in RIPA buffer to solubilize proteins. Fluorescence proteins were immunoprecipitated using anti-GFP monoclonal antibody (Roche) covalently coupled to protein G-Sepharose, as previously described. Samples were divided in to two and for input samples RNA was isolated from one half of each nuclear lysate. RNA was isolated by the TRIzol method with DNase I treatment, according to manufacturer’s instruction (Invitrogen). RT–PCR was performed to detect immunoprecipitated RNAs. Reverse transcription and PCR were performed with the following gene-specific primers:
U3: 5′-AGAGGTAGCGTTTTCTCCTGAGCG-3′ and 5′-ACCACTCAGACCGCGTTCTC-3′;
pre-GAPDH: 5′-CGCATCTTCTTTTGCGTCGCCAG-3′ and 5′-GGTCAATGAAGGGGTCATTGATGGC-3′,
U1: 5′-TACCTGGCAGGGGAGATACCATGATC-3′ and 5′-GCAGTCGAGTTTCCCACATTTGGGG-3′;
5S: 5′-ACGCGCCCGATCTCGTCTGAT-3′ and 5′-GCCTACAGCACCCGGTATTCCC-3′;
E2: 5′-GGAGTTGAGGCTACTGACTGGC-3′ and 5′-CCACTCATTGGGCCAGAGACCC-3′;
HBII-239 (hsa-miR-768 precursor): 5′-AGGATGAAAGTACGGAGTGAT-3′ and 5′-TCAGCAGTTTGAGTGTCAGCA-3′;
hsa-let-7g precursor: 5′-CGCTCCGTTTCCTTTTGCCTG-3′ and 5′-TACAGTTATCTCCTGTACCGG-3′;
hsa-miR16-1 precursor: 5′-CTTATGATAGCAATGTCAGCAGT-3′ and 5′-GATAATTTTAGAATCTTAACGCC-3′;
hsa-miR27b precursor: 5′-GTGATTGGTTTCCGCTTTGTTCA-3′ and 5′-CCCATCTCACCTTCTCTTCAGGT-3′;
hsa-miR28 precursor: 5′-TGAGTTACCTTTCTGACTTTCCC-3′ and 5′-TCAGATGAACGAAAGTGCCTGC-3′;
hsa-miR31 precursor: 5′-TGAATAGTCATAGTATTCTCC-3′ and 5′-CAGGTTCCCAGTTCAACAGCTAT-3′] using the SuperScript one-step RT-PCR kit (Invitrogen). To decide linearity of cycles, we used real time PCR using the Superscript III Platinum SYBR Green one-step qRT-PCR Kit (Invitrogen) and Rotor-Gene RG-3000 system (Corbett Research). The same amount of RNA for input and immunoprecipitated RNA (IP) was used as templates for RT-PCR reactions. Each experiment was repeated three times independently.
Scoring of the box C/D snoRNA features
To determine whether known human snoRNAs and miRNAs overlap in the genome, the genomic position of miRNAs and snoRNAs was retrieved from the UCSC Genome Browser (30), wgRNA table (17,31) from the March 2006 assembly of the human genome. Using a genetic algorithm described below, we investigated the possible relationship between miRNAs and box C/D snoRNAs by analysing the flanking sequences of miRNA precursors for evidence of previously unnoticed snoRNA-related features. To define the exact features that characterize M-box generating snoRNAs, we initially considered a region comprising 120 bases upstream and 120 bases downstream of known mature miRNAs found in introns to identify the possible positions of characteristic box C/D snoRNA elements in flanking sequences surrounding these miRNAs. This includes the presence of boxes C, D, D′ as well as regions of complementarity to rRNA. More precisely, we searched for the presence of sequences resembling box C (canonical motif: UGAUGA), box D (canonical motif: CUGA) and box D′ (canonical motif: CUGA). If the box D′ was absent, we searched for an eleven base region of complementarity to rRNA adjacent and immediately upstream of box D. If a box D′ was present, this region of complementarity to rRNA was searched for adjacent and immediately upstream of box D′ but was not required adjacent to box D. Thus, to identify box C/D snoRNA-like regions, we required the presence of boxes C and D as well as a region of complementarity to rRNA. The presence of a box D′ was optional (but scored more highly, see below).
The different features were assigned scores representing the number of matches to the respective canonical motif. The minimum score required for each region was:
Box C: scoreC ≥5 bases,
Box D: scoreD ≥3 bases,
Box D′ and adjacent rRNA complementarity region: scoreD’comp ≥12 bases,
Box D and adjacent rRNA complementarity region: scoreDcomp ≥12 bases.
Two types of hits were defined to reflect the diversity that exists among snoRNA molecules:
Type I hits that comprise a box C and a box D with adjacent rRNA complementarity region but no box D′, such that scoreC + scoreDcomp ≥ 18 bases.
Type II hits that comprise boxes C, D and a box D′ with adjacent rRNA complementarity region such that scoreC + scoreD + scoreD′comp ≥ 21 bases.
The rRNA regions considered for complementarity were the following:
between positions 1200 and 1900 of the 28S rRNA,
between positions 2250 and 2950 of the 28S rRNA,
between positions 3550 and 5035 of the 28S rRNA,
all of the 18 S and 5.8 S rRNA.
These regions were chosen because they contain the highest number of known 2′-O-ribose methylation sites in rRNA molecules. It has been suggested that it is the number and general distribution of methylation sites in rRNA rather than the exact position that has remained constant throughout evolution (32).
The scores for the different box C/D features and combinations thereof were deliberately chosen to allow deviation from the canonical sequences to ensure the detection of molecules that are evolving from functional snoRNAs to functional miRNAs and might not display exact matches to classical box C/D snoRNA elements.
Genetic algorithm
We used a genetic algorithm (33) to learn the positions of the different characteristic box C/D snoRNA features, both with respect to each other and to the mature miRNA. Ten parameters were considered and optimized to define snoRNA-like miRNA precursor features:
The range of acceptable positions for the 5′-end of the box C, with respect to the position of the miRNA (represented by parameters a1 and a2). Together, they define the region in which the box C can be located.
The range of acceptable positions for the 5′-end of box D, with respect to the position of the miRNA (represented by parameters b1 and b2).
The range of acceptable positions for the 5′-end of box D′, with respect to the position of the miRNA (represented by parameters c1 and c2).
The minimum and maximum distance (x) between the 5′-end of box C and the 3′-end of box D.
The minimum distance between the 3′ end of the box C and the 5′-end of the box D′ region (referred to as y).
The minimum value between the 3′-end of the box D′ and the 5′-end of the box D region (refered to as z).
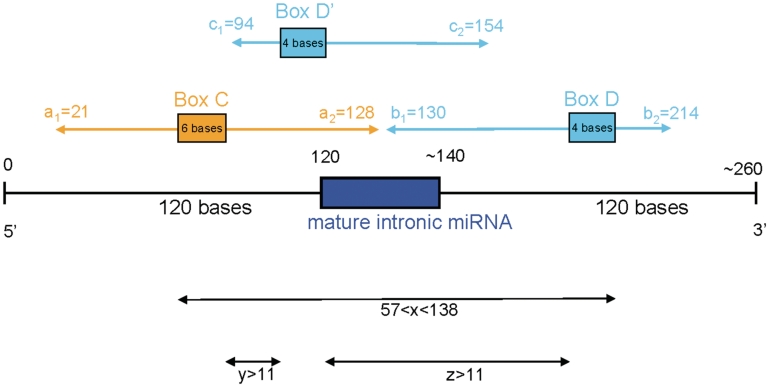
The parameters are illustrated in Figure 5.
Figure 5.
Positional characterization of the snoRNA–miRNA molecules. Ten positional parameters (a1, a2, b1, b2, c1, c2, xmin, xmax, y and z) were chosen to describe the position of snoRNA features with respect to the position of the mature miRNA. The values learnt by the genetic algorithm are indicated.
Human miRNAs from miRBase version 13 (34) were filtered to identify those found in introns according to genomic positions downloaded from the UCSC Genome Browser (30), using the March 2006 assembly of the human genome. In total, 333 miRNAs were considered by the genetic algorithm and scanned for the presence of snoRNA features using the 10 parameters defined above.
Multiple starting populations for the genetic algorithm were investigated and were generated by considering known examples of snoRNAs that contain smaller RNA fragments identified experimentally. These include HBII-180A, B, C and HBII-239 as described in the results section, as well as U95, U83, HBII-251, HBII-99 and HBII-202, as described in ref. (35). Each individual is defined by the values of the 10 parameters described above. The fitness of an individual was estimated by calculating a P-value for the proportion of intronic miRNAs identified as having snoRNA-like features using the individual’s parameter values. More precisely, for a given individual, all 333 human intronic miRNAs were scanned for the presence of snoRNA features using the individual’s parameter values. The number (N) of such predicted box C/D snoRNA-like miRNA precursors was counted. The P-value was calculated by determining the proportion of randomly chosen groups of 333 intronic regions (chosen at random from all human intronic sequences) that contain at least N hits as predicted using the individual’s parameter values.
To explore the parameter space, the starting populations were allowed to evolve and create new members of the population by crossover and mutation, as well as elimination of the least fit individuals. This was carried out over tens of generations. Crossover was implemented by randomly choosing two of the fittest individuals from the current generation and randomly assigning the value from one of the parents for each of the 10 parameters. Mutation was implemented by duplicating an individual from the current generation and allowing the value of up to three of its parameters chosen randomly to increase or decrease by an amount inversely proportional to the generation number. The fittest individuals as well as the new individuals generated by crossover and mutation formed the next generation.
Many of the initial populations converged onto a few individuals whose parameter values were very similar. One such individual with very high fitness was chosen to define the values of snoRNA-like miRNA precursor features. Its values are shown in Figure 5.
Of the 333 human miRNA precursors considered, 84 obtained at least one hit regardless of the type. In other words, 84 of the 333 miRNAs considered were predicted to be surrounded by snoRNA-like features. Eighteen thousand three hundred seventy-seven groups of 333 randomly chosen human intronic sequences were also scanned. Sixty-three of these groups contained at least 84 sequences that obtained at least one hit and thus a P-value of 0.003 was calculated.
Furthermore, if the presence of a box D′ is required and only type II hits are considered (see definition above), 74 of the 333 human intronic miRNA precursors obtained at least one hit, with a P-value of 0.0007.
Sequences and visualization
All sequences and alignments were retrieved from the UCSC Genome Browser using the March 2006 assembly of the human genome (30,36).
Secondary structure
All RNA secondary structures were predicted by RNAstructure 4.6 (37) and annotated using RnaViz 2.0 (38).
RESULTS
HBII-180 snoRNAs are processed into smaller fragments
The HBII-180 family of box C/D snoRNAs comprises three members encoded in the introns of the C19orf48 gene (22). All members of this family have well-conserved boxes C and D as well as an identical guide sequence complementary to positions 3677–3686 in the 28 S human rRNA, which contains a known 2′-O-methylation site (Figure 1). The HBII-180 molecules also have an M-box region, which is not conserved between the separate HBII-180 snoRNAs but is complementary to endogenous protein coding transcripts as previously described (22). We showed that the M-box region of snoRNA HBII-180C is capable of suppressing target gene mRNA and protein levels based on complementary base pairing interactions (22). Because the size of the HBII-180 M-boxes and their function are reminiscent of miRNAs, we tested whether RNA fragments corresponding to excised HBII-180 snoRNA M-box sequences could be detected in HeLa cells (Figure 2). First, an RNase protection assay using a HBII-180C snoRNA probe and total HeLa cell RNA revealed endogenous snoRNA fragments of ∼20 nt in length (Figure 2a). Second, a plasmid vector containing a minigene with the HBII-180C snoRNA was transiently transfected into HeLa cells (Figure 2b). A short snoRNA fragment under 20 nt was detected using a probe to the HBII-180C M-box region, but not with a probe to the 3′-end of the snoRNA, even after long exposure (Figure 2c). Similar results were found for HBII-180A and B (data not shown). These data suggested that M-box fragments are not non-specific degradation products, even though the levels of M-box fragments are lower than snoRNA levels, as judged by comparing expression levels between precursors and mature miRNAs (Figures 2 and 8). Endogenous forms of both full-length HBII-180 snoRNAs and M-box snoRNA fragments were also detected in extracts from untransfected HeLa cells (Figure 3). Interestingly, the snoRNA fragments derived from transiently expressed HBII-180C was specifically located in the nucleus and not detected in the cytoplasm (Figure 2b).
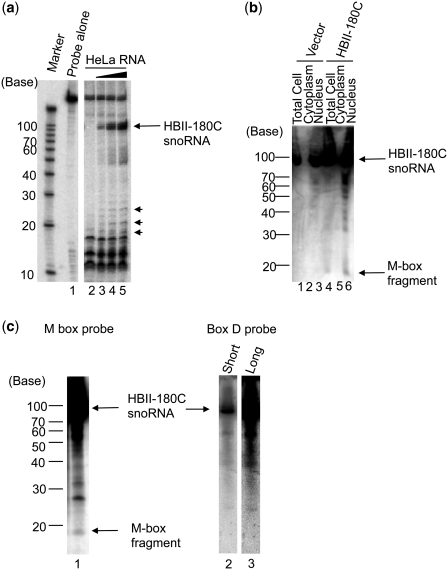
Figure 2.
Generation of M-box RNA Fragments from HBII-180 snoRNAs. (a) Detection of endogenous HBII-180C M-box fragment by RNase A/T1 protection assay. As a control, the diluted anti-sense probe against HBII-180C was loaded without RNase digestion (Probe lane). The probes were incubated with different amounts of HeLa cell total RNA (0, 1, 5, 10 µg for each, lanes 2–5). Both the mature HBII-180C snoRNA (arrow) and shorter fragments (arrow heads) were protected from RNase A/T1 digestion. (b) Detection of exogenously expressed HBII-180C M-box fragment by northern blotting. The same amount of HeLa cell cytoplasmic RNA (lanes 2 and 5), total RNA (lanes 1 and 4) and nuclear RNA (lanes 3 and 6) were compared with either a transiently transfected HBII-180C expression mini-gene or empty vector (lanes 1–3: Vector, lanes 4–6: HBII-180C). (c) Detection of endogenous HBII-180C M-box fragment by high sensitivity northern blotting. Fractionated HeLa cell nucleolar RNA was blotted on a nylon membrane and HBII-180C M-box fragment detected by hybridization with a radio-labelled RNA oligonucleotide probe (lane 1). The same filter was re-probed using a probe to the 3′ region (box D probe) of snoRNA HBII-180C. The result showed that 3′ region probe detected HBII-180C snoRNA (lane 2) but not any M-box fragment bands even after long exposure time (lane 3).
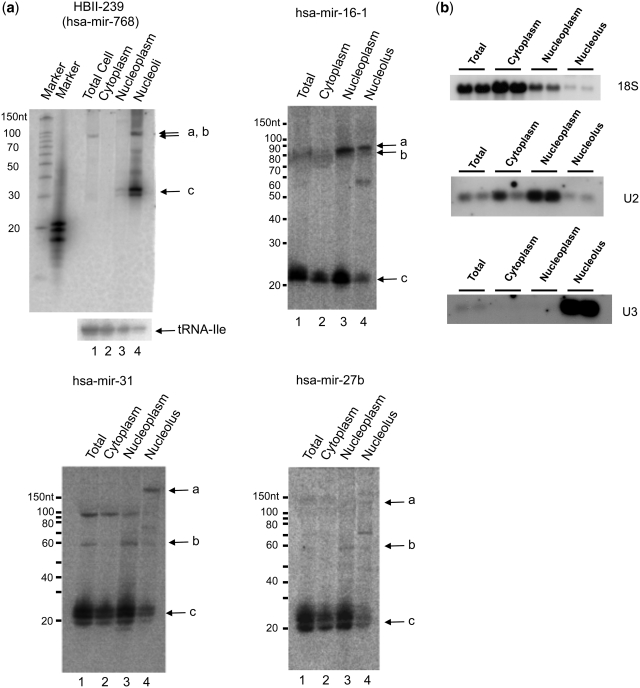
Figure 8.
Subcellular localization of box C/D snoRNA-like miRNA precursors. (a) Northern blots of HeLa cell extracts fractionated into cytoplasmic, nucleoplasmic and nucleolar fractions were probed for the presence of HBII-239, mir-16-1, mir-27b and mir-31 encoding molecules. In all panels, bands labelled with ‘a’ represent the expected size of the predicted snoRNAs, those labelled with ‘b’ represent the expected size for the miRNA hairpins and ‘c’ represents the expected size of the mature miRNA. (b) The fractionation was further controlled using U3 snoRNA, U2 snRNA and 18S rRNA. Blots show the results of two independent fractionations.
Figure 3.
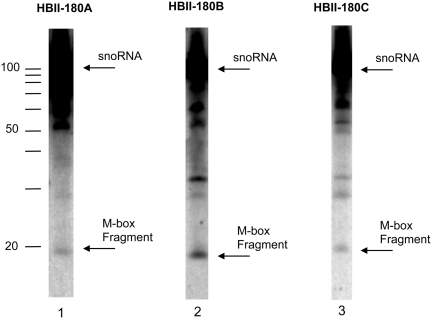
Detection of endogenous M-box RNA fragments of HBII-180 snoRNAs. Equivalent amounts of total HeLa cell RNA were analysed using oligonucleotide probes specific for the respective M-box regions (lanes 1–3).
Analysis of fractionated cell extracts confirmed that the full-length HBII-180C snoRNA was concentrated in the nucleolus, while the M-box snoRNA fragment was detected in both the nucleoplasmic and nucleolar fractions, but not in the cytoplasm (Figure 4a). This is consistent with the localization observed upon transient expression of HBII-180 minigenes. Parallel analysis showed that mature tRNA accumulates specifically in the cytoplasm, providing a positive control for cell fractionation (Figure 4b). These data suggest that HBII-180 snoRNAs can give rise to a population of ∼20 nt RNA fragments that include sequences complementary to mRNA. These M-box fragments accumulate in the nucleus but not in the cytoplasm.
Figure 4.
Detailed analysis of HBII-180C M-box fragments. (a) Equivalent amounts of total HeLa cell RNA (i), cytoplasmic RNA (ii), nucleoplasmic RNA (iii) and nucleolar RNA (iv) were separated by PAGE and transferred to a membrane using chemical cross-linking to facilitate high-sensitivity RNA detection as described. HBII-180C M-box RNA fragment sequences were detected using radiolabelled RNA oligonucleotide probes. The relative signal levels that were calculated by imaging software (ImageGauge v4.21, FUJI Photo Film co. Ltd.) are indicated graphically below each blot. The signal-back ground ratio between each fraction was normalized to set the highest signal at 100%. (b) The tRNA sequences were detected using radiolabelled RNA oligonucleotide probes as in (a).
Box C/D snoRNA-related precursors to known miRNAs
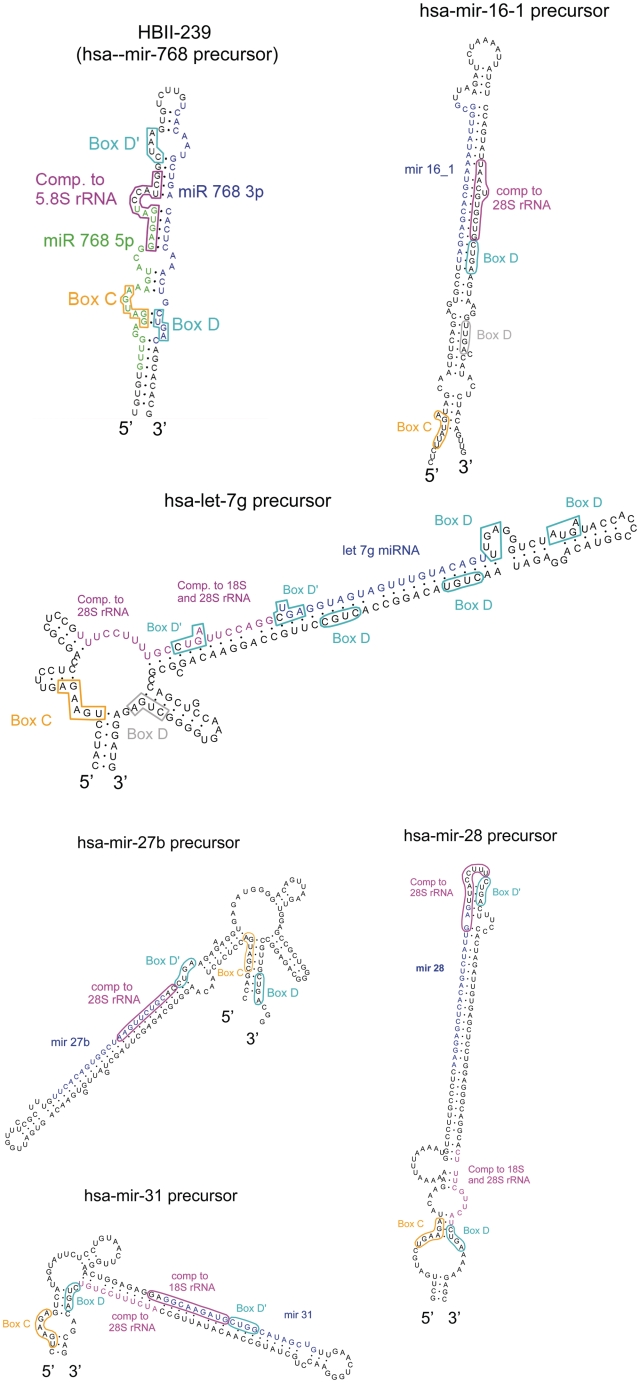
The observation that HBII-180 snoRNAs produce M-box fragments led us to investigate whether the structural relationship between box C/D snoRNAs and miRNA precursors is widespread. A comparison of published box C/D snoRNA and miRNA sequences in public databases reveals that the previously described miRNAs miR-768-3p and miR-768-5p (39), both are located within the sequence of the snoRNA HBII-239 (Figure 6). These miRNA entries were deleted from miRBase in 2008, because of their overlap with HBII-239 (31). However, we found that HBII-239 produces an ∼30 base-long RNA fragment which includes the previously reported miR768-3p sequence (Figure 8). The precursor for miR-768-3p and -5p corresponds to a putative box C/D snoRNA, whose existence was predicted by homology to the experimentally identified mouse snoRNA MBII-239 (40). This human HBII-239 snoRNA has not, however, been previously detected or characterized in human cells. Therefore, we analysed HeLa cell RNA and identified that HBII-239 is indeed expressed as a fibrillarin-associated snoRNA (Figure 7).
Figure 6.
Secondary-structure prediction of the HBII-239 box C/D snoRNA which contains the reported miR-768 miRNAs and predicted box C/D snoRNAs encoding known miRNAs. Mature miRNAs are drawn in blue and green. C and D boxes are shown, respectively, in orange and cyan while guide regions are shown in pink. Extra box D motifs not detected by our algorithm are shown in grey.
Figure 7.
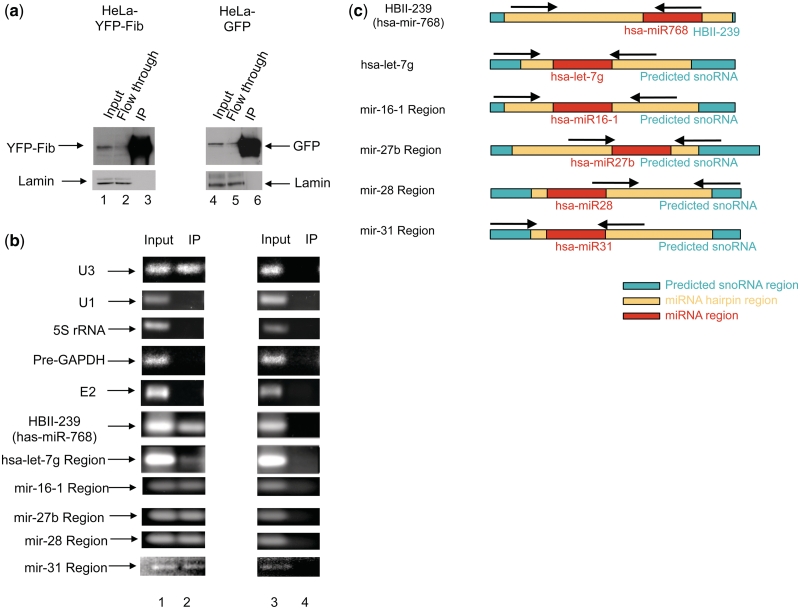
Co-immunoprecipitation of miRNA precursors with fibrillarin. (a) Western blot confirming specificity of the immunoprecipitation using an anti-GFP antibody. Nuclear extracts were prepared from HeLa cells stably expressing either free GFP or YFP-Fibrillarin (YFP-Fib) and immunoprecipitated using an anti-GFP antibody as previously described (22,27). The same membrane was reprobed with an antibody against lamin as a loading control. (b) RT–PCR used to detect co-precipitated HBII-239, hsa-mir-let-7g, hsa-mir-16-1, hsa-mir-27b, has-mir-28 and has-mir-31 miRNA precursors, with U3 box C/D snoRNA as positive control and, U1 snRNA, 5 S rRNA, GAPDH pre-mRNA and E2 box H/ACA snoRNA as negative controls for fibrillarin-associated RNAs. (c) Position of the primers used to detect the specified miRNA extended regions is shown by arrows.
To investigate the putative snoRNA–miRNA relationship further we employed an in silico approach. If certain miRNAs evolved through selection for the production of RNA fragments derived from snoRNA M-box sequences that can affect the expression of complementary mRNA targets, we reasoned that analysis of the flanking sequences of miRNA precursors may reveal evidence for previously unnoticed box C/D snoRNA-related features. Such snoRNA features are likely to be degenerate, however, assuming that dedicated miRNA precursors no longer need to function also as snoRNAs. Because most box C/D snoRNAs are encoded within introns, we examined the sequences flanking miRNAs found in introns, using a genetic algorithm (33) designed to be learn characteristic features of box C/D snoRNAs, including the position of C, D or D′ boxes with respect to the mature miRNA and a guide sequence complementary to rRNA (described in ‘Materials and Methods’ section). Because we predict that these molecules might be evolving from functional snoRNAs to functional miRNAs, we allowed a degree of deviation from the canonical box C/D features as described in the ‘Materials and Methods’ section. Using the parameters learnt by our algorithm (Figure 5), 84 human intronic miRNAs were detected showing features of box C/D snoRNAs in their flanking regions including 74 miRNA precursors displaying boxes C, D′, D and a region of complementarity to rRNA (type II hits, see Table 1 and Supplementary Table S1). When compared with randomly chosen intronic sequences, a P-value of 0.003 was calculated. This includes RNAs with complementary sequences to 5.8 S, 18 S or 28 S rRNA, combined with matches to the conserved snoRNA box C and D motifs.
Table 1.
Prevalence of the box C/D snoRNA–miRNA relationship (top 30 box C/D snoRNA-like miRNA hits)
| miRNA | Number of box C/D snoRNAhitsa | Box C score (best hit) | Box D′ score (best hit) | Box D score (best hit) | Total score of best hit | Presence of region of complementarity to rRNA |
||
|---|---|---|---|---|---|---|---|---|
| 5 S | 18 S | 28 S | ||||||
| mir-1201 | 233 | 6 | 14 | 4 | 24 | • | • | • |
| mir-27b | 20 | 5 | 14 | 4 | 23 | • | • | |
| mir-618 | 20 | 6 | 13 | 4 | 23 | • | • | • |
| mir-28 | 41 | 5 | 13 | 4 | 22 | • | • | |
| mir-423 | 5 | 5 | 13 | 4 | 22 | • | • | |
| mir-449a | 12 | 6 | 13 | 3 | 22 | • | • | |
| mir-500 | 32 | 5 | 13 | 4 | 22 | • | • | |
| mir-504 | 5 | 5 | 14 | 3 | 22 | • | • | |
| mir-582 | 13 | 5 | 13 | 4 | 22 | • | • | |
| mir-608 | 1 | 5 | 14 | 3 | 22 | • | ||
| mir-621 | 10 | 5 | 14 | 3 | 22 | • | • | |
| mir-765 | 7 | 6 | 12 | 4 | 22 | • | • | |
| mir-1259 | 13 | 6 | 12 | 4 | 22 | • | • | |
| mir-1272 | 15 | 6 | 13 | 3 | 22 | • | • | |
| mir-1288 | 45 | 5 | 13 | 4 | 22 | • | • | |
| mir-1294 | 8 | 5 | 13 | 4 | 22 | • | • | |
| let-7g | 18 | 5 | 13 | 3 | 21 | • | • | |
| mir-7-3 | 4 | 5 | 12 | 4 | 21 | • | ||
| mir-16-1 | 3 | 5 | 12 | 4 | 21 | • | ||
| mir-18a | 4 | 5 | 13 | 3 | 21 | • | ||
| mir-25 | 2 | 5 | 12 | 4 | 21 | • | • | |
| mir-26b | 2 | 5 | 13 | 3 | 21 | • | ||
| mir-29a | 9 | 5 | 12 | 4 | 21 | • | • | |
| mir-30c-1 | 3 | 5 | 12 | 4 | 21 | • | • | |
| mir-31 | 4 | 5 | 12 | 4 | 21 | • | • | |
| mir-95 | 3 | 5 | 12 | 4 | 21 | • | ||
| mir-101-2 | 3 | 5 | 12 | 4 | 21 | • | • | |
| mir-105-2 | 19 | 6 | 12 | 3 | 21 | • | • | |
| mir-142 | 6 | 5 | 13 | 3 | 21 | • | ||
| mir-151 | 4 | 5 | 12 | 4 | 21 | • | ||
aHits represent different arrangements of snoRNA characteristics around a given miRNA. Hits are considered distinct if there is a difference of positioning of at least one of boxes C, D or D′ and/or a different position of the rRNA target sequence.
A recent report identified 11 box C/D snoRNAs that are processed into miRNA-like fragments with gene-silencing capabilities (21). To test our genetic algorithm, we investigated whether these 11 box C/D snoRNAs are predicted as miRNA-like hits given the position of their derived small fragments. Nine of these 11 sno-miRNAs (21) (snR39B, HBII-336, HBII-142, U27, HBII-429, U83A, U15A, U74, U78) were predicted as hits by our genetic algorithm. The remaining two sno-miRNAs, U3 and U3-4, are much larger than most typical box C/D snoRNAs and thus do not fit the constraint we imposed in our initial hypothesis of the snoRNA precursor being at most only 120 nt longer than the 5′-end of the derived small fragment.
To test our algorithm further, we investigated two different control data sets. All known box C/D snoRNAs were used as a positive control, regardless of whether they have been found to generate smaller fragments or not. And as a negative control, we scanned all known box H/ACA snoRNAs, which as described in the ‘Introduction’ section, are intronic elements that are functionally related to box C/D snoRNAs but do not display the same characteristic features. Of the 253 box C/D snoRNAs tested, 233 (92%) were identified as hits by our genetic algorithm. In contrast, only 22 of the 94 box H/ACA snoRNAs were identified as box C/D snoRNA-like by our genetic algorithm suggesting a false positive rate of 23%. All snoRNA sequences were downloaded from snoRNAbase (17).
As described in the ‘Materials and Methods’ section, to perform our search systematically, regions of 120 nt flanking the mature miRNA sequences upstream and downstream were scanned. While the positional parameters of our genetic algorithm guarantee that the mature miRNA is included within the predicted snoRNA (Figure 5), the position of the miRNA hairpin was not considered when creating the genetic algorithm. And while it is possible that snoRNAs are processed to generate miRNA harpins which are further processed to yield the mature miRNAs, the simplest processing mechanism would be that the molecule corresponding to the snoRNA is directly processed to generate the mature miRNA (and thus the predicted snoRNA would correspond to the pre-miRNA). A comparison between the positions of the predicted box C/D snoRNA features and the miRNA hairpins predicted by miRBase, as available from the UCSC Genome Browser miRNA track (30,31), shows that 20 of the 84 predicted snoRNA-like miRNA precursors have all their predicted characteristic box C/D snoRNA features (boxes C, D′, D and region of complementarity to rRNA) entirely contained within the predicted miRNA hairpin. We note that the hairpin sequences available from miRBase (31) are computationally predicted and often do not match up exactly with miRNA precursors detected experimentally, such as those reported in ref. (41). In addition, the miRNA hairpins are likely to be predicted to end where the complementarity decreases, which would exclude the region corresponding to the bulge containing the boxes C and D at the base of the predicted snoRNA. Indeed, if extended by only 10 nt, 31 of the 84 predicted snoRNA-like miRNA hairpins predicted by miRBase contain all the characteristic snoRNA features considered (boxes C, D′, D and the region of complementarity to rRNA). Supplementary Table S1 indicates whether the characteristic box C/D snoRNA features are contained within the predicted miRNA hairpin which could be useful to prioritize the experimental characterization of these molecules.
Structures of the predicted snoRNA-like miRNA molecules
Box C/D snoRNAs have distinct and well-characterized features. Their main sequence elements consist of conserved boxes C (UGAUGA) and D (CUGA), which form a kink-turn in the folded molecule (14). They often contain a second box C/D pair referred to as the boxes C′ and D′. Regions of complementarity to rRNA, which guide 2′-O-ribose methylation, are found immediately upstream of boxes D′ (12–14,17). To investigate whether the predicted snoRNA-like miRNA precursors display typical box C/D snoRNA elements of secondary structure, all predicted snoRNA sequences were folded using RNAstructure and five examples were tested further for the ability to bind specifically to fibrillarin (Figures 6 and 7).
Predicted box C/D snoRNA-like miRNA precursors that bind fibrillarin
We investigated whether the predicted box C/D snoRNA-like miRNA precursors have retained some functional snoRNA characteristics by experimentally testing if they bind specifically to fibrillarin, a core component of box C/D snoRNPs. Fibrillarin functions as the 2′-O-ribose methylase and binds the box C and D motif (12–14,17). Five of the 84 snoRNA-like miRNAs, mir-16-1, mir-31, mir-27b, let-7g and mir-28 were selected for this analysis because they are mostly well-characterized regulatory RNAs (41–45) that are all expressed in HeLa cells. Nuclei from HeLa cells expressing either YFP-fibrillarin, or GFP, as a control, were purified and immunoprecipitated using an antibody against the fluorescent proteins (see ‘Materials and Methods’ section). The RNA was isolated from these samples and analysed by qRT-PCR (see ‘Materials and Methods’ section). As shown in Figure 7a and b, in addition to U3 (a well-characterized box C/D snoRNA), all five snoRNA-like miRNA precursors tested (i.e. the extended regions of let-7g, mir-16-1, mir-31, mir-27b and mir-28 as described in Figure 7c) as well as HBII-239, co-purify specifically with fibrillarin. In contrast, the precursor of the box H/ACA snoRNA E2 is not pulled down by fibrillarin. Other abundant nuclear RNAs, including GAPDH pre-mRNA, U1 snRNA and 5 S rRNA were used as negative controls and are not immunoprecipitated by fibrillarin. These fibrillarin binding experiments strongly support the in silico prediction that a subset of miRNA precursors are related to box C/D snoRNAs.
Predicted box C/D snoRNA-like miRNA precursors localized in the nucleolus
The subcellular localization of five of the fibrillarin-bound snoRNA-like miRNA precursors, along with the smaller fragments derived from them, was further investigated by cell fractionation and northern blot analysis (Figure 8a and b, Supplementary Figure S1). As shown in Figure 8a, bands of the predicted size or larger for box C/D snoRNA full-length molecules encoding mir-16-1, mir-31 and mir-27b are detected in the nucleolus (bands labelled with ‘a’). Similar results were also shown for both mir-28 and let-7g (Supplementary Figure S1). In contrast, miRNA hairpin sized bands are detected in the nucleoplasmic fraction (bands labelled with ‘b’). Bands corresponding in size to the mir-16-1, mir-31 and mir-27b mature forms are also detected in all three fractions and mainly found in the nucleoplasmic and cytoplasmic fractions (bands labelled with ‘c’). A band slightly larger than the mir-768 mature form was also detected, localized mainly in the nucleolus. These fractionation experiments support the in silico predictions and fibrillarin pull-down experiments and support the view that the hsa-mir-768 (HBII-239), hsa-mir-16-1, hsa-mir-27b, hsa-mir-28, hsa-let-7g and hsa-mir-31 precursors differ from classical miRNA precursors and are consistent with a snoRNA origin.
DISCUSSION
The combined computational and biochemical data presented here shows that certain mammalian miRNA precursors are closely related to box C/D snoRNAs. A subset of miRNA precursors and box C/D snoRNAs have similarities in their primary structure including the presence of characteristic box C/D snoRNA features (as detected by our genetic algorithm) as well as in their secondary structures (Figures 1 and 6). In addition, all six box C/D snoRNA-like miRNA precursors tested, bind specifically to fibrillarin (Figure 7) and display a predominant subcellular localization pattern similar to snoRNPs, consistent with a snoRNA origin (Figure 8). These observations provide additional complementary evidence of the extensive relationship between snoRNA and miRNA precursors, which has been uncovered recently. Indeed, we have also identified known miRNAs encoded within box H/ACA snoRNA-like molecules (10). While we have described here and previously (10) known miRNA precursors with snoRNA-like features, others have investigated known snoRNAs that are further processed into miRNA-like fragments (7,11,21,35), demonstrating the widespread nature of this relationship.
Several aspects of the possible snoRNA–miRNA relationship have now been explored independently, either directly, or indirectly, by different groups. The length of the most abundant form of the mature miRNAs encoded in snoRNA-like precursors as predicted by our genetic algorithm varies between 19 and 27 nt although most (75 out of 84) are between 21 and 23 nt in length. While many small box C/D snoRNA-derived fragments were previously found to be between 17 and 19 nt in length, a substantial number are of typical miRNA size (20–24 nt) or even larger (30–31 nt) (11). Thus the miRNA precursors with box C/D snoRNA features described here resemble a subpopulation of previously described small-RNA generating box C/D snoRNAs. Recently, 11 box C/D snoRNAs were shown to display gene silencing activity and generate smaller fragments (21) with a similar length distribution as the box C/D snoRNA-like miRNAs reported here.
The localization of the mature miRNAs derived from snoRNA-like precursors also agrees with recent reports characterizing miRNAs. Contrary to the miRNA biogenesis pathway originally described, many mature miRNAs have now been experimentally shown to be present in the nucleus in levels at least as high as in the cytoplasm (46) and several nucleolar mature miRNAs have been described (20). Furthermore, box C/D snoRNA-derived small RNAs have also recently been shown to localize predominantly in the nucleus (47). Additional research will be important to determine whether the function and/or biogenesis pathway including their maturation of nucleolar, nucleoplasmic and cytoplasmic miRNAs and snoRNA-like miRNA precursors differ and thus perhaps to subclassify miRNAs based on these properties. For example, they could either be added to miRBase (34) or put in a separate database describing the wealth of new miRNA-like molecules that have been identified recently. We note that mir-768, mir-1259 and mir-1201 were removed from miRBase (34) specifically because they overlap with a conserved snoRNA. The data from this study and other recent work highlight the possibility that miRNAs could be generated from snoRNAs. This questions the wisdom of removing such molecules from miRNA repositories. While the snoRNA-like miRNAs do not display all features of prototypical miRNAs (41), this may reflect the fact that additional non-canonical pathways for miRNA biogenesis exist (6).
Mammalian snoRNAs have recently been described as mobile genetic elements (18). The duplication of C/D snoRNA genes would allow retention of the rRNA 2′-O-methylation activity, while the duplicated copy may gradually evolve into a dedicated small regulatory RNA, providing an ongoing source of newly evolving small regulatory RNA molecules. A role for snoRNAs as evolutionary precursors of small regulatory RNAs is consistent with the known prevalence of large families of abundant snoRNAs in all known eukaryotic cells, including species such as budding yeast that apparently do not possess miRNAs. The evidence provided here and elsewhere supports the hypothesis that snoRNAs and/or their copies can serve as sources of precursors of miRNAs and thus are not only involved in the chemical modification of rRNAs and other nuclear RNAs, targeted through complementary base pairing, but possibly also in the regulation of gene expression, again targeted through complementary base pairing.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Wellcome Trust Programme Grant (ref: 073980/Z/03/Z to A.I.L.); MRC Milstein Award (ref: G0801738 to A.I.L.); BBSRC RASOR (Radical Solutions for Researching the proteome) (Network grant to A.I.L.); Scottish Funding Council grant from the Scottish Bioinformatics Research Network. Wellcome Trust (grant WT083481); Caledonian Research Foundation, post-doctoral fellowship (to M.S.S.). Funding for open access charge: Wellcome Trust Programme Grant (ref: 073980/Z/03/Z to A.I.L.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues for helpful discussions and suggestions. A.I.L. is a Wellcome Trust Principal Research Fellow.
REFERENCES
- 1.Lai EC. microRNAs: runts of the genome assert themselves. Curr. Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 2.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai EC. miRNAs: whys and wherefores of miRNA-mediated regulation. Curr. Biol. 2005;15:R458–R460. doi: 10.1016/j.cub.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 6.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol. Genet. Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 7.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 13.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 15.Weinstein LB, Steitz JA. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 1999;11:378–384. doi: 10.1016/S0955-0674(99)80053-2. [DOI] [PubMed] [Google Scholar]
- 16.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 17.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber MJ. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo Y, Li S. Genome-wide analyses of retrogenes derived from the human box H/ACA snoRNAs. Nucleic Acids Res. 2007;35:559–571. doi: 10.1093/nar/gkl1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Politz JC, Hogan EM, Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono M, Yamada K, Avolio F, Scott MS, van Koningsbruggen S, Barton GJ, Lamond AI. Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol. Biol. Cell. 2010;21:1569–1584. doi: 10.1091/mbc.E10-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 24.Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- 25.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 2007;17:749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pall GS, Codony-Servat C, Byrne J, Ritchie L, Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007;35:e60. doi: 10.1093/nar/gkm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trinkle-Mulcahy L, Andersen J, Lam YW, Moorhead G, Mann M, Lamond AI. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 2008;183:223–239. doi: 10.1083/jcb.200805092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung AK, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 2004;166:787–800. doi: 10.1083/jcb.200405013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennis PP, Omer A, Lowe T. A guided tour: small RNA function in Archaea. Mol. Microbiol. 2001;40:509–519. doi: 10.1046/j.1365-2958.2001.02381.x. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell TM. Machine Learning. New York: McGraw-Hill; 1997. [Google Scholar]
- 34.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1209s29. Chapter 12, Unit 12.19.11-10. [DOI] [PubMed] [Google Scholar]
- 35.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Rijk P, Wuyts J, De Wachter R. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- 39.Berezikov E, van Tetering G, Verheul M, van de Belt J, van Laake L, Vos J, Verloop R, van de Wetering M, Guryev V, Takada S, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16:1289–1298. doi: 10.1101/gr.5159906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bushati N, Cohen SM. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 43.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 44.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67:6031–6043. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 45.Michael MZ, O'Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 46.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 2010;17:1030–1034. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.