Abstract
We determined the size and shape of full-length avian sarcoma virus (ASV) integrase (IN) monomers and dimers in solution using small angle x-ray scattering. The low resolution data obtained establish constraints for the relative arrangements of the three component domains in both forms. Domain organization within the small angle x-ray envelopes was determined by combining available atomic resolution data for individual domains with results from cross-linking coupled with mass spectrometry. The full-length dimer architecture so revealed is unequivocally different from that proposed from x-ray crystallographic analyses of two-domain fragments, in which interactions between the catalytic core domains play a prominent role. Core-core interactions are detected only in cross-linked IN tetramers and are required for concerted integration. The solution dimer is stabilized by C-terminal domain (CTD-CTD) interactions and by interactions of the N-terminal domain in one subunit with the core and CTD in the second subunit. These results suggest a pathway for formation of functional IN-DNA complexes that has not previously been considered and possible strategies for preventing such assembly.
Keywords: DNA-binding Protein, Integrase, Protein Cross-linking, Protein Domains, Protein Self-assembly, Protein-Protein Interactions, Viral Protein, SAXS, Apo-integrase, Solution Structures
Introduction
Retroviral integrase (IN)3 catalyzes the insertion of viral DNA into the DNA of the infected host cell. IN is one of three retrovirally encoded enzymes that are essential for retroviral replication and therefore is an important target for drugs to treat HIV/AIDS. One IN active site-directed inhibitor, raltegravir, is approved by the Food and Drug Administration for this purpose, and a second, elvitegravir, is in advanced clinical trials. The availability of such drugs offer hope to HIV-positive individuals, especially those who have developed resistance to therapies that target the other two viral enzymes, reverse transcriptase and protease. Nevertheless, the inevitable development of drug-resistant HIV mutants drives a continuing need for additional strategies to block the activity of this viral enzyme. Unfortunately, progress has been limited by a lack of critical details concerning the molecular structure of full-length HIV IN protein and its functional multimers. Much of the difficulty arises from the fact that the HIV protein is relatively insoluble and therefore difficult to study using biophysical methods. As retroviral INs are likely to share architectures for active complexes (1), we have focused our studies on avian sarcoma virus (ASV) IN (2), which is more soluble than HIV IN and, as we show here, suitable for structural analyses.
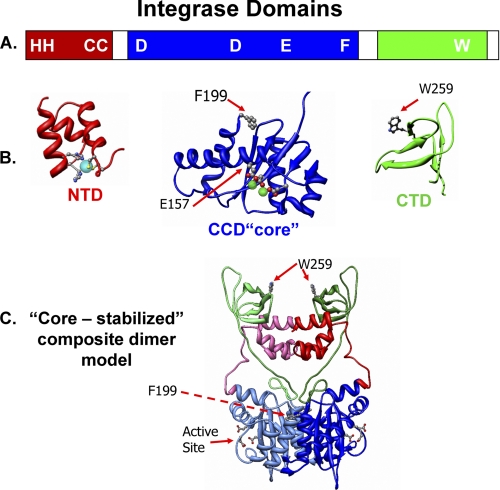
IN proteins are composed of three distinct structural domains (Fig. 1, A and B). The largest (amino acids 50–207 in ASV IN) is the central catalytic core domain (CCD or “core”). This domain contains the DD(35)E motif of acidic residues that coordinate the required divalent metal ions, Mg2+ or Mn2+ (3–6). The isolated HIV IN core domain forms a dimer in solution (7), and the three-dimensional structure of the core from several retroviral IN proteins has been solved by x-ray crystallography of either the isolated domain or two-domain fragments that include the N- (NTD) or C-terminal (CTD) domains (8–14). The same extensive interface of two core domains (e.g. Fig. 1C) has been observed in every crystal structure analyzed so far, and consequently, it is thought to be physiologically relevant. The isolated, Zn2+-binding NTD (amino acids 1–50) and the Src homology 3-like CTD (amino acids 210–286) of HIV IN also form dimers in solution (15–20), but the significance of these interactions is uncertain. For example, the spatial relationships between the CTDs and cores are different in each of the two-domain crystal structures that have been determined, and the crystal structure of the NTD + core, two-domain fragment shows a different NTD-NTD interface than that observed in the NMR structure of the NTD alone. One might ask if these interactions reflect alternative, physiologically relevant assemblies.
FIGURE 1.
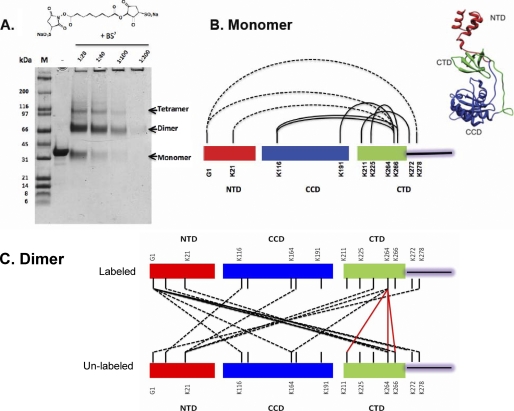
Integrase protein domains and a core-stabilized dimer model. A, linear representation of IN, indicating the borders of the three domains and the locations of several important residues. The color code, red for the NTD, blue for the CCD (core), and green for the CTD, is used throughout; linkers and the unstructured CTD tail are white. B, ribbon models of the isolated core and CTD of ASV IN are based on published data (PDB code 1C1A). The helical NTD is modeled from the HIV-1 IN domain (PDB code 1K6Y). The conserved HHCC residues, in ball-and-stick representation, bind a zinc ion (cyan sphere). The three conserved active site residues in the CCD (core) are shown in ball-and-stick coordinating the metal co-factors (green spheres) required for catalysis. Two additional residues of relevance to the present studies (Phe-199 and Trp-259) are also shown in ball-and-stick. C, a core-core stabilized ASV IN dimer model based on the HIV-1 IN model of Wang et al. (14). One subunit is depicted in muted colors.
Full-length IN proteins are known to exist as monomers, dimers, and tetramers in solution (21–26), and complementation experiments indicate that IN functions as a multimer (27–30). An IN dimer appears to be the most catalytically active form for the endonucleolytic processing of a single end viral DNA substrate in vitro (31). However, as two processed DNA ends must be joined by IN to host DNA in a concerted fashion in vivo, a tetramer is assumed to be the minimal functional multimer for this step. Indeed, analysis of ASV IN-DNA complexes imaged by atomic force microscopy revealed that assembly of a tetramer is induced upon interaction with a “disintegration” substrate, which represents a viral-host DNA integration intermediate, and that four IN monomers are required for a single catalytic turnover with this substrate (32). Purification and analysis of covalently cross-linked multimers of HIV-1 IN showed that although a dimer could process and join a single viral DNA end substrate, only a tetramer was capable of catalyzing the concerted integration of two viral ends into a target DNA (33). Analyses of in vitro assembled HIV IN synaptic complexes containing viral and target DNA substrates also indicate that concerted integration is catalyzed by an IN tetramer (34).
Models for IN dimers (Fig. 1C) and tetramers have been derived from consolidation of the crystal structures of two-domain protein fragments and other studies (13, 35–37). However, their validity has been difficult to evaluate, as experimental knowledge has been lacking concerning the disposition of the three domains with respect to each other, in the intact full-length monomer or multimers. The crystal structure of full-length IN from the human prototype foamy virus (PFV) was recently solved in complex with viral DNA (38) revealing two different dimer interfaces for IN within the “intasome” structures. Here, we report the use of small angle x-ray scattering (SAXS) and biochemical cross-linking analyses to determine the architecture of full-length ASV IN monomers and dimers, in the absence of DNA substrates. Our results show that the solution dimer interface is distinct from that previously proposed in models derived from two-domain crystal structures and more closely resembles that in the DNA-binding “inner” dimer in PFV intasome (38, 39). Analyses of the ASV apo-IN architecture described herein highlight key domain interactions and the specific conformational changes that may occur upon DNA substrate binding.
EXPERIMENTAL PROCEDURES
Light Scattering Analysis
Measurements were made with a Protein Solutions DynaPro temperature-controlled microsampler. Samples were adjusted to the desired concentration, and particulates were removed by filtration through a 0.2-μm Millipore filter and subsequent clearing by brief centrifugation at 14,000 × g at 4 °C. The protein concentration was then determined directly using absorbance at 280 nm and a calculated molar extinction coefficient, taking the average of three readings. All samples were analyzed under conditions of 10 °C in a buffer of 25 mm BisTris, pH 6.1, 500 mm NaCl, 1 mm DTT, 0.1 mm EDTA, 5% glycerol. The apparent molecular mass (MW-I) was calculated from the static light scattering measurements (at least 300 acquisitions per protein sample) using the DynaPro software.
SAXS and ab Initio Shape Modeling Methods
X-ray scattering experiments were performed at the Advanced Photon Source at Argonne National Laboratories, 5ID-D beamline. Data were collected at 10 keV (1.24 Å) with the SAXS detector at a distance of 2.584 m and simultaneous WAXS detector at 291 cm, which produced an accessible q range of 0.005 to 1.8 Å−1 (where q = 4πsinθ/λ, where 2θ is the scattering angle). To minimize protein damage, four 10-s exposures were typically taken at 10 °C with sample flowing at 4 μl/s using a 0.3 × 0.3 mm2 collimated x-ray beam. Exactly matched dialysates were sampled under the same conditions to subtract from protein samples, which were tested in the range of 0.8–3 mg/ml in the same buffer conditions used for light scattering. Samples were filtered and cleared by centrifugation at 16,000 × g just prior to placement in the sampler. Combining of SAXS and WAXS data and subsequent data reduction were as published previously (40). Although initial Rg estimates were made at APS by a linear fit of a typical Guinier plot in the q range of 0.5–1.2/Rg, subsequent data analysis using Irena software developed at APS (41) was used for data in the broader q range of 0.01–0.4 to determine Rg, I(0), as well as to calculate a paired distance distribution function (or P(r) function) and Dmax, either by Fourier transform by the method of Moore or conventional regularization. Goodness of fit was assessed with the reduced χ2 parameter. In all cases, equivalent results were obtained by regularization with the program GNOM (42). A dilution series was performed with the wild type ASV IN protein and, using the ATSAS program PRIMUS (43), these data were extrapolated to infinite dilution. The results (not included) showed no significant variation in Rg, confirming the absence of any concentration-dependent effects. Accordingly, our data analyses assume a single component ideal behavior under the concentrations tested.
Subsequent ab initio shape modeling was performed with both DAMMIN and GASBOR programs (43), with and without P2 symmetry when appropriate. In each case, several qmax cutoff values were sampled in the range of 0.3–0.9, with the standard final processing using a qmax of 0.4. These produced dummy atom output files that were then used to generate the final envelopes with the Situs software (44). To test for uniqueness, 10 shape reconstructions were performed with the program GASBOR for wild type ASV IN dimers and monomers (supplemental Fig. S1). The results showed a high degree of stability and convergence in the shape modeling for both monomer (normalized spatial discrepancies (NSDs) of 1.0 to 1.19) and dimer (NSDs of 1.0 to 1.34) envelopes, with ranges comparable with those reported in other SAXS analyses (45).
Protein Cross-linking and In-gel Digestion
A mixture of 1:1 unlabeled and isotopically labeled ASV IN proteins (6.5 μm each) was equilibrated overnight (46) and dialyzed in 20 mm HEPES, pH 7.8, 0.5 m NaCl, 2 mm DTT, 10% glycerol. Freshly prepared BS3 (Pierce) homo-bifunctional cross-linker was used at increasing concentrations. After addition of the cross-linker, the reaction was allowed to continue at 37 °C for 5 min and then quenched by addition of 20 μl of 2 m glycine and left on ice for 30 min. The reactants were precipitated with acetone and resuspended in 20 mm HEPES, pH 7.8, 0.5 m NaCl, 2 mm DTT, 10% glycerol. The products were separated on a denaturing NuPAGE 4–12% BisTris gel using MES running buffer and Coomassie Blue stain. We confirmed that sample recovery was unaffected by acetone precipitation. Furthermore, treatment with a different reagent, EDC/NHS (Pierce), produced a similar distribution of cross-linked products (data not shown). Monomer, dimer, and tetramer bands from a reaction in which the molar ratio of protein to BS3 was 1:20 were excised and destained (50% MeOH, 5% HOAC in water) overnight, after which they were dehydrated completely using 100% acetonitrile. Reduction and alkylation were performed by adding 20 mm dithiothreitol (DTT) and 50 mm iodoacetamide. After a second dehydration, gel bands were rehydrated at 4 °C for 45 min in trypsin solution (10 ng/μl, Promega sequencing grade modified trypsin, 10 mm NH4HCO3, 10% acetonitrile). Proteins were digested overnight at 34 °C.
Mass Spectrometry and Data Base Searching
The digested samples were acidified with 0.3% formic acid before being injected into an LC/MS/MS instrument QSTAR (Applied Biosystems/MDS Sciex, Foster City, CA). An Agilent nano-HPLC (Agilent, Wilmington, DE) was equipped to interface the Q-TOF mass spectrometer. Samples were automatically loaded onto a C-18 trap column (ZORBAX 300SB-C18, 0.3 × 5 mm, 5 mm) and then eluted to a reversed-phase C-18 analytical column (ZORBAX 300SB-C18, 100 × 150 mm, 3.5 mm). A typical HPLC gradient for the tryptic mixture of peptides was 5–80% organic solvent over a period of about 85 min, followed by 80–100% organic solvent for the next 15 min and 100 to 5% in the last 15 min. The 300-nl/min flow from the column elution was sprayed through a coated emitter (FS360-50-5-CE, New Objective Inc., Woburn, CA) into a mass spectrometer with a set voltage of +2.5 kV. The system was equilibrated for 15 min at the end of the gradient. The acquisition method of QSTAR was set at a 2-s TOF-MS “survey” scan followed by three MS/MS scans (3s, 4s, and 5s, respectively). Parent ions with charge state of +2 and +3 or intensity above 15 counts were fragmented. The mass range for survey scan was 400–1000 atomic mass units and was 100–2000 atomic mass units for MS/MS scan.
The MS wiff files were processed into MGF files using Mascot Distiller with default parameters. Data were searched with MassMatrix PC suite 1.1.3 program (47), and search parameters were as follows: MS accuracy, 10 ppm; MS/MS accuracy, 0.8 Da (at this level of search stringency, no peptide adducts were identified that are inconsistent with the reaching dimer); enzyme, trypsin; specificity, fully tryptic; allowed number of missed cleavages, four; fixed modifications, carbamidomethylation on cysteine. Further allowed variable modifications were K+8 for lysine; R+10 for arginine; oxidation of methionine, tryptophan, and histidine; deamidation of asparagine and glutamine. End products of BS3 mono-cross-linked adducts with lysine and N termini were allowed with water or glycine. Results of the cross-linked peptides were also manually validated using GPAMW program (48).
RESULTS
Light Scattering Analyses Reveal Homogeneous ASV IN Dimers
Static light scattering provided a direct measure of the MW-I of our proteins and protein complexes in solution. The molecular uniformity of these preparations in the concentration range appropriate for SAXS analysis, 1–4 mg/ml (32–128 μm), was also evaluated by use of dynamic light scattering. As summarized in Table 1, we obtained an MW-I of 69 kDa for wild type IN, only in slight excess of the calculated mass of a dimer, 64 kDa. This difference could reflect the presence of a minor amount of higher order multimers in the preparation. However, the values calculated from static light scattering can also differ somewhat from the theoretical because of the dynamic exchange of subunits in multimeric complexes (46). Enzymatic activity assays confirmed that this wild type protein preparation catalyzes single end cutting and joining of viral DNA as well as concerted integration (Table 1). Among the other ASV IN derivatives prepared and analyzed, several contain an F199K substitution (Fig. 1, B and C). Structural alignments show that residue Phe-199 in ASV IN is adjacent to that of Phe-185 in HIV-1 IN (1), and as with HIV, its replacement enhances protein solubility, a feature that was required for successful crystallization of the respective two-domain IN fragments of ASV and HIV-1 IN. To examine the effects of this substitution on ASV IN multimerization, we analyzed full-length derivatives containing the F199K substitution alone, or in combination with other substitutions. The MW-I values observed, 71 and 72 kDa, were not appreciably different from the value for wild type IN, indicating that these preparations also contained primarily dimers under the conditions tested. These results are noteworthy, as Phe-199 lies at the core-core interface in crystals of the isolated core domain or the core + CTD of ASV IN, and substitution of this large hydrophobic side chain is predicted to reduce the stability of this interface, as illustrated with HIV-1 IN (49). Although our data (Table 1) show that the F199K substitution in full-length ASV IN does not compromise either dimerization or single end cutting and joining of viral DNA, a role in formation of higher order IN complexes (i.e. a tetramer) is likely, as the F199K derivative is unable to catalyze concerted integration.
TABLE 1.
Light scattering and activity summary
| Protein | Concentration | MW-Ia | Apparent multimerb | Single endc |
Concerted integrationc | |
|---|---|---|---|---|---|---|
| Cutting | Joining | |||||
| mg/ml | kDa | |||||
| ASV IN(1–286) | 2.3 | 69 (3.6) | Dimer (32) | +++ | +++ | +++ |
| Wild type | ||||||
| ASV IN(1–286) | 2.5 | 71 (3.6) | Dimer (34) | +++ | +++ | − |
| F199K | ||||||
| ASV IN(1–286) | 2.2 | 72 (1.8) | Dimer (34) | − | − | − |
| E157C/F199K | ||||||
| ASV IN(49–286) | 2.0 | 54 (5.2) | Dimer (27) | +++ | +++ | − |
| F199K | ||||||
| ASV IN(1–207) | 1.5 | 28 (1.9) | Monomer (23) | +(−3)d | − | − |
| ASV IN(1–286) | 1.0 | 34 (6.8) | Monomer (34) | +(−3) | − | − |
| C23S/C125S/F199K/W259A | ||||||
| ASV IN(1–286) | 2.5 | 37 (3.8) | Monomer (34) | +(−3) | − | − |
| W259A | ||||||
| PFV-IN(1–402) | 1.0 | 57 (8.8) | Monomer (45) | NDe | +++ | +++ |
| Wild type | ||||||
a Apparent molecular mass was determined by static light scattering. The number in parentheses is the percent standard error (%).
b The numbers in parentheses are values for the mass of a monomer calculated from the amino acid sequence and includes N-terminal tag residues where appropriate.
c Activities are expressed relative to wild type.
d Cleavage is observed at the −3 position rather than the expected −2 position. Similar −3 activity is observed with the ASV IN isolated core.
e ND means not tested.
The importance of the ASV CTD for IN multimerization is illustrated by comparison of the molecular mass of IN fragments in which either the NTD or CTD is absent. The MW-I of ASV IN(49–286), which lacks the NTD, is 54 kDa, exactly twice the mass calculated from the amino acid sequence of a respective monomer, 27 kDa. In contrast, under comparable conditions the MW-I of the ASV IN(1–207), which lacks the CTD, is 28 kDa, a value close to the calculated monomer mass of 23 kDa. These light scattering data are consistent with previously published results from size exclusion chromatography of these same IN fragments (50).
Shapes and Lengths of IN Proteins in Solution Determined by SAXS
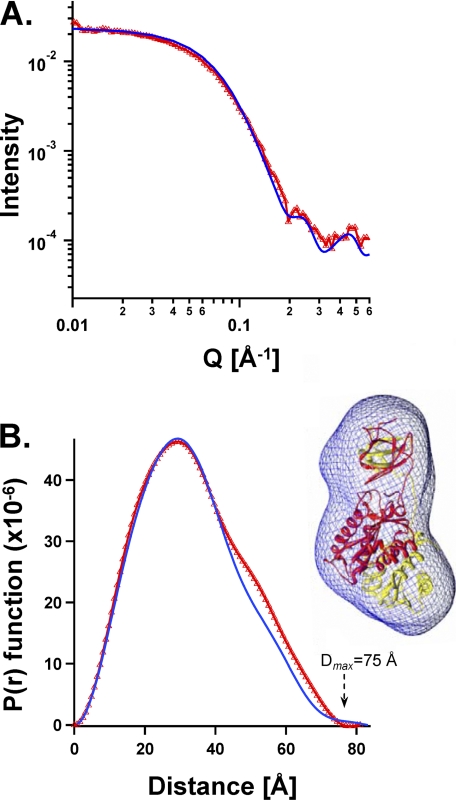
SAXS analyses provide a rotationally averaged version of the scattering of a single particle, from which size and shape can be determined. Certain features can be established unambiguously, i.e. the radius of gyration (Rg) and the longest dimension of the particle (Dmax). As verification of our methods, we performed SAXS on a preparation of the two-domain fragment lacking the NTD, ASV IN(49–286) F199K, and we compared the results with the shape and size determined from the published crystal structure of the same fragment (13). Fig. 2 shows the light scattering data and the P(r) function for this fragment from which we determined the Dmax to be 75 Å, close to the maximum of 81 Å calculated from the coordinates of the crystal structure. A low resolution shape of the dimer was derived from the SAXS data (51). Computational methods were used in reverse to calculate the expected scattering and P(r) function from the published atomic coordinates of the dimer. As shown in Fig. 2, these results were nearly super-imposable on the experimental data, and the SAXS-derived envelope was found to accommodate the atomic model of the crystal dimer neatly within its borders (Fig. 2B, right).
FIGURE 2.
Comparison of the crystal structure with solution SAXS dimensions and shapes of the same NTD-lacking two-domain ASV IN fragment (IN(49–286) F199K at 1.1 mg/ml). A, experimentally determined SAXS scattering is shown in red triangles. Values calculated from the crystal structure of the same IN fragment (PDB coordinate file 1C1A) using the Crysol program are represented by a blue line. B, experimentally derived plot of P(r) function for the SAXS data is compared with values calculated from the same crystal structure. Color code is the same as in A. Right, SAXS envelope shape derived from the experimental data is portrayed as a blue wire mesh, and the atomic resolution coordinates of 1C1A are shown within the SAXS envelope, with one monomer colored red and the other yellow.
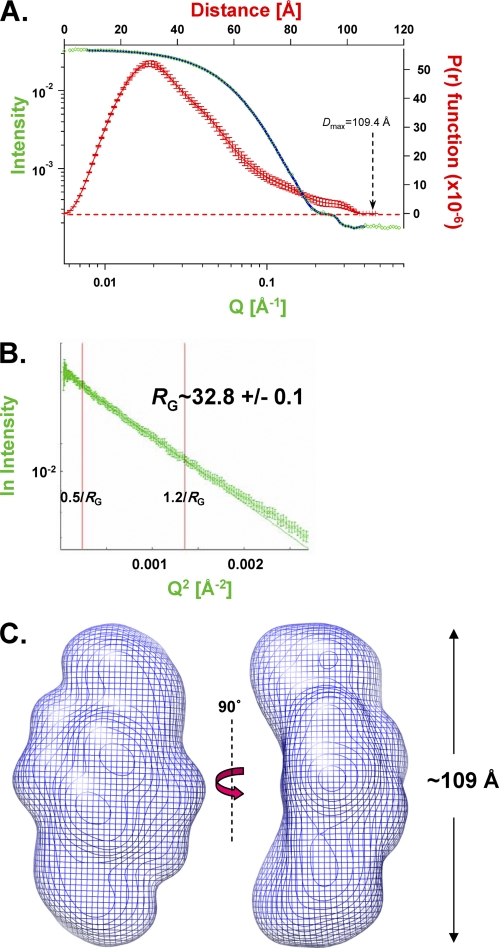
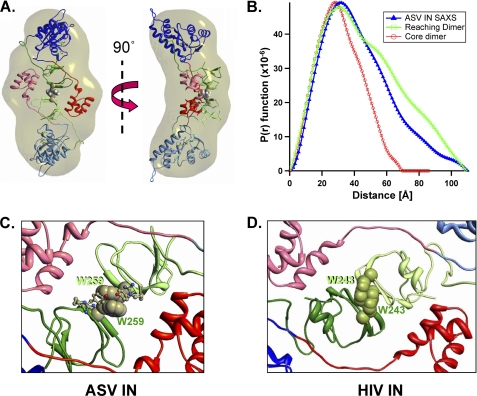
SAXS was then applied to the full-length wild type ASV IN protein, which is a homogeneous dimer at the relevant concentrations (Table 1). From the results in Fig. 3A, a Dmax of 109.4 Å was established for this dimer. Fig. 3B shows a plot of the scattering intensity (I(q)) versus Q2 (a “Guinier plot”) for this protein; an Rg of ∼32.8 Å was calculated from the slope of a linear fit of these data in the Q·Rg <1.2 region. A similar value (Rg = 33.1 ± 0.6 Å) was obtained from a nonlinear regression fitting (41). The linearity of the data at low angles verifies that the preparation was free of aggregates. A SAXs-derived envelope for the ASV IN dimer is shown in Fig. 3C (see also supplemental Fig. S1A).
FIGURE 3.
SAXS analyses of the full-length, wild type ASV IN dimer. A, experimentally determined SAXS data for ASV IN at 2.3 mg/ml, combining scattering data (green data points and blue fit line) and P(r) function (red points with error bars) into a four axis plot. B, Guinier plot of the natural log of the scattering intensity I(Q) versus Q2. A linear fit with data from 0.5/Rg to 1.2/Rg was used to approximate a radius of gyration (Rg) of 32.8 Å. C, shape of the wild type ASV IN dimer in solution, based on the SAXS data and modeled with the program GASBOR. The dimer envelope is shown in a blue mesh representation and two views.
The SAXS parameters obtained for full-length ASV IN and several other IN derivatives are summarized in Table 2. We note that, as with light scattering (Table 1), data obtained with the IN fragment that lacks the CTD (IN(1–207)) are as expected for a monomer, confirming that important determinants of dimerization reside in the CTD of ASV IN. Therefore, whereas core-core interactions can facilitate dimerization of the isolated CCD under crystallization conditions (9), under our conditions these interactions are not sufficient to allow dimerization of a protein that lacks the CTD. Furthermore, because a full-length derivative with an alanine substitution for residue Trp-259 in the CTD also displays the parameters of a monomer in solution (Table 2), we conclude that this tryptophan residue plays a key role in the dimerization interface of full-length ASV IN in solution.
TABLE 2.
Parameters derived from SAXS experiments
| Sample | Concentrationa | Q rangeb | Rgc | Dmaxc | I(0)c | χ2d |
|---|---|---|---|---|---|---|
| mg/ml | Å | Å | a.u. | |||
| ASV-IN(1–286)e | 2.3 | 0.007–0.4 | 33.1 ± 0.6 | 109.4 ± 1.3 | 0.033 | 0.8 |
| (wild type) | ||||||
| ASV-IN(1–286)e,f | 2.4 | 0.01–0.4 | 36.8 ± 0.2 | 119.7 ± 0.8 | 0.034 | 3.4 |
| (E157C/F199K) | ||||||
| ASV-IN(1–286)e | 0.7 | 0.01–0.4 | 31.5 ± 0.2 | 92.8 ± 1.0 | 0.024 | 1.7 |
| (C23S/C125S/F199K/W259A) | ||||||
| trxA-ASV-IN(1–286)e | 2.3 | 0.01–0.4 | 39.0 ± 0.4 | 114.9 ± 1.1 | 0.030 | 1.1 |
| (C23S/C125S/F199K/W259A) | ||||||
| ASV-IN(49–286)e,f | 1.1 | 0.01–0.42 | 26.0 ± 0.8 | 75.0 ± 2.2 | 0.022 | 2.2 |
| (F199K) | ||||||
| ASV-IN(1–207)e,f | 3.6 | 0.01–0.41 | 20.0 ± 0.1 | 61.0 ± 0.4 | 0.011 | 0.8 |
| PFV-IN(1–402)d | 4.3 | 0.007–0.42 | 38.5 ± 0.2 | 117.9 ± 0.6 | 0.031 | 2.7 |
| (wild type) |
a Data were determined by absorbance at 280 nm with a calculated molar extinction coefficient.
b Q = 4πsinθ/λ, where 2θ is the scattering angle; recorded data in this range were used for P(r) analysis and subsequent ab initio shape reconstructions.
c This was determined using the program IRENA. Comparable results were obtained using the program GNOM and by Guinier analysis with Auto Rg (65).
d Goodness of fit was assessed by reduced χ2 analysis.
e Data were collected at APS beamline DND-CAT 5ID-D.
f Data were also collected at local sources.
SAXS-determined Shape of Monomeric IN Establishes Constraints for the Relative Arrangement of the Three Domains
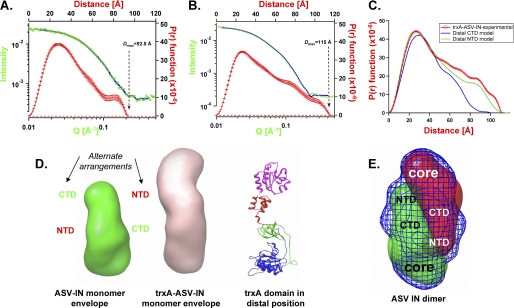
To determine how the subunits and their respective domains could be arranged within the experimentally determined IN dimer envelope, we performed SAXS analysis on a full-length ASV IN derivative that includes the W259A substitution. This protein contained three additional substitutions (C23S/C125S/F199K) that improve solubility but have no affect on single end cutting or joining activity (data not shown). The data obtained with this monomer (Fig. 4A), and its predicted elongated shape (Fig. 4D; supplemental Fig. S1B), are consistent with a structure containing the IN core domain (at the base in the figure) and the two smaller terminal domains, one close and one distal to the core. To determine whether the distal domain corresponds to the NTD or CTD, we produced a chimeric protein in which thioredoxin (trxA) is fused to the NTD of the W259A derivative (trxA-IN-W259A). The SAXS parameters for this derivative are summarized in Table 2, and the scattering data are shown in Fig. 4B. The envelope derived for the chimeric protein is considerably longer than that of the monomer lacking the N-terminal trxA domain, consistent with a distal placement for the NTD (Fig. 4D). Furthermore, the theoretical curve for a structure, in which the CTD of this derivative is the distal domain, produces parameters and an envelope that are inconsistent with the data in Fig. 4B (Fig. 4C). We therefore conclude that the distal domain in the ASV IN monomer is the NTD. A provisional model consistent with this conclusion is shown to the right of the trxA-ASV-IN envelope in Fig. 4D. This model is also supported by results from our SAXS analysis of wild type PFV IN, which contains a natural N-terminal extension called NED and is a monomer under the conditions of analysis; like the chimeric protein, the PFV monomer is longer than the ASV IN monomer (Table 2) and has a shape consistent with an NTD extension (envelope not shown). Fig. 4E shows how two monomer envelopes of ASV IN might fit within the dimeric envelope of wild type ASV IN with the approximate positions of each domain noted. We call this arrangement a “reaching dimer.”
FIGURE 4.
SAXS analyses of monomeric ASV IN proteins and relative positioning of the terminal domains. A, SAXS data obtained with the ASV IN C23S/C125S/F199K/W259A monomer at 0.7 mg/ml. Four axis plot is as described in Fig. 3. B, SAXS data obtained with the thioredoxin-ASV IN chimeric derivative, trxA-IN-C23S/C125S/F199K/W259A at 2.3 mg/ml. C, comparison of experimentally derived P(r) function for trxA-IN protein in B (red circles) with two possible P(r) plots calculated from alternate arrangements of a distal NTD (green line) or distal CTD domain (blue line). D, left, ASV IN monomer envelope indicating possible alternate arrangements of the NTD and CTD. Middle, trxA-IN monomer envelope, Right, ribbon model of a trxA-IN chimeric protein with the trxA domain (magenta) in a distal position from the core domain; IN coloring as in Fig. 1. E, envelope of the dimeric wild type ASV IN is shown in blue mesh representation, with two monomeric envelopes positioned (red and green mesh representations) to fit within the dimer envelope.
Strategy for Identifying Amino Acid Proximities in the IN Monomer and Multimers
To identify their regions of proximity within a dimer, it is necessary to be able to distinguish the two subunits. To do so, we prepared wild type ASV IN protein that was isotopically labeled with 13C and 15N in lysine and arginine residues (supplemental Fig. S2). The doubly labeled IN was then equilibrated with an equal amount of unlabeled IN using conditions described by Kessl et al. (46). After equilibration, half of the dimers are expected to be “mixed dimers,” containing one labeled and one unlabeled monomer, and the remainder either fully labeled or fully unlabeled. The mixture was then treated with BS3, a reagent that forms covalent cross-links between primary amines in lysine side chains and also with protein N termini that lie within 11.4 Å of each other. Samples were then subjected to electrophoresis in a denaturing polyacrylamide gel to determine the optimal concentration of BS3 (Fig. 5A). Cross-linked monomer, dimer, and tetramer bands were excised from the 1:20 lane, and the proteins eluted for identification of intra- and intersubunit cross-links, respectively, using trypsin digestion followed by mass spectrometry (see examples in supplemental Fig. S3).
FIGURE 5.
Monomer and dimer proximities uncovered by protein cross-linking coupled with mass spectrometry. A, SDS-PAGE showing the separation of ASV multimers after cross-linking with increasing concentrations of BS3. Positions of cross-linked monomers, dimers, and tetramers, which migrate slightly faster than the non cross-linked forms, are indicated at the right of the gel. B, cross-link map of the ASV IN monomer and a model structure. Residues involved in cross-linking between NTD and CTD in labeled wild type monomers are joined with dashed lines; solid lines identify cross-links within CTD residues or between CTD and CCD residues. Similar cross-links were observed with unlabeled monomers (data not shown). Right, HADDOCK-generated monomer IN structure, using the monomer cross-linking data and the SAXS envelope derived from the W259A IN derivative. C, map of dimer cross-links between labeled and unlabeled IN subunits. CTD to CTD links are shown with red lines; some included in supplemental Table S2 are omitted here for clarity. NTD to core or NTD to CTD links are denoted with dashed black lines.
Proximities Determined from Analysis of Cross-linked Monomeric IN
MS/MS analysis of protein excised from the monomer band, which contained both labeled and unlabeled IN protein, showed extensive intra-protein cross-linking (Fig. 5B and supplemental Table S1). However, no peptides corresponding to chemical cross-linking between the labeled and unlabeled IN proteins were detected in the isolated monomers. Demonstrating the uniform accessibility of side chains by this methodology, 15 of the total 20 surface-accessible lysine residues in all three domains of IN, as well as the N termini, were found to be mono-modified by the BS3, with dead ends comprising glycine or water (supplemental Table S1). As summarized in Fig. 5B, 5 of the 10 lysine residues in the CTD are within ∼11 Å of lysines in the NTD and the core; CTD tail residue Lys-278 was cross-linked to NTD Gly-1, and residues in the core domain, Lys-116 and Lys-191 were cross-linked to CTD Lys-264. In the CTD linker region, residues Lys-211 and Lys-225 were cross-linked to Lys-266 and Lys-272, respectively (Fig. 5B and supplemental Table S1), consistent with the Src homology 3-like fold of the CTD.
A monomer structure of IN that satisfies the observed cross-link constraints would place the NTD close to the C-terminal tail region of the CTD. In addition, the observed cross-links between lysine residues in the CTD with those in the NTD and core domains places the CTD in a position proximal to both. A structure consistent with all of the cross-linking data (Fig. 5B, right) has an extended NTD, which points away from the core domain, and a CTD in the cleft between the NTD core-linker region. This independently derived arrangement is consistent with the SAXS data summarized in Fig. 4.
Identification of Inter-subunit Proximities in the IN Dimer
Protein excised from the cross-linked dimer band was then analyzed. In this sample, inter-subunit proximities in mixed dimers can be identified unambiguously by mass spectrometry because of the hybrid mass of cross-linked peptides. Results from analysis of such peptides revealed an extensive network of interactions with a total of 21 cross-links between lysine residues in all domains of both subunits (Fig. 5C and supplemental Table S2). For example, NTD residue Lys-6 in the unlabeled IN monomer cross-linked with core domain Lys-116 in the labeled IN, and NTD Lys-21 in the unlabeled subunit formed cross-links with Lys-166 in the core and the CTD Lys-264. In addition, the amino group of the N-terminal glycine in the labeled IN subunit formed cross-links with core domain residues Lys-116 and Lys-164 and CTD residues Lys-264, Lys-266, and Lys-278 near the base of the tail in the unlabeled subunit. Reciprocal cross-links were identified between the core domain residue Lys-164 in the unlabeled subunit to the N-terminal Gly-1 and CTD residue Lys-264 in the labeled subunit. At least five lysine residues in the CTD of the unlabeled IN were found to cross-link with six residues in the labeled IN, and the identified cross-link adduct pairs were as follows: Lys-211:Lys-264, Lys-264:Lys-6, Lys-264:Gly-1, Lys-264:Lys-264, Lys-164:Lys-264, Lys-21:Lys-278, Lys-21:Lys-166, Lys-6:Lys-116, and Lys-266:Lys-264, respectively. Representative mass spectrometry data from the analysis of cross-linked peptides with hybrid mass are shown in supplemental Fig. S3. In summary, a total of 16 unique cross-links were uncovered, of which 8 of the 20 lysine residues in the unlabeled IN subunit formed cross-links with lysines in the labeled IN subunit, and 7 of the 20 lysine residues in the labeled IN subunit formed cross-links with unlabeled IN subunit (Fig. 5C and supplemental Table S2). The failure to identify completely reciprocal adducts could be due to incomplete detection or to minor asymmetry of interactions between the dimer interfaces, perhaps reflecting some flexibility in the subunit domains. With greater than 95% sequence coverage, we favor the latter interpretation.
The proximity data obtained from our analyses of cross-linked IN monomers and dimers support a dimer model that includes the following notable features. (a) In the dimer interface, CTD domains from each monomer come into close enough contact (i.e. ≤11Å) to form the following cross-links: Lys-264:Lys-211, Lys-264:Lys-264, and Lys-264:Lys-266, and others not included in Fig. 5C (see supplemental Table S2). (b) No cross-links between the two core domains were detected in the dimer. Consequently, the position of this domain in each subunit is sufficiently remote to exclude such interaction. As no cross-links were observed between NTDs in the mixed dimers, a similar constraint applies to this domain. (c) The NTD from one subunit is sufficiently close to the core domain and CTD of the other subunit to permit the following cross-link interactions between the subunits: Gly-1:Lys-116, Gly-1:Lys-164, Gly-1:Lys-264, Lys-116:Lys-6, Lys-166:Lys-21, and Lys-264:Lys-21. Additional experiments with the zero length protein cross-linker EDC confirmed NTD-core proximities (data not shown.)
The features delineated above are uniformly inconsistent with the core-core dimer model proposed from the two-domain crystal structures (14). The full-length dimer deduced from our results is stabilized by CTD-CTD interactions between both subunits and by interactions of the NTD of one IN subunit with the core domain and the CTD of the second subunit.
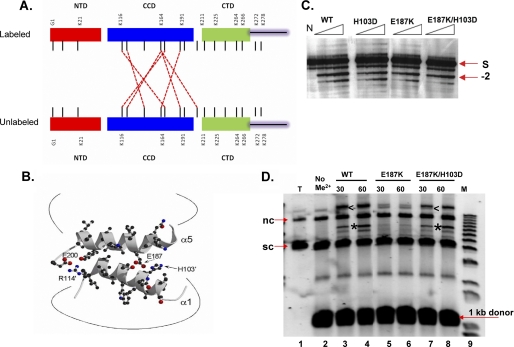
Identification of Core-Core Interactions in the IN Tetramer
MS/MS analysis of protein from the IN tetramer band (Fig. 5) revealed core-core cross-links in addition to the novel cross-links identified in protein from the dimer band (supplemental Table S3). Cross-links were observed with five of the seven lysine residues in this domain, of which reciprocal adducts of Lys-164:Lys-184 were observed between the labeled and unlabeled subunits (Fig. 6A). The remaining cross-links between these subunits were as follows: Lys-116:Lys-166, Lys-119:Lys-164, Lys-129:Lys-116, Lys-164: Lys-116 and Lys-211:Lys-164. These interactions are consistent with the interface observed in crystals of the isolated ASV IN core domain and those of the core + CTD two domain fragment (3, 13).
FIGURE 6.
Cross-linking evidence for core-core interactions in tetramers and their functional relevance. A, summary of core-core cross-link data. Red dashed lines show cross-links that were unique to protein in the tetramer band. B, reciprocal interactions in the core-core dimer interface in the crystal structure of the isolated ASV domain (PDB code 1VSH) are mediated predominantly by side chains in α-helices 1 and 5 of this domain; potential electrostatic interactions between Arg-114′ and Glu-200, as well as His-103′ and Glu-187 are highlighted; the prime designation distinguishes subunits. C, single end processing assays. Times of incubation were 5, 10, 20, and 30 min. The arrow labeled −2 shows the position of the normal processing product. The position of the 5′-32P-labeled viral DNA end substrate is indicated by S at the right of the gel; the control reaction in the lane marked N contained no IN protein. D, concerted integration assays. An arrowhead marks the position of a half-site reaction, in which a single end is joined to the plasmid target; the product of concerted integration is identified with an asterisk. Minutes of incubation are shown above each lane. The reaction in lane T contained no donor DNA, and in lane No Me2+, the divalent metal cofactor, Mg2+, was omitted. Lane M contains molecular markers, and positions of the supercoiled and nicked circular forms of the target DNA are marked sc and nc, respectively.
As illustrated in Fig. 6B, reciprocal interactions in the core-core dimer interface of ASV IN are mediated predominantly by side chains from α-helices 1 and 5; potential electrostatic interactions between Arg-114 and Glu-200 and His-103 and Glu-187 are highlighted. To investigate the functional importance of these interactions, we made charge-reversing single substitutions, E187K and H103D, and a compensatory double substitution, E187K/H103D. Comparison of the single end processing activity as a function of time showed no significant differences; each derivative was capable of −2 cleavage (Fig. 6C). In contrast, an assay for concerted integration activity showed that the protein with a single substitution, E187K, is defective in this reaction (Fig. 6D, lanes 5 and 6). However, this function is restored in the derivative with the compensatory substitutions, which exhibits activity similar to that of the wild type protein (Fig. 6D, lanes 7 and 8). We conclude that stability of a core-core interface, which is detected only in the cross-linked tetramers, is required for concerted integration but not single end processing.
ASV IN Solution Dimer Derived from Data-driven Docking
To gain more detailed insight into the architecture of the IN dimer, we employed the HADDOCK 2.0 docking program (52) with distance constraints established by our cross-linking data (Fig. 5C). These data-driven runs were performed on superimposed monomers constructed from the coordinates of the ASV core + CTD crystal structure (1C1A) and HIV core + NTD crystal structure (1K6Y) maintaining a minimum distance of 2.5 Å to a maximum reach of 11 Å between the defined cross-linked lysines across both monomers. Iterative runs were performed until Rg values from the docked structures were in close approximation to our experimentally determined SAXS envelope (supplemental Fig. S4). Rigid body fitting of these models within the SAXS envelope was performed by steepest descent local optimization, which converges on an orientation that minimizes the number of atoms lying outside the envelope. The resulting minimized symmetrical arrangement, shown in Fig. 7A, is stabilized by face-to-face hydrophobic interaction between Trp-259 from each of the monomers (see also supplemental Movie 1).
FIGURE 7.
A reaching dimer model of the ASV IN apoprotein. A, dimer model of ASV IN that satisfies both distance constraints from the cross-linking experiments and the envelope shape and dimensions from SAXS experiments. Two orthogonal views are shown with our standard color coding for the three IN domains in one monomer, and the second IN monomer in muted colors. B, comparison of the experimentally determined P(r) function (blue line) of dimeric wild type ASV IN, with a P(r) function calculated from the core-core stabilized dimer model shown in Fig. 1C (red line) and the reaching dimer structure in A (green line). C, details of the CTD-CTD interface in the ASV IN dimer showing how stacking between proximal Trp-259 side chains from each CTD is a prominent feature of this interface. D, model of a reaching dimer of HIV-1 IN reveals the potential for conservation of CTD-CTD interface interactions.
Fig. 7B shows the P(r) function derived from our SAXS analysis of the wild type ASV IN dimer and theoretical curves calculated from the core-stabilized dimer model (Fig. 1C) and the reaching dimer in Fig. 7A. This comparison shows that the core-stabilized dimer model possesses a significantly shorter Dmax and is less elongated and more spherical in shape than that deduced from our experimental SAXS data. The theoretical curve for the reaching dimer matches the experimental data more closely, and it shows the same Dmax. These results, together with the observation that the IN W259A derivative behaves as a monomer, are consistent with a subunit arrangement in which the CTDs, rather than the core domains, play a critical role in dimerization. Fig. 7C shows a close-up view of potential stacking interactions between Trp-259 residues in the CTDs of the reaching dimer.
A Reaching Dimer Model for HIV-1 IN
Although sequence identity between ASV and HIV IN proteins is less than 20%, they have very similar domain structures (1). Consequently, we constructed a reaching dimer model for HIV IN, based on the ASV IN dimer, to uncover any conserved features and evaluate the correlation with previous mutagenesis data. A comparison of the two reaching dimers shows that the CTD interfaces of both can be stabilized by face-to-face interactions between aromatic residues as follows: Trp-259 residues as described above for ASV IN and Trp-243 residues for HIV-1 (Fig. 7, C and D). As noted in Table 1, replacement of Trp-259 with alanine abolishes both dimer formation by ASV IN and proper cleavage at −2 in the viral DNA substrates. The comparable substitution in HIV IN (W243A) also results in the loss of single end processing activity (53), as would be expected if Trp-243 played a similar role in HIV-1 IN.
Further inspection of the reaching dimer interfaces of ASV IN and HIV-1 IN revealed a network of potential hydrogen bonds between the NTD from one monomer to both of the linkers and the CTD in the second monomer (summarized in supplemental Fig. S5). With ASV IN, we have observed that interruption of the proposed hydrogen bonding between Asn-24 and Arg-53 in the linker region (supplemental Fig. S5A) by replacement of the latter residue with alanine resulted in loss of single end joining activity (data not shown). Potential interactions between the CTD from one monomer with the CTD and NTD in the second monomer include buried hydrophobic interactions involving Trp-259 in ASV IN and Trp-243 in the HIV IN. In the ASV IN structure, side chains from residues 244 to 246 can stabilize the dimer interface further through formation of inter-molecular hydrogen bonds between the two tyrosine side chains (supplemental Fig. S5A). We have observed that substitution of alanine for Tyr-246 results in a 50% decrease in single end joining activity (data not shown). These results are consistent with a role for such hydrogen bonds in the architecture of a functional dimer. The potential hydrogen bond interactions in the proposed reaching dimer interface of HIV IN (supplemental Fig. S5B) includes residues that are highly conserved in the HIV genome, with less than 1% variance observed in the genomes of viruses isolated from 488 inhibitor-naive patients in a recent study (54). Such conservation would be expected from stringent evolutionary pressure for assembly of a functional form of the apoenzyme.
DISCUSSION
Here, we describe the use of two complementary approaches to elucidate the architecture of both monomeric and multimeric forms of a full-length retroviral IN protein, in the absence of its DNA substrates. These are the first experimentally derived full-length apoprotein solution structures of IN to be reported. Others have characterized the solution size and shape of HIV-1 IN but in the presence of the LEDGF protein fragments, which are known to modulate IN multimerization (45). Although of relatively low resolution, the use of SAXS with wild type IN and IN derivatives provided valuable insight into the length, shape, and domain organizations in full-length monomers and dimers. Protein cross-linking, which tethers all dynamically involved lysines separated by ≤11 Å coupled with mass spectrometry, provided independent constraints for docking within the SAXS-derived envelopes. After equilibrating an equal mixture of unlabeled and labeled IN proteins, inter-molecular cross-links could be identified unambiguously by the isolation of adducts with hybrid mass. As no hybrid adducts were observed in our analyses of cross-linked monomers isolated from the mixture, we conclude that the native structure was conserved within the cross-linked proteins.
In the IN monomers, the CTD was found to cross-link with the core and the NTD, and the NTD with the CTD “tail” (residues 270–289). A model for the full-length IN monomer structure that combines our SAXS and cross-linking data (Fig. 5B) shows the core and NTDs at distal poles and the NTD in close proximity to the extended tail of the centrally located CTD. The solution dimer interface revealed in our studies is noteworthy for the absence of any core-core domain interactions, which had previously been thought to stabilize this multimeric form. Analysis of our cross-linked dimers uncovered a cluster of hybrid adducts formed between two IN monomers. The unanticipated architecture so revealed shows a reciprocal arrangement in which the CTD from one monomer anchors into the CTD of the second monomer, and the NTD from one monomer interacts with core and CTD of the second monomer. The absence of any core-core and NTD-NTD cross-links between the subunits implies that these domains are distantly separated in the two subunits. A model consistent with results from the SAXS and cross-linking studies places the two core domains at opposite ends, with the association of subunits stabilized by interactions between opposing NTDs and CTDs, which reach out to each other (Figs. 4E and 7A).
Cross-links corresponding to the core-core interface observed in crystals of the isolated core, and two-domain fragments were detected only in full-length ASV IN tetramers (Fig. 6A). Results from our mutational studies suggest that the stability of this interface is required for concerted integration, but it is not essential for catalysis of single end processing or joining by IN, which can be accomplished by IN dimers (33). Consequently, we conclude that the primary role of the core-core interface is in assembly of a tetrameric synaptic complex, which can catalyze the concerted joining of two 3′ viral DNA ends into a target DNA (33, 34). This interpretation is supported by previously published effects of other substitutions in this core-core interface (55).
A detailed structural model of the reaching dimer of ASV IN was obtained by combining the observed chemical cross-linking distance constraints with data-driven docking (Fig. 7A and supplemental Fig. S4). In the iterative docking runs, the protein-protein buried surface area increased from 1100 to 2200 Å2, whereas Rg was reduced from 47 to 34 in the final minimum structure. Increase in the buried surface area and decrease in Rg implies a structure that is considerably more compact than the sum of the initial docking monomers. In studies with other proteins, values within the range of 1600 ± 400 Å2 have been observed for the buried surfaces in complexes that undergo minimal conformational change during their assembly, whereas values in the range of 2000–4400 Å2 are typical for complexes that undergo large conformational changes during formation (56).
As the SAXS envelope of the ASV monomer was only observed with an IN W259A derivative, we cannot be definitive about the orientation of the CTD in this structure. We note that our original rationale for substituting Trp-259 stemmed from a comparison of the IN CTD to other Src homology 3-like domains in proteins that bind DNA, such as the Sso7 chromatin-binding protein (57). Alignment, modeling, and identification of potential DNA-binding residues in the ASV IN CTD suggested a likely role for Trp-259 in binding to DNA substrates. It was only after purification and analysis of this protein that we noticed its deficiency in dimerization, implying a potential dual role for this particular residue in both dimerization and DNA binding.
The interface in the reaching dimer model is dominated by aromatic interactions between a cluster of residues in the CTDs, which represent a unique hot spot for the maintenance of dimer stability. Results from our mutational studies indicate that the tryptophan residues, Trp-259 in ASV IN and Trp-243 in HIV IN, play a critical role in both catalytic activity and stability of an intersubunit interface (Table 1 and Fig. 7, C and D) (53). Amino acids surrounding such hot spots, classified as O-ring residues, are predicted to protect the hydrophobic residues from solvent and to stabilize their interaction via hydrogen bonds and salt bridges (58). Close analysis of reaching dimers of ASV-IN and modeled HIV-IN reveals that the majority of potential variable O-ring residues at the CTD-CTD interfaces in both structures can form hydrogen bonds or electrostatic interactions. The potential for similar interactions can be observed in a number of other retroviral IN proteins.
Previous studies on the mechanism of inhibition by monoclonal antibodies that are specific to the HIV-IN NTD and CTD have uncovered critical epitope residues in these domains (59, 60). The inhibitory activity of mAb17 can be explained by distortion of the NTD helix-turn-helix motif via binding to residues 25–35. The epitope of mAb33 includes CTD residues Phe-223, Arg-224, Tyr-226, Lys-224, Ile-267, and Ile-268 and expression of a mAb33 sFv fragment in host cells blocks HIV-1 infectivity prior to the integration step (61). Our HIV-1 IN model predicts that binding of either one of these antibodies would interfere with the assembly of a functional reaching dimer. Furthermore, alanine substitution of any one of these epitope residues drastically reduces the single end joining activity of HIV-1 IN (53, 60). This result is consistent with the derived reaching dimer interface of HIV-1 IN in which Lys-34 is predicted to hydrogen bond with Glu-246; buried Arg-262 is predicted to interact with the backbones of Pro-30 and Val-31 and Arg-263 to hydrogen bond with Glu-33. Substitution of the conserved, buried Arg-262 with alanine was also found to abolish catalytic activity (53), suggesting that this residue might represent a second hot spot in the dimer interface, in addition to the buried Trp-243. Taken together, these data indicate that the dimer interfaces of both ASV and HIV-1 IN apoproteins are characterized by fully buried tryptophan hot spots as well as O-ring residues that are partly accessible to solvent. The deleterious effects of alanine substitution for some of the partly accessible residues can be modulated by water molecules. In contrast, substitution of buried residues that are close-packed, optimizing van der Waals interactions, cannot be tolerated without significant structural and functional cost. The buried hydrophobic interactions in our model at the CTD-CTD interface likely shield the hydrophobic patch prior to interaction with DNA.
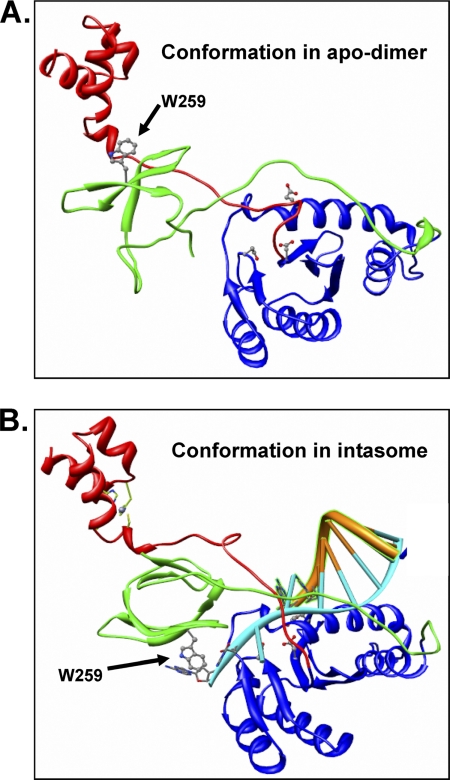
The organization of the reaching ASV IN dimer that forms in the absence of DNA substrate bares a striking resemblance to the “inner” dimers observed to bind DNA substrates in the recently reported crystal structure of the PFV intasome (38, 39). The major difference between the two is in the position of the CTD (Fig. 8 and supplemental movie 2). However, the reaching dimer is in a conformation that is energetically favorable to accommodate association with viral DNA ends, simply by unpairing of the conserved CTD tryptophans (Trp-259 for ASV and Trp-243 for HIV IN) and rotating via the linker region. A DNA-induced conformational change that involves the linker of HIV-1 IN has been reported previously (62). A stable intasome would then be formed by association of the viral DNA strand to be processed with the catalytic triad and stabilization of the nontransferred DNA strand by hydrophobic interactions between the terminal bases and the now translocated conserved tryptophans. Stabilizing or disrupting alternate multimeric assemblies to modulate enzyme activity in an allosteric manner has been suggested as an alternative to active site inhibition (63). Although such an approach has been proposed for HIV-1 integrase (64), these studies targeted only the core-core dimer interface. Disruption of the critical CTD-CTD interface interactions could represent a novel strategy for development of anti-HIV drugs.
FIGURE 8.
Conformational change required for the transition from a reaching dimer to the intasome complex with DNA. A, conformation of a single ASV IN subunit in the apo-IN reaching dimer. B, conformation predicted from the inner dimer of an intasome complex that includes the viral substrate DNA. The subunit structure is modeled from the PFV intasome (PDB code 3OYA (38)). The change in orientation of the CTD residue Trp-259, shown in ball-and-stick, is highlighted with arrows. Active site residues are also shown in ball-and-stick fashion. A supplemental movie that simulates the conformational change between these two states is provided.
Supplementary Material
Acknowledgments
We thank Drs. Richard Katz, Eileen Jaffe, and Roland Dunbrack for critical comments and helpful discussions and Marie Estes for valuable help in preparing our manuscript. Dr. Peter Cherepanov generously provided a plasmid that expresses PFV IN protein, advice for its preparation, and PFV intasome coordinates prior to publication. Dr. Zimei Bu performed some preliminary SAXS analyses at the Fox Chase Cancer Center, and Dr. S. Seeholzer provided advice for our mass spectrometry analyses. Molecular modeling was performed with the help of the Fox Chase Molecular Modeling Facility and mass spectrometry analyses benefited from assistance of Dr. Yibai Chen in the Fox Chase Biotechnology and Biochemistry Facility. Sequences of all expression plasmid constructs were verified by the Fox Chase DNA Sequencing Facility. Use of APS was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences Grant DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grants AI40385, CA71515, and CA006927. This work was also supported by an appropriation from the Commonwealth of Pennsylvania.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3, Figs. S1–S5, Movies 1 and 2, and additional references.
- IN
- integrase
- SAXS
- small angle x-ray scattering
- SAX
- small angle x-ray
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- ASV
- avian sarcoma virus
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- BS3
- bis(sulfosuccinimidyl) suberate
- PDB
- Protein Data Bank
- CCD
- catalytic core domain
- PFV
- prototype foamy virus
- trxA
- thioredoxin A
- MW-I
- apparent molecular mass
- APS
- Advanced Photon Source.
REFERENCES
- 1. Jaskolski M., Alexandratos J. N., Bujacz G., Wlodawer A. (2009) FEBS J. 276, 2926–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katz R. A., Merkel G., Kulkosky J., Leis J., Skalka A. M. (1990) Cell 63, 87–95 [DOI] [PubMed] [Google Scholar]
- 3. Bujacz G., Jaskólski M., Alexandratos J., Wlodawer A., Merkel G., Katz R. A., Skalka A. M. (1996) Structure 4, 89–96 [DOI] [PubMed] [Google Scholar]
- 4. Bujacz G., Alexandratos J., Wlodawer A., Merkel G., Andrake M., Katz R. A., Skalka A. M. (1997) J. Biol. Chem. 272, 18161–18168 [DOI] [PubMed] [Google Scholar]
- 5. Maignan S., Guilloteau J. P., Zhou-Liu Q., Clément-Mella C., Mikol V. (1998) J. Mol. Biol. 282, 359–368 [DOI] [PubMed] [Google Scholar]
- 6. Lins R. D., Straatsma T. P., Briggs J. M. (2000) Biopolymers 53, 308–315 [DOI] [PubMed] [Google Scholar]
- 7. Jenkins T. M., Hickman A. B., Dyda F., Ghirlando R., Davies D. R., Craigie R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6057–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dyda F., Hickman A. B., Jenkins T. M., Engelman A., Craigie R., Davies D. R. (1994) Science 266, 1981–1986 [DOI] [PubMed] [Google Scholar]
- 9. Bujacz G., Jaskólski M., Alexandratos J., Wlodawer A., Merkel G., Katz R. A., Skalka A. M. (1995) J. Mol. Biol. 253, 333–346 [DOI] [PubMed] [Google Scholar]
- 10. Goldgur Y., Dyda F., Hickman A. B., Jenkins T. M., Craigie R., Davies D. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9150–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen J. C., Krucinski J., Miercke L. J., Finer-Moore J. S., Tang A. H., Leavitt A. D., Stroud R. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Z., Yan Y., Munshi S., Li Y., Zugay-Murphy J., Xu B., Witmer M., Felock P., Wolfe A., Sardana V., Emini E. A., Hazuda D., Kuo L. C. (2000) J. Mol. Biol. 296, 521–533 [DOI] [PubMed] [Google Scholar]
- 13. Yang Z. N., Mueser T. C., Bushman F. D., Hyde C. C. (2000) J. Mol. Biol. 296, 535–548 [DOI] [PubMed] [Google Scholar]
- 14. Wang J. Y., Ling H., Yang W., Craigie R. (2001) EMBO J. 20, 7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lodi P. J., Ernst J. A., Kuszewski J., Hickman A. B., Engelman A., Craigie R., Clore G. M., Gronenborn A. M. (1995) Biochemistry 34, 9826–9833 [DOI] [PubMed] [Google Scholar]
- 16. Eijkelenboom A. P., Sprangers R., Hård K., Puras Lutzke R. A., Plasterk R. H., Boelens R., Kaptein R. (1999) Proteins 36, 556–564 [DOI] [PubMed] [Google Scholar]
- 17. Cai M., Zheng R., Caffrey M., Craigie R., Clore G. M., Gronenborn A. M. (1997) Nat. Struct. Biol. 4, 567–577 [DOI] [PubMed] [Google Scholar]
- 18. Eijkelenboom A. P., Lutzke R. A., Boelens R., Plasterk R. H., Kaptein R., Hård K. (1995) Nat. Struct. Biol. 2, 807–810 [DOI] [PubMed] [Google Scholar]
- 19. Eijkelenboom A. P., van den Ent F. M., Vos A., Doreleijers J. F., Hård K., Tullius T. D., Plasterk R. H., Kaptein R., Boelens R. (1997) Curr. Biol. 7, 739–746 [DOI] [PubMed] [Google Scholar]
- 20. Eijkelenboom A. P., van den Ent F. M., Wechselberger R., Plasterk R. H., Kaptein R., Boelens R. (2000) J. Biomol. NMR 18, 119–128 [DOI] [PubMed] [Google Scholar]
- 21. Coleman J., Eaton S., Merkel G., Skalka A. M., Laue T. (1999) J. Biol. Chem. 274, 32842–32846 [DOI] [PubMed] [Google Scholar]
- 22. Deprez E., Tauc P., Leh H., Mouscadet J. F., Auclair C., Brochon J. C. (2000) Biochemistry 39, 9275–9284 [DOI] [PubMed] [Google Scholar]
- 23. Jenkins T. M., Engelman A., Ghirlando R., Craigie R. (1996) J. Biol. Chem. 271, 7712–7718 [DOI] [PubMed] [Google Scholar]
- 24. Jones K. S., Coleman J., Merkel G. W., Laue T. M., Skalka A. M. (1992) J. Biol. Chem. 267, 16037–16040 [PubMed] [Google Scholar]
- 25. Lee S. P., Xiao J., Knutson J. R., Lewis M. S., Han M. K. (1997) Biochemistry 36, 173–180 [DOI] [PubMed] [Google Scholar]
- 26. Zheng R., Jenkins T. M., Craigie R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13659–13664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bushman F. D., Engelman A., Palmer I., Wingfield P., Craigie R. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3428–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ellison V., Gerton J., Vincent K. A., Brown P. O. (1995) J. Biol. Chem. 270, 3320–3326 [DOI] [PubMed] [Google Scholar]
- 29. Engelman A., Bushman F. D., Craigie R. (1993) EMBO J. 12, 3269–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van Gent D. C., Vink C., Groeneger A. A., Plasterk R. H. (1993) EMBO J. 12, 3261–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guiot E., Carayon K., Delelis O., Simon F., Tauc P., Zubin E., Gottikh M., Mouscadet J. F., Brochon J. C., Deprez E. (2006) J. Biol. Chem. 281, 22707–22719 [DOI] [PubMed] [Google Scholar]
- 32. Bao K. K., Wang H., Miller J. K., Erie D. A., Skalka A. M., Wong I. (2003) J. Biol. Chem. 278, 1323–1327 [DOI] [PubMed] [Google Scholar]
- 33. Faure A., Calmels C., Desjobert C., Castroviejo M., Caumont-Sarcos A., Tarrago-Litvak L., Litvak S., Parissi V. (2005) Nucleic Acids Res. 33, 977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li M., Mizuuchi M., Burke T. R., Jr., Craigie R. (2006) EMBO J. 25, 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao K., Butler S. L., Bushman F. (2001) EMBO J. 20, 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Karki R. G., Tang Y., Burke T. R., Jr., Nicklaus M. C. (2004) J. Comput. Aided Mol. Des. 18, 739–760 [DOI] [PubMed] [Google Scholar]
- 37. Podtelezhnikov A. A., Gao K., Bushman F. D., McCammon J. A. (2003) Biopolymers 68, 110–120 [DOI] [PubMed] [Google Scholar]
- 38. Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. (2010) Nature 464, 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maertens G. N., Hare S., Cherepanov P. (2010) Nature 468, 326–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baker N. M., Weigand S., Maar-Mathias S., Mondragón A. (2011) Nucleic Acids Res. 39, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ilavsky J., Jemian P. R. (2009) J. Appl. Crystallogr. 42, 347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Semenyuk A. V., Svergun D. I. (1991) J. Appl. Crystallogr. 24, 537–540 [Google Scholar]
- 43. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wriggers W., Birmanns S. (2001) J. Struct. Biol. 133, 193–202 [DOI] [PubMed] [Google Scholar]
- 45. Gupta K., Diamond T., Hwang Y., Bushman F., Van Duyne G. D. (2010) J. Biol. Chem. 285, 20303–20315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kessl J. J., Eidahl J. O., Shkriabai N., Zhao Z., McKee C. J., Hess S., Burke T. R., Jr., Kvaratskhelia M. (2009) Mol. Pharmacol. 76, 824–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu H., Freitas M. A. (2009) Proteomics 9, 1548–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hojrup P. (1990) in General Protein Mass Analysis (GPMA), a Convenient Program in Studies of Proteins by Mass Analysis (Hedin A., et al., eds) pp. 61–66, John Wiley & Sons Ltd., Chichester, UK [Google Scholar]
- 49. Al-Mawsawi L. Q., Hombrouck A., Dayam R., Debyser Z., Neamati N. (2008) Virology 377, 355–363 [DOI] [PubMed] [Google Scholar]
- 50. Andrake M. D., Skalka A. M. (1995) J. Biol. Chem. 270, 29299–29306 [DOI] [PubMed] [Google Scholar]
- 51. Svergun D. I. (1999) Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. de Vries S. J., van Dijk M., Bonvin A. M. (2010) Nat. Protoc. 5, 883–897 [DOI] [PubMed] [Google Scholar]
- 53. Ramcharan J., Colleluori D. M., Merkel G., Andrake M. D., Skalka A. M. (2006) Retrovirology 3, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ceccherini-Silberstein F., Malet I., D'Arrigo R., Antinori A., Marcelin A. G., Perno C. F. (2009) AIDS Rev. 11, 17–29 [PubMed] [Google Scholar]
- 55. Moreau K., Faure C., Violot S., Verdier G., Ronfort C. (2003) Eur. J. Biochem. 270, 4426–4438 [DOI] [PubMed] [Google Scholar]
- 56. Lo Conte L., Chothia C., Janin J. (1999) J. Mol. Biol. 285, 2177–2198 [DOI] [PubMed] [Google Scholar]
- 57. Gao Y. G., Su S. Y., Robinson H., Padmanabhan S., Lim L., McCrary B. S., Edmondson S. P., Shriver J. W., Wang A. H. (1998) Nat. Struct. Biol. 5, 782–786 [DOI] [PubMed] [Google Scholar]
- 58. Liu Q., Li J. (2010) BMC Bioinformatics 11, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yi J., Arthur J. W., Dunbrack R. L., Jr., Skalka A. M. (2000) J. Biol. Chem. 275, 38739–38748 [DOI] [PubMed] [Google Scholar]
- 60. Yi J., Cheng H., Andrake M. D., Dunbrack R. L., Jr., Roder H., Skalka A. M. (2002) J. Biol. Chem. 277, 12164–12174 [DOI] [PubMed] [Google Scholar]
- 61. Levy-Mintz P., Duan L., Zhang H., Hu B., Dornadula G., Zhu M., Kulkosky J., Bizub-Bender D., Skalka A. M., Pomerantz R. J. (1996) J. Virol. 70, 8821–8832 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 62. Zhao Z., McKee C. J., Kessl J. J., Santos W. L., Daigle J. E., Engelman A., Verdine G., Kvaratskhelia M. (2008) J. Biol. Chem. 283, 5632–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lawrence S. H., Ramirez U. D., Tang L., Fazliyez F., Kundrat L., Markham G. D., Jaffe E. K. (2008) Chem. Biol. 15, 586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hayouka Z., Rosenbluh J., Levin A., Loya S., Lebendiker M., Veprintsev D., Kotler M., Hizi A., Loyter A., Friedler A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Petoukhov M. V., Konarev P. V., Kikhney A. G., Svergun D. I. (2007) J. Appl. Crystallogr. 40, S223–S228 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.