Abstract
Mutations in the three genes encoding the heterotrimeric RNase H2 complex cause Aicardi-Goutières Syndrome (AGS). Our mouse RNase H2 structure revealed that the catalytic RNase H2A subunit interfaces mostly with the RNase H2C subunit that is intricately interwoven with the RNase H2B subunit. We mapped the positions of AGS-causing RNase H2A mutations using the mouse RNase H2 structure and proposed that these mutations cause varied effects on catalytic potential. To determine the functional consequences of these mutations, heterotrimeric human RNase H2 complexes containing the RNase H2A subunit mutations were prepared, and catalytic efficiencies and nucleic acid binding properties were compared with the wild-type (WT) complex. These analyses reveal a dramatic range of effects with mutations at conserved positions G37S, R186W, and R235Q, reducing enzymatic activities and substrate binding affinities by as much as a 1000-fold, whereas mutations at non-conserved positions R108W, N212I, F230L, T240M, and R291H reduced activities and binding modestly or not at all. All mutants purify as three-subunit complexes, further supporting the required heterotrimeric structure in eukaryotic RNase H2. These kinetic properties reveal varied functional consequences of AGS-causing mutations in the catalytic RNase H2A subunit and reflect the complex mechanisms of nuclease dysfunction that include catalytic deficiencies and altered protein-nucleic acid interactions relevant in AGS.
Keywords: DNA-binding Protein, Enzyme Kinetics, Genetic Diseases, Innate Immunity, Nucleic Acid Enzymology, RNA-binding Protein, RNA Catalysis
Introduction
Aicardi-Goutières Syndrome (AGS)2 is a rare genetic neurological disorder that mimics congenital viral infection with evidence of high interferon-α levels in serum and cerebrospinal fluid (1, 2). Mutations in the five genes RNASEH2A, RNASEH2B, RNASEH2C, TREX1, and SAMHD1 are known to cause AGS (3–5). The RNASEH2A, RNASEH2B, and RNASEH2C genes encode the RNase H2 heterotrimeric complex that recognizes and cleaves ribonucleotides present in RNA/DNA duplexes (4, 6). The TREX1 gene encodes a homodimeric 3′ → 5′ DNA exonuclease (7–12), and the SAMHD1 gene encodes an enzyme of unknown function. The identification of disease-causing mutations in genes encoding the nucleases RNase H2 and TREX1 provides evidence for the concept that failure to properly process endogenous nucleic acids triggers an interferon-mediated immune response, helping to explain the phenotypic overlap of AGS with congenital infection (13). Furthermore, there is emerging evidence for a clinical, biochemical, and genetic connection between AGS and related autoimmune disorders including systemic lupus erythematosus, indicating that dysfunctional nucleic acid metabolism leads to immune activation and an autoimmune phenotype (14).
RNase H2 provides the majority of ribonuclease H activity in human cells (15). All ribonuclease H enzymes specifically recognize RNA/DNA hybrids and hydrolyze the ribonucleotide-containing polynucleotide strand. These enzymes are divided into two major families, type 1 and type 2, based on amino acid sequence similarities and distinguishing biochemical properties that suggest unique in vivo substrates (16). RNase H2 belongs to the type 2 family, which is biochemically characterized by its ability to recognize and cleave a single ribonucleotide within a DNA duplex (17). The heterotrimeric nature of eukaryotic RNase H2 distinguishes it from other type 2 family members like those found in bacteria and archaea that exist as single polypeptides (18–20). Active eukaryotic RNase H2 enzymes have only been recovered when all three subunits are present, highlighting the intimate relationship between these proteins in the complex (6, 19, 21).
The mammalian RNase H2 complex has a critical function in nucleic acid metabolism to prevent immune activation with potential roles in multiple biological pathways. Ribonucleotides that are misincorporated by DNA polymerases during genomic replication create potential adducts that are removed in an RNase H2-dependent repair process, thereby preventing replicative stress and genomic instability (22). RNase H2 and endonuclease FEN-1 cooperate to remove RNA primers from an RNA-DNA duplex, similar to what might occur during Okazaki fragment processing (23–25). The direct interaction of RNase H2 with proliferating cell nuclear antigen in the replication apparatus could facilitate either ribonucleotide removal or Okazaki fragment processing (26). Stable RNA/DNA hybrids between nascent RNA transcripts and the DNA template (R-loops) are generated by RNA polymerases and can promote genomic instability that is resolved by RNase H2 (27, 28). Additionally, RNase H2 may function to degrade RNA/DNA hybrids during cell death to avoid nucleic acid-mediated immune activation (4).
Two distinguishable clinical presentations of AGS have been described in patients with mutations in the three genes encoding RNase H2 (13, 29). An early-onset neonatal form with clinical features similar to congenital infection is seen mostly in patients with mutations in RNASEH2A and RNASEH2C, as is also the case for patients carrying mutations in TREX1 and SAMHD1. A later-onset form of presentation after a significant period of normal development is most frequently seen in patients with RNASEH2B mutations. Attempts to correlate diminished RNase H2 activities to AGS-causing mutations in heterotrimeric complexes indicate normal levels of ribonuclease H activity in the human (6) and yeast (16, 19) enzymes containing RNase H2B and H2C subunit mutations. In contrast, activity measurements on the RNase H2A G37S mutant indicate altered nuclease activities dependent upon the specific nucleic acid substrate (6, 30). Our RNase H2 structure shows the interwoven architecture of the RNase H2B and RNase H2C proteins that interface with the RNase H2A catalytic subunit at a position on the opposite face from the proposed nucleic acid binding site (Fig. 1) (30). Thus, full catalytic activities in the RNase H2B and H2C mutant enzymes likely point to mechanisms of dysfunction mediated through altered protein-protein interactions rather than catalytic inactivity (6, 16, 19, 30).
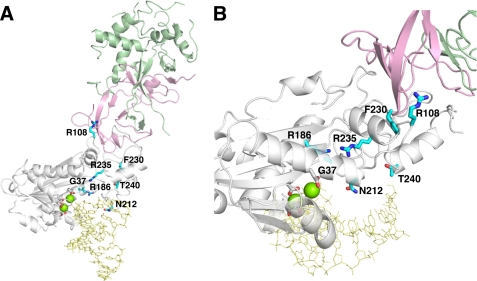
FIGURE 1.
The RNase H2A subunit AGS-causing mutations. A, the mouse RNase H2 heterotrimer with a model RNA-DNA/DNA (yellow) (PDB ID 3KIO (30)) consists of the H2B (green), H2C (pink), and H2A (white) subunits is shown. The human RNase H2A subunit AGS-causing mutations (cyan) were mapped onto the mouse structure. The four conserved acidic residues (white) that likely coordinate two divalent metal ions (green spheres) for catalysis are shown. The indicated residue numbers are of the human H2A protein (actual numbers are mouse:human; Gly-37:Gly-37, Arg-109:Arg-R108, Arg-187:Arg-186, Asn-213:Asn-212, Phe-231:Phe-230, Arg-236:Arg-235, Thr-241:Thr-240). B, the H2A G37S, R186W, and R235Q mutations are in close proximity to the active site with dramatic consequences to catalytic function. The H2A R108W, N212I, F230L, and T240M mutant residues are located more distant from the active site with corresponding reduced effects on nucleic acid binding and catalysis.
The AGS-causing missense mutations in RNASEH2A encoding the catalytic subunit are distributed throughout the gene (14, 29). Mutations in RNASEH2A are autosomal recessive with homozygous and compound heterozygous patients exhibiting early-onset neonatal clinical presentation. To determine the functional consequences of these RNASEH2A mutations, we expressed and purified the human RNase H2 mutant heterotrimeric complexes and compared nuclease activities to the WT enzyme. We first performed a detailed kinetic analysis to measure the catalytic and nucleic acid binding properties of the RNase H2 WT enzyme. These studies revealed the strikingly different individual kinetic constants that govern the RNase H2 cleavage reaction, dependent upon the number of ribonucleotides present in the polynucleotide substrate. The AGS-causing RNase H2A mutations at phylogenically conserved positions exhibit the greatest reductions in enzymatic activities and nucleic acid binding affinities relative to the RNase H2 WT with all substrates tested. In contrast, RNase H2A subunit mutations at positions that are only conserved in the eukaryotic RNase H2 three-subunit complexes exhibit modest or no decreases in enzymatic activities or binding affinities. These results show that AGS-causing RNase H2A subunit mutations exhibit a spectrum of effects on the catalytic competency of this enzyme. Furthermore, multiple mechanisms of RNase H2 dysfunction that include loss of ribonuclease H activity as well as dysfunctional protein-nucleic acid interactions essential to avoid activation of the immune system are indicated.
EXPERIMENTAL PROCEDURES
Materials
The oligonucleotides 5′-FAM-ACCTATCTAACAACACTCAcTCCATCATAT (1-ribo), 5′-FAM-ACCTATCTAACAACACucacTCCATCATAT (4-ribo), and 5′-FAM-acctatctaacaacactcacTCCATCATAT (20-ribo) were purchased from Bio-Synthesis, Inc. Ribonucleotides are indicated with lowercase letters. Oligonucleotides were purified on urea-polyacrylamide gels, recovered by ethanol precipitation, and resuspended in 2 mm Tris-HCl (pH 8.0). The deoxynucleotide 5′-ATATGATGGAGTGAGTGTTGTTAGATAGG (DNA) complementary to all ribo-containing oligonucleotides was from Operon. Duplex polynucleotide substrates were prepared by incubating equal molar amounts of complementary oligonucleotides in 20 mm Tris-HCl (pH 7.5), 50 mm NaCl in a 1-liter boiling water bath and allowing the bath to cool to room temperature.
Enzyme Preparations
The human RNase H2 WT and mutant heterotrimer complexes were expressed and purified as previously described (6). Site-directed mutations were introduced into the RNASEH2A gene using QuikChange (31) with the pET-A plasmid, and constructs were confirmed by sequencing. The RNase H2A N212I mutation was cloned directly into the pET-A expression plasmid after PCR amplification of mRNA from peripheral blood leukocytes of an AGS patient carrying this allele.3 Purified enzymes were concentrated and stored at −80 °C in 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, and 10% glycerol (supplemental Fig. 2). Enzyme concentrations were determined by A280 using the molar extinction coefficient: 83,030 m−1cm−1.
Ribonuclease H Assays
Relative ribonuclease H activities were measured in reactions (30 μl) containing 20 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 2 mm dithiothreitol, 50 μg/ml bovine serum albumin, 200 nm 5′-FAM-DNA19-RNA1-DNA10/DNA30 (1-ribo), 5′-FAM-DNA16-RNA4-DNA10/DNA30 (4-ribo), or 5′-FAM-RNA20-DNA10/DNA30 (20-ribo) duplex (Table 1) and the amounts of RNase H2 or mutant enzyme indicated in the figure and table legends (Fig. 4, Table 2). After incubation at 25 °C for 20 min, reactions were quenched with 3 volumes of 100% ethanol and dried in vacuo. Pellets were resuspended in 9 μl of formamide and heated at 95 °C for 5 min. Reaction products from a sample (5 μl) were separated on 23% urea-polyacrylamide gels, visualized using a Storm PhosphorImager (GE Healthcare), and quantified with ImageQuant 5.2. Reported relative activities and S.E. are the averages of triplicate reactions using four different enzyme concentrations in which less than 30% of substrate was converted to product.
TABLE 1.
DNA-RNA hybrids used in this study
| Name | Sequencea |
|---|---|
| 1-ribo | 5′FAM-ACCTATCTAACAACACTCAcTCCATCATAT-3′ |
| 3′-TGGATAGATTGTTGTGAGTGAGGTAGTATA-5′ | |
| 4-ribo | 5′FAM-ACCTATCTAACAACACucacTCCATCATAT-3′ |
| 3′-TGGATAGATTGTTGTGAGTGAGGTAGTATA-5′ | |
| 20-ribo | 5′FAM-acctatctaacaacactcacTCCATCATAT-3′ |
| 3′-TGGATAGATTGTTGTGAGTGAGGTAGTATA-5′ |
a Ribonucleotides are indicated in lowercase. All single-stranded oligonucleotides were annealed to the reverse complement at a1:1 molar ratio. Duplex formation was confirmed by visualization using a non-denaturing PAGE.
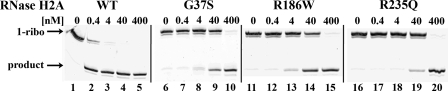
FIGURE 4.
The ribonuclease H activities of RNase H2A mutants. Reactions containing RNase H2A WT (lane 2, 0.4 nm; lane 3, 4 nm; lane 4, 40 nm; lane 5, 400 nm), RNase H2A G37S (lane 7, 0.4 nm; lane 8, 4 nm; lane 9, 40 nm; lane 10, 400 nm), RNase H2A R186W (lane 12, 0.4 nm; lane 13, 4 nm; lane 14, 40 nm; lane 15, 400 nm), or RNase H2A R235Q (lane 17, 0.4 nm; lane 18, 4 nm; lane 19, 40 nm; lane 20, 400 nm) were incubated with the 1-ribo RNA-DNA (200 nm). Lanes 1, 6, 11, and 16 contain no RNase H2. Reaction products were visualized by denaturing PAGE and quantified as described under “Experimental Procedures.”
TABLE 2.
Relative activities of the RNase H2A AGS-causing mutants
| RNase H2A | 1-ribo |
4-ribo |
20-ribo |
|||
|---|---|---|---|---|---|---|
| Activitya | Relative activityb | Activitya | Relative activityb | Activitya | Relative activityb | |
| WT | 9.5 ± 1.0 | 1 | 7.8 ± 1.6 | 1 | 8.3 ± 1.6 | 1 |
| G37S | 0.046 ± 0.006 | 1/210 | 2.6 ± 0.4 | 1/3 | 2.5 ± 0.4 | 1/3 |
| R186W | 0.056 ± 0.01 | 1/160 | 0.90 ± 0.2 | 1/9 | 1.0 ± 0.2 | 1/8 |
| R235Q | 0.026 ± 0.004 | 1/370 | 0.013 ± 0.003 | 1/600 | 0.011 ± 0.003 | 1/750 |
| R108W | 3.8 ± 0.3 | 1/3 | 5.3 ± 0.8 | 1/2 | 5.8 ± 0.5 | 1/1 |
| N212I | 10 ± 1 | 1/1 | 7.5 ± 2 | 1/1 | 8.5 ± 0.9 | 1/1 |
| F230L | 0.81 ± 0.1 | 1/12 | 2.3 ± 0.4 | 1/3 | 2.1 ± 0.3 | 1/4 |
| T240M | 1.1 ± 0.2 | 1/9 | 7.2 ± 2 | 1/1 | 16 ± 2 | 2/1 |
| R291H | 21 ± 3 | 2/1 | 17 ± 2 | 2/1 | 18 ± 2 | 2/1 |
a To precisely quantify activities, each enzyme was assayed as described under “Experimental Procedures” in triplicate at four different concentrations that converted less than 30% of substrate to product. Plots of activity versus enzyme concentrations confirmed linearity of the assays and generated the enzyme activity values. The average activities (pmol of product/min/pmol of enzyme) and S.E. for RNase H2 WT and mutants are shown.
b The relative activity was calculated as: relative activity = 1/[(pmol of product/min/pmol of WT enzyme)/(pmol of product/min/pmol of mutant enzyme)].
Steady-state Reactions
The ribonuclease H reactions to determine Km and kcat values for the RNase H2 WT and mutant enzymes were as described above with the concentrations of FAM-labeled RNA/DNA duplex substrate and enzymes indicated in the figure and table legends (Figs. 2 and 5, and Table 3). Reactions were initiated by the addition of substrate. The kinetic data were fit to a hyperbolic equation to calculate the Km and kcat values using Sigma Plot 8.02 (SPSS Science, Inc.).
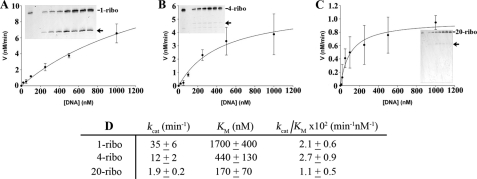
FIGURE 2.
Catalytic efficiency of RNase H2 WT using various nucleic acid substrates. The RNase H2 WT (500 pm) was incubated in reactions with increased concentrations of 1-ribo at 0, 10, 20, 50, 100, 250, 500, 1000 nm (A), 4-ribo at 0, 20, 50, 100, 250, 500, 1000 nm (B), and 20-ribo at 0, 10, 20, 50, 100, 250, 500, 1000 nm (C) RNA-DNA substrates. Reaction products were subjected to electrophoresis on 23% polyacrylamide gels (insets), indicating that only the initial cleavage products (arrow) are detected under these conditions. The products were quantified, and reaction rates (nm/min) were plotted against RNA-DNA concentrations. The data were fit to a single hyperbola to calculate the kinetic constants kcat and Km. The RNase H2 WT steady-state kinetic data (D) for triplicate reactions on each substrate indicate variations in kcat and Km yield overall efficiencies (kcat/Km) that are similar for each substrate.
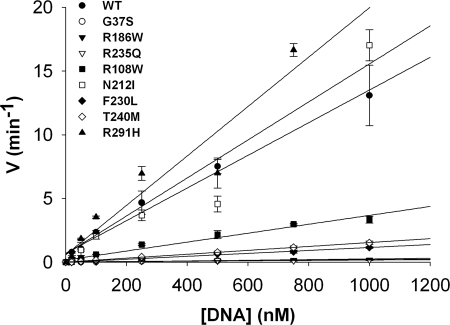
FIGURE 5.
Catalytic efficiencies of the RNase H2A mutants. Reactions containing RNase H2A WT (▴), G37S (●), R186W (▾), R235Q (▿), R108W (■), N212I (□), F230L (⧫), T240M (◊), and R291H (▴) were incubated with increased concentrations of the 1-ribo RNA-DNA substrate. Reaction products were subjected to electrophoresis on 23% polyacrylamide gels, and the products were quantified as described under “Experimental Procedures.” Reaction rates (min−1) were plotted against 1-ribo substrate concentrations. The second-order rate constant (kcat/Km) was estimated from the slope of the linear portion of the steady-state reaction that could be clearly defined for all RNase H2A mutants (Table 3).
TABLE 3.
Catalytic efficiencies of RNase H2A AGS-causing mutants
| RNase H2A | kcat/Km × 102a | Fraction of WT efficiency |
|---|---|---|
| min−1nm−1 | ||
| WT | 1.28 ± 0.06 | 1 |
| G37S | 0.026 ± 0.001 | 1/49 |
| R186W | 0.023 ± 0.001 | 1/56 |
| R235Q | 0.0016 ± 0.0004 | 1/800 |
| R108W | 0.44 ± 0.04 | 1/3 |
| N212I | 1.5 ± 0.1 | 1/1 |
| F230L | 0.12 ± 0.004 | 1/11 |
| T240M | 0.15 ± 0.002 | 1/8 |
| R291H | 2.8 ± 0.2 | 2/1 |
a The second order rate constant (kcat/Km) was estimated from the slope of the linear portion of the steady state reaction using the data from Fig. 5.
Equilibrium Binding Analyses
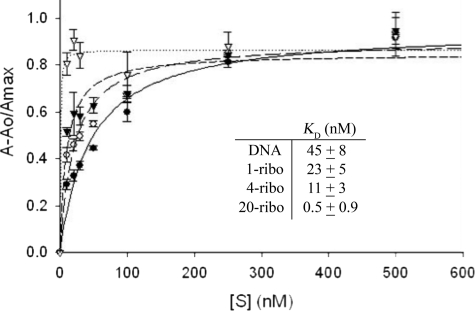
The RNA/DNA substrate binding reactions (30 μl) were as described for the activity assays except Ca2+ was substituted for Mg2+ to prevent catalysis. Binding reactions were prepared using RNase H2 WT and mutant enzymes with the 1-, 4-, and 20-ribo DNA duplex substrates (1 nm) and varied concentrations of enzyme indicated in the figure and table legends (Fig. 3, Table 4). Fluorescence anisotropy measurements to quantify RNase H2 binding to the FAM-labeled RNA/DNA duplex substrates were obtained at 25 °C in microtiter plates using a Tecan Safire2TM with fluorescence polarization module using excitation and emission wavelengths of 470 and 525 nm, respectively. The observed relative fluorescence anisotropy (Aobs) for each enzyme concentration was calculated using the equation Aobs = A − Ao/Amax, where A is the anisotropy at the given enzyme concentration, Ao is the anisotropy at 0 enzyme concentration, and Amax is the maximum change in anisotropy. Results were plotted in SigmaPlot 8.0 and fit to a rectangular hyperbola to derive KD values for each substrate. Reactions were performed at least in triplicate.
FIGURE 3.
Equilibrium substrate binding by RNase H2 WT. The FAM-labeled duplexes (1 nm) were incubated with increased concentrations of RNase H2 WT and the observed relative fluorescence anisotropy Aobs = A − Ao/Amax was determined as described under “Experimental Procedures” for the 1-ribo (○, long dashes), 4-ribo (▴, short dashes), 20-ribo (△, dots), and DNA (●, solid line). The fluorescence anisotropy data were fit to a single hyperbola to derive KD values (inset).
TABLE 4.
Equilibrium binding analyses of RNase H2A AGS-causing mutants
| RNase H2A | 1-ribo |
4-ribo |
20-ribo |
|||
|---|---|---|---|---|---|---|
| KDa | Fraction of WT affinityb | KDa | Fraction of WT affinityb | KDa | Fraction of WT affinityb | |
| nm | nm | nm | ||||
| WT | 23 ± 5 | 1 | 11 ± 3 | 1 | ≤0.5 ± 0.9 | 1 |
| G37S | 120 ± 20 | 1/5 | 21 ± 3 | 1/2 | 7 ± 2 | 1/14 |
| R186W | 100 ± 10 | 1/4 | 40 ± 10 | 1/4 | 5 ± 2 | 1/10 |
| R235Q | 190 ± 40 | 1/8 | 280 ± 60 | 1/25 | 50 ± 20 | 1/100 |
| R108W | 27 ± 7 | 1/1 | 26 ± 8 | 1/2 | ≤3 ± 1 | 1/6 |
| N212I | 26 ± 5 | 1/1 | 25 ± 4 | 1/2 | ≤1.6 ± 0.8 | 1/3 |
| F230L | 40 ± 10 | 1/2 | 14 ± 4 | 1/1 | 5.8 ± 0.8 | 1/12 |
| T240M | 48 ± 4 | 1/2 | 70 ± 10 | 1/6 | 29 ± 5 | 1/58 |
| R291H | 64 ± 10 | 1/3 | 70 ± 10 | 1/6 | ≤2.6 ± 0.7 | 1/5 |
a The KD values were determined for RNase H2 WT and mutants with the three substrates described in Table 1 using fluorescence anisotropy as described under “Experimental Procedures.” The RNase H2 WT and mutants demonstrate an increased affinity for the RNA-DNA hybrid as the number of ribonucleotides increased. The RNase H2A mutants show moderate decreases in affinity with the 1-ribo and 4-ribo substrates and large decreases in affinity with the 20-ribo substrate.
b The fraction of WT affinity was calculated as: Fraction WT affinity = 1/[WT KD/mutant KD].
RESULTS
Catalytic Efficiency of RNase H2 WT
The human RNase H2 recognizes and cleaves ribonucleotides within a deoxyribonucleotide polymer by distinct mechanisms dependent upon the number of ribonucleotides present in the heteropolymer substrate. This result is apparent by the different kinetic parameters that govern catalysis as revealed in a steady-state kinetic analysis of the human RNase H2 WT using polynucleotide substrates containing 1, 4, and 20 ribonucleotides (Fig. 2). RNase H2 enzymes are unique in their ability to cleave at the 5′ side of a ribonucleotide positioned at the RNA-DNA junction in a DNA-RNA-DNA/DNA duplex whether single or multiple ribonucleotides are present (6, 21, 30). Incubation of RNase H2 WT with increased concentrations of the 20-ribo substrate using steady-state kinetic conditions demonstrates preferential cleavage at the RNA-DNA junction. This is apparent by detection of a predominant single band product corresponding to phosphodiester cleavage of the RNA20-DNA10 between the 19th and 20th ribonucleotides (rAde-rCyt) near the RNA-DNA junction (Fig. 2C). Furthermore, the RNase H2 three-subunit complex exhibits saturation kinetics indicative of its Michaelis-Menten behavior. Incubation of RNase H2 with increased concentrations of the 1-ribo (Fig. 2A) and 4-ribo (Fig. 2B) substrates shows that higher substrate concentrations are required to saturate the RNase H2, indicating lower binding affinities as the number of ribonucleotides within the DNA-RNA-DNA heteropolymer is decreased. Quantification of these kinetic data shows that Km values for RNase H2 increase from 170 nm using the 20-ribo substrate to 440 nm using the 4-ribo and to >1500 nm with the 1-ribo substrate (Fig. 2D). In contrast, the kcat values decrease as the number of ribonucleotides increase (Fig. 2D). Cleavage of the 20- and 4-ribo substrates results from hydrolysis between two ribonucleotides, and cleavage of the 1-ribo substrate results from hydrolysis between a deoxy- and a ribonucleotide. These data indicate that cleavage of the preferred DNA-RNA bond using the 1-ribo substrate (kcat value = 35 min−1) is ∼20 times faster than cleavage of the RNA-RNA bond using the 20-ribo substrate (kcat value = 1.9 min−1). Overall, the catalytic efficiencies (kcat/Km) for RNase H2 cleavage of the three substrates tested are similar.
Equilibrium Substrate Binding by RNase H2 WT
Equilibrium binding analysis reveals distinct binding mechanisms by RNase H2 WT dependent upon the number of ribonucleotides present in the heteropolymer duplex. A series of FAM-labeled nucleic acid polymer duplexes (Table 1) was incubated with increased concentrations of RNase H2 WT in the presence of non-activating Ca2+ ion, and binding was measured by fluorescence anisotropy (Fig. 3). The observed changes in fluorescence anisotropy indicate binding to the 1-, 4-, and 20-ribo duplexes with very different affinities. Quantification of these binding isotherms indicates that RNase H2 WT binds the 20-ribo duplex (KD value ∼0.5 nm) with an ∼50-fold greater affinity than to the 1-ribo duplex (KD value = 23 nm). The binding affinity of RNase H2 measured for the 1-ribo duplex was only slightly greater than that for a DNA duplex (KD value = 45 nm). These RNase H2 binding data parallel the steady-state kinetic data, demonstrating that the number of ribonucleotides contained within the heteropolymer substrate directly impacts the nucleic acid binding affinities as reflected in the KD and Km values.
Positions of the AGS-causing RNase H2A Subunit Mutations
The RNase H2A subunit AGS-causing mutations were mapped onto our mouse RNase H2 structure (30) showing that these altered residues are dispersed throughout the protein (Fig. 1). Structural determination of the mouse RNase H2 identified the four conserved acidic residues in the active site and provided the basis for a model of the proposed two-metal ion mechanism of phosphodiester bond cleavage at an RNA/DNA junction within a polynucleotide. Location of the 10 currently known AGS-causing mutations in the RNase H2A protein relative to the catalytic center and proposed substrate binding site suggested that some mutations were likely to have a large impact on catalytic function, whereas others were much less likely to affect activity. This hypothesis was further supported by a structure-based sequence alignment of RNase H2/HII enzymes from diverse species using the mouse RNase H2A to determine the level of conservation at sites of AGS-causing mutations (supplemental Fig. 1). Together, our structural model and sequence alignment analyses suggested that varied functional consequences on the catalytic potential of the RNase H2 enzyme were likely, but the precise effect of these AGS-causing mutations on catalytic function had not been established.
The Ribonuclease H Activities of RNase H2A Mutants
Eight AGS-causing RNase H2A missense mutants were purified as heterotrimeric complexes, and activities were first examined using the 1-ribo substrate (Table 1, Fig. 4). Incubations with increased enzyme concentrations up to 400 nm demonstrate measurable ribonuclease H activity for all the RNase H2A AGS-causing mutants. This result contrasts the complete loss of enzymatic activity exhibited upon mutation of the four acidic residues that coordinate divalent metal ions required for catalysis (30). Mutations at highly conserved positions in RNase H2A G37S, R186W, and R235Q are dramatically reduced in enzyme activity (Fig. 4), whereas mutations at nonconserved positions R108W, N212I, F230L, T240M, and R291H are only moderately reduced or exhibit activities indistinguishable from WT (data not shown).
The relative activities of the eight AGS-causing RNase H2A missense mutants were then precisely quantified using the 1-, 4-, and 20-ribo substrates (Table 2). These analyses show that the mutants exhibit a spectrum of ribonuclease H activities ranging from nearly 1000-fold reduced levels to WT activity. The magnitude of the activity reduction is dependent upon the substrate used for analysis. The RNase H2A mutants that exhibit the most dramatic reduction in activity relative to WT reveal this loss of catalytic function most prominently using the 1-ribo substrate. For example, G37S and R186W exhibit 1/210 and 1/160 the levels of WT activity using the 1-ribo substrate, respectively, and 1/3 and 1/8 the levels of WT activity using the 20-ribo substrate. R235Q exhibits the most dramatic reduction in activity, and this loss of catalytic function is apparent using all substrates tested. The RNase H2A mutations at non-conserved positions R108W, N212I, F230L, T240M, and R291H exhibit moderately lower or equivalent activities relative to WT. Reduced activities detected in the AGS-causing mutations at non-conserved residues were only detected using the 1-ribo substrate.
Catalytic Efficiencies of RNase H2A Mutants
The catalytic efficiencies of the RNase H2A mutants were determined to reveal the specific mechanism of enzyme dysfunction. A steady-state kinetic analysis demonstrates the large variations in catalytic efficiencies exhibited by the AGS-causing RNase H2A subunit mutants using the 1-ribo substrate (Fig. 5). The catalytic efficiencies (kcat/Km) of AGS-causing mutants were estimated from the linear portions of the Michaelis-Menten curves and directly compared with WT (Table 3). Mutations at non-conserved positions N212I and R291H exhibit kinetic behavior indistinguishable from WT, whereas mutations at conserved positions G37S, R186W, and R235Q exhibit the greatest variation from WT with nearly undetectable activities. The RNase H2A F230L and T240M exhibited catalytic efficiencies 10-fold lower than WT. The varied catalytic efficiencies exhibited by these AGS-causing mutations are consistent with the positions of these mutant residues relative to the active site, proposed nucleic acid binding site, and levels of amino acid conservation in the RNase H2A subunit.
Equilibrium Binding Analysis of RNase H2A Mutants
Equilibrium binding analysis reveals altered binding affinities by the RNase H2A mutants. The FAM-labeled 1-, 4-, and 20-ribo nucleic acid polymer duplexes were incubated with increased concentrations of the WT and mutant enzymes in the presence of Ca2+, and binding was measured by fluorescence anisotropy. Quantification of these binding data is summarized in Table 4. Similar to the WT, all RNase H2A mutants exhibit a greater binding affinity for substrates containing more ribonucleotides, as indicated by the lower KD values measured as the number of ribonucleotides within the RNA-DNA hybrid is increased. For example, the RNase H2A G37S exhibits an ∼20-fold greater affinity for the 20-ribo substrate (KD value = 7 nm) compared with the 1-ribo substrate (KD value = 120 nm). Thus, RNase H2A WT and mutants exhibit greater binding affinities for heteropolymer substrates containing more ribonucleotides, further confirming the different binding mechanisms exhibited were dependent upon the number of ribonucleotides present in the substrate. A comparison of the relative binding affinities exhibited by WT and mutants on these three substrates indicates that the RNase H2A mutants exhibit the greatest levels of reduced binding using the 20-ribo substrate. For example, the RNase H2A R235Q exhibits an 8-fold lower binding affinity using the 1-ribo substrate compared with WT, and this same mutant exhibits a 100-fold lower binding affinity using the 20-ribo substrate. Thus, these equilibrium binding analyses reveal dramatically different relative binding affinities in the RNase H2 mutant enzymes dependent upon the composition of the heteropolymer.
DISCUSSION
RNase H2 is a heterotrimeric enzyme complex. Mutations in any one of the three genes encoding the human RNase H2 cause the neurological disorder AGS, prompting our investigations into the functional consequences of these mutations. In previous studies we generated the human and mouse RNase H2 heterotrimeric complexes by co-expression of the RNASEH2A, RNASEH2B, and RNASEH2C genes in bacterial cells and demonstrated the ribonuclease H activity in vitro (6, 30). Most AGS-causing RNase H2 mutations are located in subunits H2B and H2C (13, 29). Attempts to correlate diminished ribonuclease H activity in AGS-causing alleles have demonstrated that mutations in subunits H2B and H2C yield heterotrimeric complexes exhibiting ribonuclease H activities indistinguishable from RNase H2 WT enzyme (6, 19, 26). Some AGS disease mutations destabilize the RNase H2 complex (32). These results suggest that the mechanisms of RNase H2 dysfunction in AGS involving the H2B and H2C subunits are mostly mediated through altered protein-protein interactions that disrupt complex formation and cellular functions. In contrast, there is evidence that AGS-causing mutations in the RNase H2A catalytic subunit alter nuclease specificity and enzymatic activity (6, 19, 26, 30). The studies presented here demonstrate that AGS-causing mutations in the RNase H2A subunit cause varied levels of catalytic dysfunction. The varied levels of biochemical dysfunction exhibited by these RNase H2A mutants are coupled with the paucity of RNASEH2A mutant alleles and similar early-onset phenotypes of these patients. Collectively, these data illustrate the challenges to explain the complex mechanisms of RNase H2 dysfunction in AGS that cannot always be simply attributed to loss of enzymatic activity but lead to nucleic acid accumulation and immune activation.
Our biochemical and structural studies of the mammalian heterotrimeric RNase H2 were the first to offer insight into the potential consequences on catalytic function of the AGS-causing mutations (6, 30). The mouse RNase H2 structure, modeled with a nucleic acid substrate indicated that the catalytic H2A subunit interfaces with the H2C/H2B subunits on the opposite face of the protein from the position of the active site and nucleic acid binding surface (Fig. 1). Thus, the AGS mutations identified in subunits H2B and H2C are located a considerable distance from the active site and the predicted nucleic acid-binding interface, helping to explain the full catalytic activity measured in these mutant complexes. When we mapped the RNase H2A subunit AGS-causing mutations onto our mouse structure, the positions of these mutations predicted varied effects on catalytic efficiencies. The RNase H2A G37S, R186W, and R235Q mutations are in close proximity to the active site (Fig. 1) and are phylogenetically conserved (supplemental Fig. 1). The flexible Gly-37 residue aids in a turn at the end of the β2 strand, and the Arg-186 and -235 residues position above and below to stabilize this turn. The recently described monomeric Thermotoga maritima RNase H2 in complex with nucleic acid (20) is nearly identical to the mouse RNase H2A structure. This closely related structure identifies Gly-37 in the “GRG motif” (residues Gly-37—Arg-38—Gly-39 in mammalian RNase H2A; Gly-21—Arg-22—Gly-23 in T. maritima RNase H2) responsible for recognition of the substrate RNA-DNA junction and confirms the critical role of the Arg-186 and -235 residues (Arg-141 and -182 in T. maritima RNase H2) in stabilizing this structural element. The G37S, R186W, and R235Q AGS mutations exhibit the most dramatic reductions in RNA-DNA hybrid cleavage activity (Table 2). This loss of catalytic activity was greatest for the G37S and R186W mutants using the 1-ribo substrate, whereas the R235S mutation was most impacted using the 20-ribo substrate. In addition, our previous work demonstrated the altered specificity exhibited by the G37S mutant (6). Thus, the greatly reduced catalytic activities and altered specificities detected in the RNase H2A G37S, R186W, and R235Q AGS mutations are explained by structural perturbations caused by these mutations near the active site.
The RNase H2A T240M and N212I AGS-causing mutations exhibit relatively modest or undetectable effects on catalytic and binding activities that are partly explained by the positions of the mutant residues and the nucleic acid substrate composition. The RNase H2A Thr-240 residue (Pro-187 in T. maritima RNase H2 (20)) is located at the end of a helix in position to make contact with the phosphodiester backbone of the non-cleaved DNA strand in the duplex substrate, directly opposite the site of cleavage. Thus, the Thr-240 residue likely contributes to recognition and binding of the RNA/DNA duplex structure characteristic of ribonuclease H enzymes. There is structural conservation in this region between the RNase H2A and T. maritima RNase H2 despite the considerable sequence variation. The RNase H2A T240M AGS-causing mutant exhibits an ∼60-fold reduction in binding affinity using the 20-ribo substrate and more modest 6-fold and 2-fold reductions in affinity for the 4- and 1-ribo substrates, respectively (Table 4). These different levels of substrate binding perturbation might be explained by the different duplex structures dependent upon the number of ribonucleotides present in the heteropolymer strand. The RNase H2A T240M mutant exhibits WT activity using the 4- and 20-ribo substrates and a ∼10-fold reduced activity using the 1-ribo substrate (Table 2). A possible explanation for these results is that an apparently modest reduction in binding affinity of RNase H2 for the 1-ribo substrate has a relatively large impact on the correct positioning of the scissile bond due to the adjacent duplex DNA structure that is instead a DNA/RNA duplex in the 4- and 20-ribo substrates.
The RNase H2A Asn-212 residue (Thr-165 in T. maritima RNase H2 (20)) is positioned on a loop that potentially contacts the minor groove in the region of duplex DNA binding. The loop containing Asn-212 in RNase H2A is structurally conserved in the T. maritima enzyme, but unlike the mammalian enzyme, there are no amino acid side chains extending toward the DNA, suggesting a potentially unique function for Asn-212 in RNase H2A. The AGS-causing RNase H2A N212I mutation has little measured effect on enzyme function. Our results show that the RNase H2A N212I retains full catalytic activity (Tables 2 and 3, Fig. 5) and exhibits only modest levels of reduced nucleic acid binding affinity (Table 4). It is possible that the RNase H2A Asn-212 residue contributes to the detection of structural distortions like ribonucleotides within duplex DNA through minor groove interactions. Asn-212 is on the same loop as Tyr-210 (Tyr-163 in T. maritima RNase H2), an important residue in binding the 2′-OH of the junction ribonucleotide (20). Our results using limited nucleic acid substrates do not reveal an obvious catalytic dysfunction. The catalytic competency of this AGS-causing mutation suggests an unidentified nucleic acid substrate or protein interaction as the mechanism for nucleic acid mediate immune activation.
The RNase H2A F230L, R108W, and R291H AGS-causing mutations are located on structural elements that are unique in the mammalian RNase H2A subunit and are located at distant sites from the active center. The RNase H2A Phe-230 is positioned in a hydrophobic center between a pair of H2A helices and appears to anchor a unique H2A extended loop that interfaces with subunit H2C. Our results show that the RNase H2A F230L exhibits modest reductions in catalytic activity (Tables 2 and 3, Fig. 5) and modest levels of reduced nucleic acid binding affinity (Table 4). Mutation of Phe-230 to Leu might contribute to reduced stability of the hydrophobic core in this region that could impact indirectly on nucleic acid binding and catalysis. The H2A helices that are stabilized by Phe-230 contain the Thr-240 and Asn-212 residues located more closely to the nucleic acid substrate. It is also possible that the F230L mutation reduces stability of the H2A:H2C interface critical for heterotrimeric complex formation.
The RNase H2A R108W and R291H complexes exhibit catalytic activities (Tables 2 and 3, Fig. 5) and nucleic acid binding affinities (Table 4) nearly equivalent to WT enzyme. The RNase H2A Arg-108 is located on a unique loop that interfaces with the H2C subunit. A recent structure of the human RNase H2 complex (PDB ID 3P56) (32, 33) confirms the Arg-108 interaction with the H2C subunit and indicates specific contacts with the H2C Leu-134 and Glu-135 residues. Also, the human RNase H2 structure locates RNase H2A Arg-291 more than 50 Å removed from the active site positioned to interface with the H2B and H2C subunits. These structural data and our kinetic studies that demonstrate the robust catalytic activities of the RNase H2A R108W and R291H complexes support a mechanism of dysfunction in AGS mediated by dysfunctional RNase H2 heterotrimer formation or by altered protein-protein interactions.
In summary, we present data that further support a complex multimechanistic view of RNase H2 dysfunction in AGS. There are now at least 30 identified AGS-causing mutations mapping to all three genes encoding the RNase H2 heterotrimeric complex. The mutants locating to the H2B and H2C subunits exhibit robust catalytic function in vitro supporting a non-catalytic mechanism of dysfunction. Our results now show that some of the catalytic H2A subunit mutants also exhibit robust activities, whereas several exhibit severely reduced levels of activities. Additional studies are required to identify the various mechanisms of RNase H2 dysfunction in AGS.
Supplementary Material
Acknowledgments
We thank G. Rice and Y. Crow for providing the unpublished RNase H2A Asn-212 clone.
This work was supported, in whole or in part, by National Institutes of Health Grant GM069962 (to F. W. P.). This work was also supported by the Alliance for Lupus Research 179222 (to F. W. P.), American Heart Association Grant 10GRNT3650033 (to T. H.), and National Institutes of Health Postdoctoral Fellowship Award F32GM095290 (to S. R. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
G. Rice and Y. Crow, unpublished information.
- AGS
- Aicardi-Goutières Syndrome
- FAM
- 6-carboxyfluorescein.
REFERENCES
- 1. Aicardi J., Goutières F. (1984) Ann. Neurol. 15, 49–54 [DOI] [PubMed] [Google Scholar]
- 2. Goutières F., Aicardi J., Barth P. G., Lebon P. (1998) Ann. Neurol. 44, 900–907 [DOI] [PubMed] [Google Scholar]
- 3. Crow Y. J., Hayward B. E., Parmar R., Robins P., Leitch A., Ali M., Black D. N., van Bokhoven H., Brunner H. G., Hamel B. C., Corry P. C., Cowan F. M., Frints S. G., Klepper J., Livingston J. H., Lynch S. A., Massey R. F., Meritet J. F., Michaud J. L., Ponsot G., Voit T., Lebon P., Bonthron D. T., Jackson A. P., Barnes D. E., Lindahl T. (2006) Nature Genetics 38, 917–920 [DOI] [PubMed] [Google Scholar]
- 4. Crow Y. J., Leitch A., Hayward B. E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., Baumann C., Baxter P., Bertini E., Chandler K. E., Chitayat D., Cau D., Déry C., Fazzi E., Goizet C., King M. D., Klepper J., Lacombe D., Lanzi G., Lyall H., Martínez-Frías M. L., Mathieu M., McKeown C., Monier A., Oade Y., Quarrell O. W., Rittey C. D., Rogers R. C., Sanchis A., Stephenson J. B., Tacke U., Till M., Tolmie J. L., Tomlin P., Voit T., Weschke B., Woods C. G., Lebon P., Bonthron D. T., Ponting C. P., Jackson A. P. (2006) Nat. Genet. 38, 910–916 [DOI] [PubMed] [Google Scholar]
- 5. Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., Fuller J. C., Jackson R. M., Lamb T., Briggs T. A., Ali M., Gornall H., Couthard L. R., Aeby A., Attard-Montalto S. P., Bertini E., Bodemer C., Brockmann K., Brueton L. A., Corry P. C., Desguerre I., Fazzi E., Cazorla A. G., Gener B., Hamel B. C., Heiberg A., Hunter M., van der Knaap M. S., Kumar R., Lagae L., Landrieu P. G., Lourenco C. M., Marom D., McDermott M. F., van der Merwe W., Orcesi S., Prendiville J. S., Rasmussen M., Shalev S. A., Soler D. M., Shinawi M., Spiegel R., Tan T. Y., Vanderver A., Wakeling E. L., Wassmer E., Whittaker E., Lebon P., Stetson D. B., Bonthron D. T., Crow Y. J. (2009) Nat. Genet. 41, 829–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perrino F. W., Harvey S., Shaban N. M., Hollis T. (2009) J. Mol. Med. 87, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mazur D. J., Perrino F. W. (1999) J. Biol. Chem. 274, 19655–19660 [DOI] [PubMed] [Google Scholar]
- 8. Höss M., Robins P., Naven T. J., Pappin D. J., Sgouros J., Lindahl T. (1999) EMBO J. 18, 3868–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mazur D. J., Perrino F. W. (2001) J. Biol. Chem. 276, 17022–17029 [DOI] [PubMed] [Google Scholar]
- 10. Mazur D. J., Perrino F. W. (2001) J. Biol. Chem. 276, 14718–14727 [DOI] [PubMed] [Google Scholar]
- 11. de Silva U., Choudhury S., Bailey S. L., Harvey S., Perrino F. W., Hollis T. (2007) J. Biol. Chem. 282, 10537–10543 [DOI] [PubMed] [Google Scholar]
- 12. Lehtinen D. A., Harvey S., Mulcahy M. J., Hollis T., Perrino F. W. (2008) J. Biol. Chem. 283, 31649–31656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crow Y. J., Rehwinkel J. (2009) Hum. Mol. Genet. 18, R130–R136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramantani G., Kohlhase J., Hertzberg C., Innes A. M., Engel K., Hunger S., Borozdin W., Mah J. K., Ungerath K., Walkenhorst H., Richardt H. H., Buckard J., Bevot A., Siegel C., von Stülpnagel C., Ikonomidou C., Thomas K., Proud V., Niemann F., Wieczorek D., Häusler M., Niggemann P., Baltaci V., Conrad K., Lebon P., Lee-Kirsch M. A. (2010) Arthritis Rheum. 62, 1469–1477 [DOI] [PubMed] [Google Scholar]
- 15. Frank P., Braunshofer-Reiter C., Wintersberger U., Grimm R., Büsen W. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12872–12877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cerritelli S. M., Crouch R. J. (2009) FEBS J. 276, 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nowotny M., Gaidamakov S. A., Crouch R. J., Yang W. (2005) Cell 121, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 18. Itaya M. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8587–8591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rohman M. S., Koga Y., Takano K., Chon H., Crouch R. J., Kanaya S. (2008) FEBS J. 275, 4836–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rychlik M. P., Chon H., Cerritelli S. M., Klimek P., Crouch R. J., Nowotny M. (2010) Mol. Cell 40, 658–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeong H. S., Backlund P. S., Chen H. C., Karavanov A. A., Crouch R. J. (2004) Nucleic Acids Res. 32, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M. (2003) J. Biol. Chem. 278, 1618–1625 [DOI] [PubMed] [Google Scholar]
- 24. Qiu J., Qian Y., Frank P., Wintersberger U., Shen B. (1999) Mol. Cell. Biol. 19, 8361–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rydberg B., Game J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16654–16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chon H., Vassilev A., DePamphilis M. L., Zhao Y., Zhang J., Burgers P. M., Crouch R. J., Cerritelli S. M. (2009) Nucleic Acids Res. 37, 96–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El Hage A., French S. L., Beyer A. L., Tollervey D. (2010) Genes Dev. 24, 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin Y., Dent S. Y., Wilson J. H., Wells R. D., Napierala M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rice G., Patrick T., Parmar R., Taylor C. F., Aeby A., Aicardi J., Artuch R., Montalto S. A., Bacino C. A., Barroso B., Baxter P., Benko W. S., Bergmann C., Bertini E., Biancheri R., Blair E. M., Blau N., Bonthron D. T., Briggs T., Brueton L. A., Brunner H. G., Burke C. J., Carr I. M., Carvalho D. R., Chandler K. E., Christen H. J., Corry P. C., Cowan F. M., Cox H., D'Arrigo S., Dean J., De Laet C., De Praeter C., Dery C., Ferrie C. D., Flintoff K., Frints S. G., Garcia-Cazorla A., Gener B., Goizet C., Goutieres F., Green A. J., Guet A., Hamel B. C., Hayward B. E., Heiberg A., Hennekam R. C., Husson M., Jackson A. P., Jayatunga R., Jiang Y. H., Kant S. G., Kao A., King M. D., Kingston H. M., Klepper J., van der Knaap M. S., Kornberg A. J., Kotzot D., Kratzer W., Lacombe D., Lagae L., Landrieu P. G., Lanzi G., Leitch A., Lim M. J., Livingston J. H, Lourenco C. M., Lyall E. G., Lynch S. A., Lyons M. J., Marom D., McClure J. P., McWilliam R., Melancon S. B., Mewasingh L. D., Moutard M. L., Nischal K. K., Ostergaard J. R., Prendiville J., Rasmussen M., Rogers R. C., Roland D., Rosser E. M., Rostasy K., Roubertie A., Sanchis A., Schiffmann R., Scholl-Burgi S., Seal S., Shalev S. A., Corcoles C. S., Sinha G. P., Soler D., Spiegel R., Stephenson J. B., Tacke U., Tan T. Y., Till M., Tolmie J. L., Tomlin P., Vagnarelli F., Valente E. M., Van Coster R. N., Van der Aa N., Vanderver A., Vles J. S., Voit T., Wassmer E., Weschke B., Whiteford M. L., Willemsen M. A., Zankl A., Zuberi S. M., Orcesi S., Fazzi E., Lebon P., Crow Y. J. (2007) Am. J. Hum. Genet. 81, 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shaban N. M., Harvey S., Perrino F. W., Hollis T. (2010) J. Biol. Chem. 285, 3617–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kunkel T. A. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 488–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reijns M. A., Bubeck D., Gibson L. C., Graham S. C., Baillie G. S., Jones E. Y., Jackson A. P. (2011) J. Biol. Chem. 286, 10530–10539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Figiel M., Chon H., Cerritelli S. M., Cybulska M., Crouch R. J., Nowotny M. (2011) J. Biol. Chem. 286, 10540–10550 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.