Abstract
Triacylglycerols are stored in eukaryotic cells within lipid droplets (LD). The LD core is enwrapped by a phospholipid monolayer with phosphatidylcholine (PC), the major phospholipid, and phosphatidylethanolamine (PE), a minor component. We demonstrate that the onset of LD formation is characterized by a change in cellular PC, PE, and phosphatidylserine (PS). With induction of differentiation of 3T3-L1 fibroblasts into adipocytes, the cellular PC/PE ratio decreased concomitant with LD formation, with the most pronounced decline between confluency and day 5. The mRNA for PS synthase-1 (forms PS from PC) and PS decarboxylase (forms PE from PS) increased after day 5. Activity and protein of PE N-methyltransferase (PEMT), which produces PC by methylation of PE, are absent in 3T3-L1 fibroblasts but were induced at day 5. High fat challenge induced PEMT expression in mouse adipose tissue. PE, produced via PS decarboxylase, was the preferred substrate for methylation to PC. A PEMT-GFP fusion protein decorated the periphery of LD. PEMT knockdown in 3T3-L1 adipocytes correlated with increased basal triacylglycerol hydrolysis. Pemt−/− mice developed desensitization against adenosine-mediated inhibition of basal hydrolysis in adipose tissue, and adipocyte hypotrophy was observed in Pemt−/− animals on a high fat diet. Knock-out of PEMT in adipose tissue down-regulated PS synthase-1 mRNA, suggesting coordination between PE supply and converting pathways during LD biosynthesis. We conclude that two consecutive processes not previously related to LD biogenesis, (i) PE production via PS and (ii) PE conversion via PEMT, are implicated in LD formation and stability.
Keywords: Lipid Droplet, Phosphatidylcholine, Phosphatidylethanolamine, Phosphatidylserine, Phospholipid Metabolism, PEMT
Introduction
Cytosolic neutral lipid droplets (LD)7 within eukaryotic cells represent intracellular storage compartments for triacylglycerols (TG) and cholesteryl esters to bridge alimentary or metabolic gaps (1). Adipocytes are the body's primary depots for efficient TG storage. Upon defined stimulation of adipocytes (2), fatty acids are released from TG droplets to supply energy or to provide essential components for the synthesis of biological membranes. Given these important functions, LD of all cell types are considered as dynamic organelles that represent fundamental components of intracellular lipid homeostasis (3).
It is generally accepted that LD originate from the cytosolic leaflet of the endoplasmic reticulum (ER), contain a core of neutral lipids, and are surrounded by a phospholipid (PL) monolayer (4, 5). The finding that cytosolic LD in adipocytes contain at their periphery minor amounts of ER proteins such as BiP (6) and calnexin might reflect their site of origin in the ER. The primary LD proteins are so-called cage proteins, which not only stabilize LD but also protect their neutral lipid cores from unregulated degradation (7). Several other proteins contributing to the regulation of intracellular vesicle trafficking or targeting (3), and components of the intermediate filament protein machinery, have also been shown to be associated with intracellular LD (8, 9).
Despite recent advances in our understanding of the LD protein shell and how newly formed TG enters LD (10), our knowledge of the nature and architecture of the LD monolayer lags far behind. Although PC is the most abundant cellular PL, PE exhibits, among all PL, the highest relative increase during fat cell differentiation (11), suggesting a specific function for PE during LD biogenesis. In addition to PC and PE, the monolayer contains a broad spectrum of minor membrane components such as ether lipids (12) and unesterified cholesterol (6).
One would expect compositional similarities between ER membranes and the LD monolayer due to the origin of the PL monolayer from ER structures. However, mass spectrometric analyses of the LD monolayer PL subspecies, PC and lyso-PC, revealed differences in the fatty acid pattern compared with the parental ER membrane (4). Whereas mono-unsaturated fatty acids are the predominant constituents of lyso-PC and PC in LD, saturated fatty acids or fatty acids with two double bonds are more highly represented in rough ER membranes (4). It is unclear whether this finding reflects the species of PL fatty acids in LD of distinct cell types or whether other glycero-PL classes (e.g. PE, phosphatidylserine (PS), and phosphatidylinositol) are also differentially distributed between LD and subcellular membranes. Potentially, PL remodeling could occur close to or at the ER membrane during biogenesis of the LD monolayer.
Several in vitro studies confirm that PL remodeling mechanisms are required for sprouting of nascent LD monolayers from the outer ER leaflet and/or subsequent budding of the nascent LD from the ER monolayer. Olofsson and co-workers (13) reconstructed some of the fundamental steps (14) of early LD biogenesis. Their results clearly demonstrate that enzyme activity of phospholipase D1 (PLD1) is a prerequisite for LD formation. Similar results underscoring the impact of PLD1 were obtained in 3T3-L1 cells (15). These findings imply that the products of this reaction, conically shaped phosphatidic acid molecules (16), might represent structural tools that initiate curvatures in the ER outer leaflet when LD budding is initiated. From other studies it is apparent that PE, also conically shaped (16), represents another component that might initiate formation of concavely bent membranes (17).
The characterization of distinct but mechanistically related intracellular processes (e.g. biogenesis of very low density lipoproteins) indirectly supports this idea. In liver, where these lipoproteins are made, PC homeostasis is maintained by two independent pathways (18) as follows: (i) the Kennedy (CDP-choline) pathway, which is the primary route for synthesis of PC from diacylglycerol and activated choline, and (ii) the PE methylation pathway in which PC is produced from PE by three sequential methylation steps in a reaction catalyzed by phosphatidylethanolamine N-methyltransferase (PEMT). Previous results have shown that normal very low density lipoprotein secretion requires not only de novo PC biosynthesis (19–21) but a delicate balance between the levels of hepatic PE and PC (22). In line with this observation, Pemt−/− mice rapidly develop hepatic steatosis when challenged by a high fat diet (23, 24). In this animal model, hepatic accumulation of neutral lipids is due to a mechanistically uncharacterized defect in very low density lipoprotein secretion. A direct or indirect role for PS in LD biogenesis has no precedent in the current literature.
The presence of specific PL such as PE and phosphatidic acid in biological membranes may be a prerequisite for induction of local membrane curvature of the ER. Given this potentially important structural function of PE during LD biogenesis, we reasoned that PE represents a primary component for the biogenesis and growth of intracellular LD in murine adipocytes. We report that several previously unrecognized PL remodeling events occur during LD formation.
EXPERIMENTAL PROCEDURES
Animals
Male Pemt+/+ and Pemt−/− mice (25) (8–16 weeks old) were fed ad libitum either regular rodent chow or a high fat diet containing 30.2% crude fat (SSNIFF® Spezialdiäten GmbH, Germany, EF R/M, E15116) for 3 weeks; adipose tissue from 45- to 50-week-old animals was used for lipolysis assays. Tissues were harvested, rinsed with ice-cold phosphate-buffered saline (PBS), and either flash-frozen in liquid nitrogen until used for protein, immunoblot, and RNA analyses or used immediately for lipolysis assays.
Cell Culture and Isolation of Lipid Droplets
3T3-L1 cells were cultured and differentiated as described previously (6). Accumulated neutral lipids were analyzed by direct in vivo staining of neutral lipids with Nile Red and fluorescence microscopy (26). OP9 mouse stromal cells (kindly provided by Dr. Toru Nakano, Osaka University) were differentiated using insulin-oleate albumin complex (27). For isolation of LD, 3T3-L1 adipocytes at different days of differentiation were scraped into 1 ml of cold water, and the lysate was layered on a 0.25 m sucrose/TKM buffer (50 mm Tris, pH 7.4, 25 mm KCl, 5 mm MgCl2) as described previously (6).
Precursor-Product Experiments and Quantitation of Labeled PL
The following radiolabeled precursors were used: [1,2-14C]ethanolamine hydrochloride (E-2388; Sigma) (55 mCi/mmol); l-[G-3H]serine (NET248; PerkinElmer Life Sciences) (24.6 Ci/mmol); and [1-14C]dimethylethanolamine (ARC1626; American Radiolabeled Chemicals) (51 mCi/mmol). For 56-cm2 dishes, 5 μCi of 14C label (10 μCi for 3H label) were used. For each well of a 6-well tray, 1 μCi of 14C label (2 μCi of 3H label) were used. Cells were incubated at 37 °C with radiolabeled precursor for 24 h, washed with phosphate-buffered saline, and incubated at 37 °C for an additional 12 or 24 h. Cells cultured on 56-cm2 dishes were scraped into 1 ml of distilled water, and three dishes were pooled for isolation of lipid droplets. Lipids from cells cultured on 6- or 24-well plates were directly extracted using hexane/isopropyl alcohol (see below). For quantitation of incorporation of radiolabel, the corresponding lipids were scraped from thin layer plates (see below), transferred into scintillation mixture, and radiolabel incorporation was measured (Tri-Carb 2700 TR, Packard Instrument Co.).
Lipid Extraction and TLC Separation of Phospholipids
Lipids from cells on 6-well trays were extracted using two consecutive extractions (30 min each) with 0.5 ml of hexane/isopropyl alcohol (3:2; v/v). Lipids from LD or pellet membrane fractions were extracted as described previously (28). The amount of TG was measured with the Triglyceride FS* kit (DiaSys, Germany), and PC was quantified with Phospholipids FS* kit (DiaSys, Germany). For quantification of phospholipids of OP9 cells, the lipid fractions were dried, solubilized in chloroform/methanol (2:1; v/v), and spotted onto Silica Gel 60 plastic sheets (20 × 20 cm) (Merck). Lipid classes were separated with methyl acetate/isopropyl alcohol/chloroform/methanol/potassium chloride (0.25% in H2O) (25:25:25:10:9; v/v) as mobile phase. TLC plates were sprinkled with a primuline (206865, Sigma) aerosol spray (28), and lipids were visualized with a UV lamp (366 nm) and scraped for quantification by gas chromatography.
Gas Chromatographic Quantitation of Phospholipids
Fatty acid composition of individual PL of OP9 cells was performed by gas chromatography. Bands corresponding to individual PL were scraped from thin layer chromatography plates under argon and subjected to direct, in situ trans-esterification with BF3/MeOH in the presence of the thin layer adsorbent (29). Pentadecanoic acid (10 μg in toluene) was added as internal standard, and trans-esterification was performed in the presence of 1 ml of toluene and 1 ml of BF3/MeOH (20%) at 110 °C for 60 min. Excess BF3 was eliminated by the addition of 2 ml of water, and fatty acid methyl esters were extracted into 200 μl of hexane. Two μl of organic phase were analyzed by gas-liquid chromatography on a CP-FFAP CB column (25 m, 0.32 mm inner diameter) with a Hewlett Packard (HP) 5890 or Thermo Scientific Trace GC ultra gas chromatograph equipped with a flame ionization detector and a split/splitless injector. Helium was used as carrier gas; the split ratio was ∼10:1. The initial temperature (150 °C) increased to 215 °C at 2.5 °C/min with a hold at 215 °C for 10 min, and the temperature was then increased to 230 °C at 10 °C/min with a hold at this temperature for 12.5 min. Detector temperature was 300 °C, and the injector temperature was 300 °C. Concentrations of fatty acids were calculated by comparison of peak areas with internal standard (30).
HPLC and Mass Spectrometry
Dried lipid extracts from 3T3-L1 cells were resuspended in 100 μl of CHCl3/MeOH (1:1, v/v) containing PC 12:0/13:0, PC 17:0/20:4, PC 17:0/14:1, PE 12:0/13:0, PE 17:0/20:4, PE 17:0/14:1, phosphatidylinositol 12:0/13:0, phosphatidylinositol 17:0/20:4, phosphatidylinositol 21:0/22:6, PS 21:0/22:6, PS 17:0/20:4, and PS 17:0/14:1 at 100 pmol each as internal standards. Chromatographic separation of lipids was performed by an Accela HPLC (Thermo Scientific) on a Thermo Hypersil GOLD C18, 100 × 1 mm, 1.9-μm column. Solvent A was an aqueous solution of 1% ammonium acetate and 0.1% formic acid, and solvent B was acetonitrile/2-propanol (5:2, v/v) supplemented with 1% ammonium acetate and 0.1% formic acid, respectively. The gradient was run from 35 to 70% solvent B for 4 min and then to 100% B for an additional 16 min with a subsequent hold at 100% for 10 min. The flow rate was 250 μl/min. PL species were determined by a TSQ Quantum Ultra (Thermo Scientific) triple quadrupole instrument in positive and negative ESI mode. The spray voltage was set to 2000 V (−5000 V) and capillary voltage to 35 V (−35 V). PC, PE, and PS species were detected in positive ionization by precursor ion scan on m/z 184 at 35 eV, neutral loss scan at Mr 141 at 25 eV, and neutral loss scan on Mr 185 at 25 eV, respectively, as described previously (31). Molar values were converted to PL mass values using 1-palmitoyl-2-oleoyl-sn-2-glycero-3-phosphocholine (Mr 761.0 for PC), 1-palmitoyl-2-oleoyl-sn-2-glycero-3-phosphoethanolamine (Mr 718.0 for PE), and 1-palmitoyl-2-oleoyl-sn-2-glycero-3-phospho-l-serine (Mr 784.0 for PS) as conversion factors.
RNA Silencing
siRNA (Qiagen, Germany) for PEMT (NM 008819) and control (AllStars Negative Control, Qiagen, Germany) were transfected to 3T3-L1 adipocytes at time points indicated in the figure legends with the Neon Transfection System (Invitrogen) according to the manufacturer's protocol. In brief, 8 × 104 3T3-L1 cells were transfected with siRNA (0.25 μm) with the following protocol: pulse voltage 1,400 V; pulse width 30 ms; pulse number 1. Cells were analyzed 72 h after transfection.
RNA Isolation and Analysis
RNA was isolated from tissues using TRIzol (Invitrogen). In brief, tissues (1 g) were homogenized in 1 ml of TRIzol in a TissueLyser for 20 s at 5000 rpm, followed by a phase separation with 0.2 ml of chloroform/ml TRIzol and precipitation with isopropyl alcohol. Total RNA from cultured cells was isolated by an RNeasy kit (Qiagen, Germany). Reverse transcription was carried out using reverse transcriptase (Invitrogen) and 1.5–3 μg of RNA after DNase I treatment with 1 unit of RQ1 (Promega) in 15 μl of reverse transcriptase reaction buffer (15 min at 37 °C). BioThermTM DNA polymerase (genXpress, Austria) was used for the PCR. Amplification of the PCR products was measured during the exponential phase of the reaction. The following primers (sequence 5′ → 3′) were used for cDNA amplification: PEMT fw (CCATTGTGTTCAACCCACTC) and PEMT rev (CTGCCCCAGTACATGGGGTT); PSS1 fw (CTCGACCTCATCCAGCCTTA) and PSS1 rev (GATCTTCCCTGTGGTGGTGT); PSS2 fw (ATTGTGGCCAGTATTTTGGT) and PSS2 rev (CTTCATGCCACAGTAGATGC); GAPDH fw (GAAGGTCGGTGTGAACGGATT TGGC) and GAPDH rev (GGTGGTGCAGGATGCATTGCTGACA); CTβ fw (CAGAAATGCAGCATGGACAAGGACG) and CTβ rev (GGCGCGTCTCTGATGACTTCATCCA); and hPEMT fw (CTACCTGGCCTGCTACTCTC) and hPEMT rev (CTCAATCAGCTCCTCTTGTG).
Quantitative Real Time PCR (qPCR)
RNA isolation and reverse transcription were performed as described above. RT-PCR analysis was performed in 384-well plates in a total volume of 4 μl containing 2 ng of original total RNA using the QuantiFast SYBR Green RT-PCR kit (Qiagen, Germany) and validated QuantiTec primer assays (Qiagen, Germany) according to the manufacturer's instructions for Light Cycler 480 instruments. In brief, after the initial 5-min heat activation step at 95 °C, cycling conditions were as follows: 40 cycles of denaturation at 95 °C for 10 s; combined annealing and extension at 60 °C for 30 s. The PCR efficiency of the target and housekeeping genes was determined by cDNA dilution series prepared from an untreated sample. Results were corrected with the LightCycler relative quantification software (Roche Diagnostics). mRNA levels of PEMT (primer assay QT00109809), PSS1 (primer assay QT00159103), PSS2 (primer assay QT00162540), PSD (primer assay QT00162071), and CTβ (primer assay QT00111433) were normalized to mouse GAPDH (primer assay QT01658692) and expressed as relative ratio (ΔΔCT). All samples were assayed in duplicate, and average values were used for quantification.
PEMT-EGFP
Full-length rat PEMT cDNA (NCBI Reference Sequence NM_013003.1) (32) was cloned in the vector pCI (Promega). PEMT cDNA (87 bp upstream of start codon plus 684-bp coding region, lacking the stop codon) was amplified by PCR using primers introducing EcoRI (5′-CGTCGAATTCAAGCTCTTCGAAGCC-3′) and NcoI (5′-GATGCCATGGCGCTCCTTTTGTGC-3′) sites at the 5′ and 3′ ends, respectively. PCR was performed with a thermal cycler (Mastercycler gradient; Eppendorf) at an annealing temperature of 68 °C (20 s), denaturation at 94 °C (10 s), and elongation at 72 °C (40 s) for 25 cycles. The PEMT fragment was cloned into vector pIRES2-DsRed2 (Clontech) together with an EGFP cDNA fragment. EGFP was excised from vector pEGFP-1 (Clontech) and replaced DsRed2 at NcoI and NotI sites. In the final construct, the fusion site of the peptide was serine (last amino acid of PEMT)-alanine-methionine (first amino acid of EGFP). Transient transfections of 3T3-L1 and OP9 cells, grown on glass coverslips in 6-well plates, using Metafectene (Biontex) were performed according to the manufacturer's protocol. In brief, 3 μg of plasmid-DNA and 5 μl of Metafectene transfection reagent (Biontex, Germany), diluted with 50 μl of serum-free cell medium each, were incubated at room temperature for 20 min. 10–20 μl of the mixture were added directly to the cell culture medium 1 h after a routine medium change. After 6 h of incubation with the plasmid-Metafectene complex, the medium was changed again. Fluorescence microscopy was performed 18–24 h post-transfections.
PEMT and Protein Assays
Protein content was determined with the Coomassie Blue protein protocol from Bio-Rad with bovine serum albumin as a standard or the BCA reagent (Pierce). PEMT assays were performed as described previously (33) using phosphatidylmonomethylethanolamine as a substrate (Avanti Polar Lipids).
Immunoblotting
Epididymal fat pads and liver were sonicated, and indicated amounts of protein were separated by electrophoresis on a 10–12.5% SDS-PAGE. Proteins were transferred to Immobilon PVDF (Millipore) membranes and probed with either rabbit anti-rat PEMT (32), rabbit anti-bovine protein-disulfide isomerase (SPA-890, StressGen), or guinea pig anti-perilipin antibody (PROGEN, Germany). Secondary antibodies were goat anti-rabbit or rabbit anti-guinea pig IgG, conjugated to horseradish peroxidase. Immunoreactive bands were visualized by enhanced chemiluminescence (GE Healthcare) (6).
Fluorescence Microscopy
Using a customized array confocal laser scanning fluorescence microscope, transiently transfected cells were alternatively illuminated at 488 and 514 nm with a 150-milliwatt argon laser (34) to excite PEMT-EGFP and Nile Red, respectively. Emission was detected at 535 nm for PEMT-EGFP and at 570 nm for Nile Red (535/30 and E570LPv2) using Metamorph 5.0 (Universal Imaging, Visitron Systems, Germany). For three-dimensional reconstruction, images were taken every 100 nm with a ×100 objective (α Plan-Fluar ×100/1.45 oil objective from Zeiss Microsystems, Germany) alternately for PEMT-EGFP and Nile Red starting at the bottom of the cell as described previously (34).
Lipolysis Assays and Quantification of Nonesterified Fatty Acids
Wild type and Pemt−/− mice (chow diet) were sacrificed and tissue samples surgically excised from visceral white adipose tissue (WAT) using three visceral fat samples from each of six animals of each genotype. Fat pads (∼2–3 mg) were washed several times with phosphate-buffered saline and incubated in Dulbecco's modified Eagle's medium (Invitrogen) containing 2% defatted BSA (Sigma) (35) either in the presence or absence of 1 μm PIA ((R)-PIA, N6-[l-2-phenylisopropyl]adenosine) (Sigma) or 0.5 units/ml adenosine deaminase (ADA) (Calbiochem) at 37 °C for 180 min. Aliquots of the medium were taken at several time points and analyzed for fatty acid content using commercial kits (NEFA C, Wako Chemicals, Germany). After 180 min, fat pads were transferred into fresh medium containing 1 μm isoproterenol (Sigma) and either 1 μm PIA or 0.5 units of ADA and incubated for another 60 min at 37 °C. Thereafter, aliquots of the medium were removed and analyzed for fatty acid content. Fat pads were delipidated in chloroform/methanol/acetic acid (66:33:1; v/v) for 1 h, and lysed in 0.3 n NaOH containing 0.1% SDS. Protein measurements were performed using the BCA reagent (Pierce). Values were normalized to protein content of the corresponding tissue piece (36). In 3T3-L1 cells, lipolysis was measured 72 h after transfection with siRNA.
Fat Cell Morphometry and Statistical Analyses
Visceral fat pads of Pemt+/+ and Pemt−/− mice (n = 4) that had been fed either a high fat diet or a chow diet were fixed in 4% buffered formaldehyde and embedded in paraffin. Hematoxylin- and eosin-stained sections (4 μm thick) were scanned using ScanScope XT/Aperio ImageScope software (Aperio). At least four different locations of visceral fat were identified using low magnification of the scanned slides. ImageJ software (37) was used to measure cell size at ×400 magnification. At least 500 fat cells were measured manually from each slide in duplicate. The following number of cells was measured: Pemt+/+/chow, 1243; Pemt−/−/chow, 2506; Pemt+/+/high fat, 2624; Pemt−/−/high fat, 2238. Statistical analysis was performed using SPSS software, and a Kruskal-Wallis test was used to compare fat cell size distribution among experimental groups.
Statistical Analyses
Experiments were performed at least three times, and values are expressed as mean ± S.D. Statistical significance was determined by Student's unpaired t test (two-tailed). Group differences were considered significant for p < 0.05 (*); p < 0.01 (**); p < 0.001 (***).
RESULTS
Phospholipid Pattern Changes Profoundly at the Onset of Adipocyte Differentiation
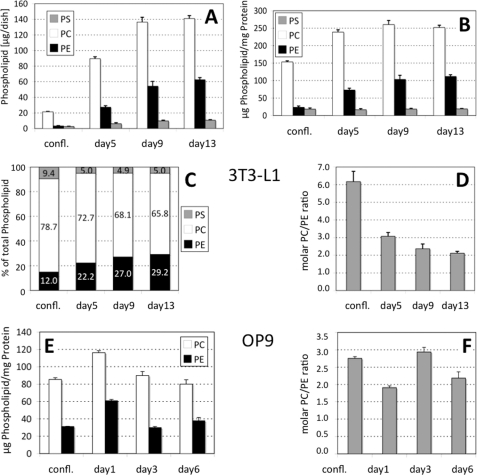
An earlier study (11) demonstrated that among the different PL classes, 32Pi label was preferentially incorporated into PC and PE during differentiation of 3T3-L1 cells. Because the significance of this finding was unclear, we determined whether or not the amounts of PC, PE, and PS change during adipocyte differentiation. We quantified the amounts of these PL in two well established adipogenic cell lines, 3T3-L1 (Fig. 1, A–D) and OP9 (Fig. 1, E and F). In 3T3-L1 cells, the absolute amounts of the three PL, PC, PE, and PS, as well as that of protein progressively increased over 2 weeks of differentiation (Fig. 1A). The most pronounced change in PL profile was observed between day 0 (confluency) and day 5. Within this time frame, PC increased from 21.4 ± 0.5 to 89.5 ± 2.4 (p < 0.001), PE from 3.3 ± 0.5 to 27.3 (p < 0.001), and PS from 2.6 to 6.2 μg/dish (p < 0.01). When normalized on the cell protein content, no significant changes were observed in the PS fraction (18.3 ± 3.2 μg/mg protein at confluency and 19.1 ± 1.3 at day 13) (Fig. 1B). In contrast, the normalized PC content increased from 153.3 ± 3.4 μg/mg protein at confluency to 238.7 ± 6.5 at day 5, and the raise in the normalized PE value at onset of differentiation was even more pronounced (23.3 ± 3.6 versus 72.9 ± 5.2 μg/mg protein). Virtually no changes in the normalized PC values were observed between day 5 and day 13, although that of PE still increased to reach a plateau at day 9 (at 103.4 ± 11.7 versus 111.7 ± 5.0 at day 13). In the time frame between confluency and day 5, the relative proportion of PE (Fig. 1C) increased from 12.0 (day 0) to 22.2% of the total cellular PL mass (day 5), although that of PC decreased from 78.7 to 72.7% and that of PS from 9.4 to 5.0%. In later stages of differentiation (days 9 and 13), the relative PS proportion remained essentially unchanged, although that of PC slightly decreased to 68.1 (day 9) and 65.8% (day 13). Remarkably, these changes in PL abundance at onset of adipocyte differentiation caused a profound reduction of the PC/PE molar ratio (Fig. 1D) from 6.18 ± 0.58 (confluent cells) to 3.08 ± 0.21 at day 5. This PC/PE ratio further declined via 2.37 ± 0.27 at day 9 to 2.12 ± 0.10 at day 13. In OP9 cells, the PC/PE ratio (2.75 ± 0.05 at confluency) was lower than in 3T3-L1 cells, but it also declined at onset of differentiation (day 1) (1.91 ± 0.05) (Fig. 1F) to increase again at day 3 (2.93 ± 0.13). At day 6, the PC/PE ratio decreased to 2.18 ± 0.18. It is important to note that the time courses of both cell lines for TG accumulation are not identical, because OP9 cells (27) accumulate TG ∼3 times faster than 3T3-L1 cells. Together, these findings indicated an unexpected and pronounced increase in PE in absolute and relative terms, likely at the expense of PC and PS, which coincides with the onset of adipocyte differentiation.
FIGURE 1.
Phospholipid composition changes profoundly at onset of 3T3-L1 and OP9 cell differentiation. A, individual PL from 3T3-L1 (A–D) and OP9 cells (E and F) are given as micrograms/dish; B and E, phospholipid values are normalized to cell protein content; C, individual PL are given as % of total PL; D and F, molar PC/PE ratios are given. At the indicated times, nondifferentiated 3T3-L1 cells were harvested, and lipids were extracted. Quantitation of PL was performed by either mass spectrometry (A–D) or gas chromatography (E and F). Values are derived from three independent experiments and represent means ± S.D. in A, B, D, E, and F or mean values in C. confl., confluent preadipocytes; days 5, 9, or 13 (A–D, 3T3-L1) and days 1, 3, or 6 (E and F, OP9) indicate the days after start of adipocyte differentiation.
Serine but Not Ethanolamine Phospholipids Is Remodeled during LD Formation
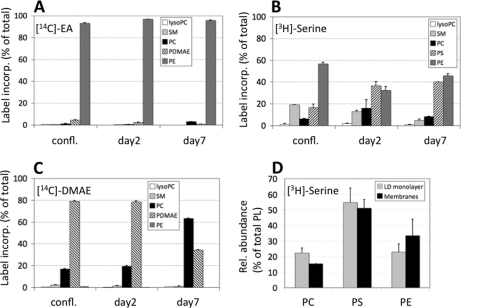
In mammalian cells, PE can be provided by two independent pathways as follows: (i) the CDP-ethanolamine (Kennedy) pathway (18), and (ii) the mitochondrial decarboxylation of PS (38, 39). To investigate if one of these biosynthetic pathways was responsible for the changes in PE content, adipocytes were incubated at different stages of differentiation with radiolabeled ethanolamine (marker of CDP-ethanolamine pathway) or serine (marker of PS decarboxylation). When 3T3-L1 cells were incubated with [14C]ethanolamine, >90% of incorporated label was recovered in PE fractions in differentiated as well as undifferentiated cells (Fig. 2A). Only traces of [14C]PE were further metabolized during differentiation suggesting a high rate of synthesis of PE but limited access to PE-modifying enzymes.
FIGURE 2.
PL remodeling processes utilize PS decarboxylation during LD formation. 3T3-L1 cells were incubated with [14C]ethanolamine (A), [3H]serine (B and D), or [14C]dimethylethanolamine (C) for 24 h. Lipids were extracted from the cells, and incorporation of radiolabel was quantified. confl., confluent preadipocytes; days 2 and 7 indicate the days after start of adipocyte differentiation. Values are means ± S.D. of three independent experiments and represent radiolabel incorporated into PL as % of total cell-associated label. SM, sphingomyelin; PDMAE, phosphatidyldimethylethanolamine. D, 3T3-L1 cells (day 6) were incubated with [3H]serine for 12 h at 37 °C. Thereafter, cells were lysed, and LD (LD monolayer) were separated from pelleting membranes (Membranes) by ultracentrifugation. Processing of lipid samples was as in A–C.
In contrast, extensive PL remodeling occurred when [3H]serine was used as radiolabeled precursor (Fig. 2B). Incorporation of [3H]serine into PS increased between confluency (16.7 ± 2.9%) and day 2 (immature adipocytes) (36.6 ± 3.8%) (p < 0.005) and remained high at day 7 (40.1 ± 0.2%) (confluence to day 7, p < 0.05). Concomitantly, the proportion of radioactivity in PE decreased from 56.7 ± 1.5% (confluence) to 32.4 ± 3.3% at day 2 (p < 0.01). At day 7, PE was at 46.0 ± 1.7% (days 2–7, p < 0.05). Notably, label incorporation into PC increased from 6.2 ± 0.5% (confluence) to 16.0 ± 8.0% (day 2) and declined at day 7 (8.2 ± 0.4%) (days 2–7, p < 0.05). Incorporation of [3H]serine into sphingomyelin decreased gradually from confluency (19.2 ± 0.0%) to 5.6 ± 0.9% at day 7 (confluence to day 2, p < 0.005, days 2–7, p < 0.05; confluence to day 7, p < 0.001). Incorporation of either of the radiolabels into lyso-PC was low at all stages of adipocyte differentiation. Together, these findings indicate an augmented requirement of serine-derived PE during onset of fat cell differentiation (day 2) and subsequent (day 7) metabolism of PE.
PEMT converts PE to PC in mammals. In liver, about one-third of PC is produced by this reaction (18). Because expression of PEMT has recently been described in differentiating 3T3-L1 adipocytes (40), this pathway was assessed during adipocyte differentiation by measuring the incorporation of [14C]dimethylethanolamine into phosphatidyldimethylethanolamine and PC (Fig. 2C). Whereas only 16.8 ± 0.5% of label was in PC of confluent preadipocytes and in immature adipocytes (day 2) (19.6 ± 0.5%), an ∼3-fold higher proportion of label (63.5 ± 0.1%) was in PC of day 7 adipocytes indicating that direct methylation of phosphatidyldimethylethanolamine to PC had occurred. Considered together, these data imply that PEMT methylates preferentially PS-derived PE rather than PE produced directly via the CDP-ethanolamine pathway.
LD Monolayer, Compared with Cellular Membranes, Contains PC Preferentially Derived via PS
Considering that PEMT may be associated with LD or localized in their close proximity, we next determined if PC, derived via PS and PE, was enriched in LD compared with cell membranes (Fig. 2D). 3T3-L1 cells were incubated with [3H]serine for 24 h at day 6, and LD were isolated. At this stage, LD are already microscopically visible. The majority of label was present in both LD-localized (54.8 ± 9.2) and membrane-localized PS (51.1 ± 5.6%). Incorporation of [3H]serine into sphingomyelin and lyso-PC was <5% of total incorporation and was not significantly different between LD and membranes (data not shown). In LD, the proportion of incorporated [3H]serine into PC was significantly higher (22.3 ± 3.2%) than in the membrane fraction (15.4 ± 0.0%) (p < 0.01), indicating that [3H]serine-derived PC was preferentially incorporated into LD compared with cell membranes. Taken together, these findings suggest that PE methylation occurs in proximity to sites of LD formation in adipocytes and allows potential interaction with PS-derived PE.
High Fat Challenge Induces PEMT Expression in Adipose Tissue
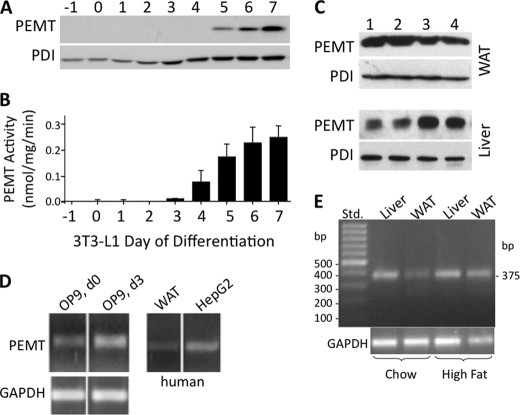
Recent results have demonstrated that PEMT expression in 3T3-L1 adipocytes gradually increases in a time-dependent manner during adipocyte differentiation, starting at day 5 and reaching a maximum at day 7 (40). Consistent with these findings, Fig. 3, A and B, demonstrates that PEMT protein and activity are undetectable until approximately day 4 and reach a plateau in mature adipocytes (day 6/7). To investigate whether this finding from cultured cells reflects the in vivo situation, we determined if PEMT is expressed in WAT of mature control mice. Immunoblotting experiments (32) demonstrated that PEMT protein is expressed in mouse WAT (Fig. 3C) at ∼20% of the level in mouse liver.
FIGURE 3.
Expression of PEMT mRNA, protein, and activity in mouse and human fat cells. Proteins from 3T3-L1 adipocytes (A), WAT from four 6-month-old mice (C, lanes 1–4, top) (50 μg each), or liver (10 μg) (C, lanes 1–4, bottom) were immunoblotted with anti-PEMT or protein-disulfide isomerase (PDI) antibodies (A, bottom) in three separate experiments; one representative experiment is shown in A and C. B, PEMT activity (nmol/mg protein/min) was quantitated in the same samples as in A. Data are from three independent experiments and represent means ± S.D. D, PEMT mRNA expression was assessed relative to glyceraldehyde-3-P dehydrogenase (GAPDH) in murine OP9 preadipocytes, human HepG2 hepatoma cells, and WAT. The OP9 cells were harvested prior to (d 0), and 3 days after onset of differentiation (d3). E, samples of liver and WAT from 6-month-old mice fed chow or high fat diet (3 weeks) were analyzed by RT-PCR for the amount of PEMT mRNA relative to GAPDH mRNA. Size of the corresponding PEMT cDNA product was 375 bp. Std, 100-bp DNA standard ladder.
Expression of PEMT was further examined in another murine adipogenic cell line, OP9, as well as in human WAT. PEMT mRNA expression increased in OP9 cells at day 3 of adipocyte differentiation (Fig. 3D) at which time OP9 cells have differentiated into mature adipocytes with similar TG accumulation as in 3T3-L1 adipocytes at day 9. Notably, a faint amplicon representing PEMT expression was also observed in human WAT. Because the PEMT expression profile in differentiating adipocytes indicated an induction of enzyme expression during TG accumulation, we tested this possibility in vivo in mice fed a high fat diet for 3 weeks. Expression of PEMT mRNA was substantially increased in WAT under these conditions (Fig. 3E). The correlation of intracellular TG deposition with PEMT expression raised the possibility that PEMT is directly or indirectly involved in LD biogenesis.
PEMT-EGFP Localizes with ER and the Periphery of LD
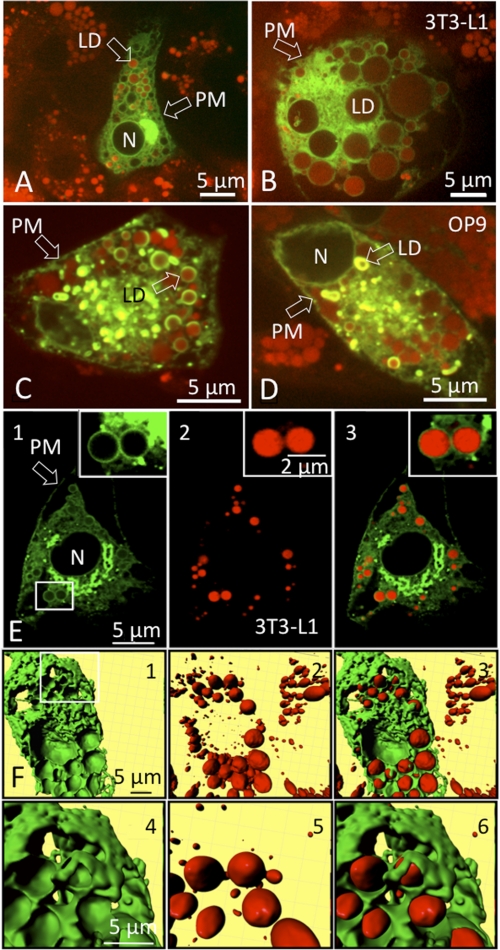
Because our results suggested that PE and its modifying enzyme, PEMT, might have a functional role during the biogenesis of intracellular LD, we studied the intracellular distribution of PEMT in differentiating adipocytes. An expression plasmid was constructed that contained PEMT cDNA fused to EGFP to allow visualization of the protein by fluorescence microscopy. 3T3-L1 cells were transfected with plasmid at day 7 (Fig. 4, A and B) and OP9 cells at day 2 of differentiation (Fig. 4, C and D). LD were visualized by Nile Red staining 48 h after transfection. Weak PEMT-EGFP staining was detected in the plasma membranes of transfected cells. However, most PEMT-EGFP fluorescence localized to intracellular membranes that most likely represented ER and MAM regions (41). Remarkably, PEMT-EGFP fluorescence was abundant at the periphery of LD of 3T3-L1 adipocytes (Fig. 4, A, B, and E) and OP9 cells (Fig. 4, C and D). A three-dimensional reconstruction of 3T3-L1 adipocytes was generated from horizontal sections obtained from EGFP (Fig. 4F, panels 1 and 4) and Nile Red fluorescence (Fig. 4F, panels 2 and 5). The three-dimensional merged pictures (Fig. 4F, panels 3 and 6) clearly demonstrated that LD are wrapped by regions of PEMT-EGFP fluorescence, presumably representing ER/MAM sites. Thus, our data indicate that fat cell LD are surrounded by ER/MAM, the previously characterized sites of PEMT expression in hepatocytes (18, 32). Therefore, physical interactions of the monolayer PL of LD with the ER/MAM are feasible, most likely via specific membrane contact sites (42, 43).
FIGURE 4.
PEMT-EGFP localizes to ER and the periphery of LD. 3T3-L1 cells at day 7 of differentiation (A, B, E, and F) and OP9 cells at day 3 of differentiation (C and D) expressing PEMT-EGFP. Images were taken 24 h after transfection using an array confocal laser scanning fluorescence microscope. Intracellular LD were stained with Nile Red prior to image acquisition. Excitation at 488 and 514 nm and emission at 535 nm was recorded for PEMT-EGFP and is depicted in green, and 570 nm for Nile Red is shown in red, respectively. The highlighted region in E, panel 1, was further magnified and shows PEMT-EGFP staining (inset in E, panels 1 and 3) on the periphery of Nile Red-stained LD (inset in E, panels 2 and 3). F, three-dimensional view of PEMT-EGFP (F, panels 1 and 4, zoom; highlighted region of F, panel 1) and Nile Red-stained LD (F, panels 2 and 5/zoom) and the respective overlay (F, panels 3 and 6, zoom) in 3T3-L1 adipocytes. Transfection with EGFP (control) resulted in unspecific cytosolic staining (data not shown). N, nucleus; PM, plasma membrane.
Absence of PEMT Activity Increases Basal Lipolysis in 3T3-L1 Adipocytes
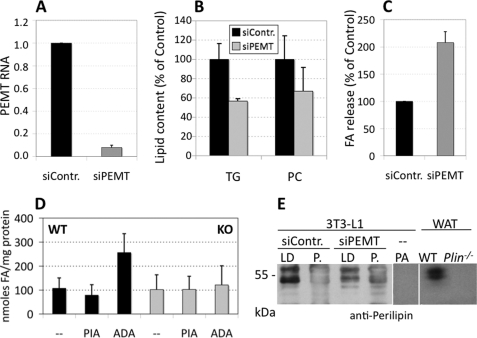
To investigate the biological significance of PEMT activity for PL synthesis in adipocytes, we investigated the stability of LD when PEMT expression and activity (data not shown) were attenuated. 3T3-L1 cells (day 8 of differentiation) were transfected with PEMT siRNA or control siRNA via electroporation. qPCR analysis demonstrated that PEMT expression was reduced by ∼90% (p < 0.001) in 3T3-L1 adipocytes transfected with PEMT siRNA compared with control siRNA (Fig. 5A). PEMT siRNA was without effect on expression of PS decarboxylase (data not shown). Strikingly, the level of TG in 3T3-L1 adipocytes was significantly diminished by ∼33% (p < 0.05) by attenuation of PEMT expression (Fig. 5B). The PC content was slightly, but not significantly, reduced. Concomitantly, PEMT RNAi caused a 2-fold higher release of fatty acids from adipocytes (p < 0.01), indicative of increased basal (nonstimulated) lipolysis (Fig. 5C). To test whether PEMT deficiency causes a comparable phenotype in vivo, we performed a similar experiment with WAT from Pemt−/− mice and Pemt+/+ littermates. Under nonstimulated conditions, release of fatty acids from fat pads after 180 min was similar in WT and Pemt−/− animals (Fig. 5D). Notably, addition of PIA, an adenosine A1 receptor agonist (44), significantly (p < 0.05) confined fatty acid release from WAT in WT but not in Pemt−/− mice. However, whereas reduction of adenosine concentrations by ADA caused an ∼3-fold increase in lipolysis in fat pads of WT animals (in the absence of isoproterenol), no response was seen in Pemt−/− mice. The difference in fatty acid release between the two genotypes under unstimulated/ADA conditions was statistically highly significant (p < 0.001). When fat pads were transferred to media where lipolysis was stimulated (each containing isoproterenol ± PIA or ADA), no significant differences were observed between both genotypes (WT/−, 616 ± 193; WT/PIA, 584 ± 164; WT/ADA, 699 ± 217; KO/−, 556 ± 248; KO/PIA, 565 ± 198; KO/ADA, 621 ± 162 nmol of fatty acids/mg of protein). Taken together, our data indicate that PEMT deficiency enhances basal lipolytic activity (in the presence of PIA) in mouse WAT moderately and in cultured 3T3-L1 adipocytes by ∼2-fold, suggesting a role for PEMT in stabilizing adipocyte LD. We reasoned that the association of the LD monolayer with perilipin A, the major LD protein constituent that regulates lipolysis in adipocytes, might be impaired under PEMT knockdown conditions (Fig. 5E). Immunoblotting demonstrated that the majority of the typical perilipin A doublet band of ∼ 60 kilodaltons (45) was detectable in the LD fraction of control cells (siContr.), with only minor staining in the pellet fraction. In cells with attenuated PEMT expression (siPEMT), perilipin A staining was reduced in the floating LD fraction.
FIGURE 5.
Absence or attenuation of PEMT activity increases basal lipolysis in cultured adipocytes and in vivo. 3T3-L1 adipocytes (A–C and E) (day 5 or 6 of differentiation) were transfected with either siRNA specific for PEMT (siPEMT) or control siRNA (siContr.) as indicated under “Experimental Procedures.” Cells were analyzed 72 h later. A, PEMT cDNA level was normalized to GAPDH, with control siRNA set at 1.0 (n = 3). Cell TG and PC after RNA silencing is expressed as % of control (siContr.) (n = 3) (B). C, quantitation of release of unesterified fatty acids (FA) from siPEMT cells into cell medium compared with cells transfected with control siRNA set at 100% (n = 3). D, fatty acid release from mouse visceral adipose tissue under nonstimulated lipolysis conditions. Lipolysis assay using fat pads of wild type (WT) and Pemt−/− (KO) mice was performed as indicated under “Experimental Procedures” for 180 min without further compounds (−) or in the presence of PIA or ADA, and fatty acids released into the medium were quantitated. Data are derived from six WT and six Pemt−/− mice (KO); three WAT fat pads from each animal were used (n = 18). Values are means ± S.D. WT/− versus WT/PIA, p < 0.05; KO/− versus KO/PIA; n.s., not significant. WT/ADA versus KO/ADA, p < 0.001. E, perilipin A immunoblot. 3T3-L1 cells were transfected with PEMT siRNA or control RNA at day 8 and harvested at day 11, and lipid droplets were isolated. 100 μg of proteins per lane were resolved on a 10% SDS-PAGE, and proteins were transferred to Immobilon PVDF membranes and immunoblotted with perilipin antibody. Immunoreactive bands were visualized by enhanced chemiluminescence. Protein extracts from preadipocytes (PA) and from WAT of wild type (WT) and Perilipin−/− (Plin) animals were loaded as positive and negative controls, respectively. P., cell pellet fraction.
PEMT-deficient Mice Are Resistant to High Fat-induced Fat Cell Hypertrophy
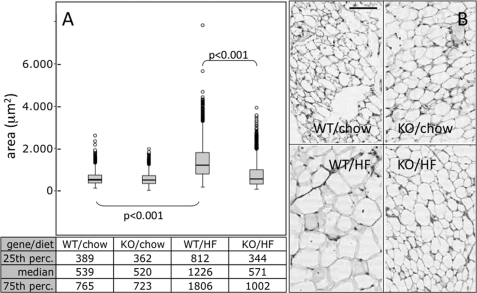
Physiological effects of enhanced basal lipolysis on morphology of WAT were investigated in Pemt−/− and control mice kept on a high fat diet or regular chow for 3 weeks. Visceral adipose tissue was collected and stained with hematoxylin-eosin. Fat cell size morphometry was performed manually on >8,000 fat cells using pictures from different sections of adipose tissue from Pemt−/− and Pemt+/+ mice (Fig. 6A). In response to high fat feeding, fat cell size increased (hypertrophy) in WT animals (Fig. 6, A and B, bottom left) (539 versus 1226 μm2; chow versus high fat diet) (p < 0.001), although it was essentially unaffected in Pemt−/− mice (520 versus 571 μm2; chow versus high fat diet). Fat cell sizes within the 25th percentile were in a similar range (340–390 μm2) for all experimental conditions except for adipose tissue from Pemt+/+mice fed the high fat diet (∼812 μm2). In the 75th percentile, fat cells from both Pemt−/− and Pemt+/+ mice fed the high fat diet (∼1000 μm2) were significantly larger than from mice of both genotypes fed the chow diet (∼750 μm2). The difference in fat cell size between Pemt−/− and Pemt+/+ mice fed the high fat diet was also statistically significant (p < 0.001). Thus, PEMT-deficient mice are resistant to high fat diet-induced hypertrophy of adipocytes in WAT. A similar protection against obesity was observed in Pemt−/− mice fed a high fat diet for 10 weeks (46).
FIGURE 6.
PEMT-KO-mice are resistant to high fat-induced fat cell hypertrophy. A, analysis of fat cell size in visceral fat from Pemt−/− mice (KO) and Pemt+/+ (WT) mice fed a high fat diet (HF) for 3 weeks compared with visceral fat from mice fed a normal chow diet (four mice per subgroup) was performed as described under “Experimental Procedures.” The Kruskal-Wallis test was used to compare fat cell size distribution among experimental groups. Results are presented as box plots with values for medians; the 25th and 75th percentiles (perc.) are indicated. B, representative pictures of adipose tissue sections from Pemt−/− mice (KO) and Pemt+/+ (WT) mice fed chow or the high fat (HF) diet. Bar, 25 μm.
Regulatory Loop between PE-synthesizing and PE-converting Enzymes
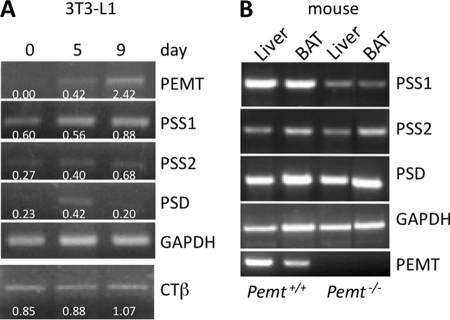
Because LD stability was reduced in both the white adipose tissue of Pemt−/− mice and in 3T3-L1 adipocytes with attenuated PEMT expression, we investigated a potential cross-talk between PE-converting (i.e. PEMT-mediated) and PE-producing (i.e. PS synthase1- and PS decarboxylase-mediated) processes. First, expressions of PEMT, PS synthase-1, PS synthase-2, and PS decarboxylase were determined during differentiation of 3T3-L1 preadipocytes to adipocytes (Fig. 7A). In line with a recent report (40), PEMT mRNA levels increased markedly between days 5 and 9 of 3T3-L1 differentiation, whereas the signal was virtually absent from undifferentiated cells (day 0). Expression of PS synthase-1 mRNA, which produces PS by exchange of choline in PC for serine (47), was unchanged at day 5 and was moderately higher at day 9. The mRNA encoding PS synthase-2, which produces PS by exchange of ethanolamine of PE for serine (48), was increased at day 9. Notably, however, PS decarboxylase mRNA increased at day 5. The expression of the mRNA encoding CTP:phosphocholine cytidylyltransferase β (CTβ), an enzyme of the CDP-choline pathway, was moderately increased during 3T3-L1 differentiation until day 9.
FIGURE 7.
Regulatory loop between PE-synthesizing and -converting enzymes. RNA was isolated from 3T3-L1 cells at days 0, 5, and 9 (A) and from mouse liver and BAT (B) and analyzed by RT-PCR and additionally by qPCR. Values derived from qPCR represent means ± S.D. (n = 4). Numbers below the amplicons indicate ΔΔCT values. B, lanes 1 and 2, Pemt+/+; lanes 3 and 4, Pemt−/− mice (all mice were 5 days old). PSS1, phosphatidylserine synthase-1; PSS2, phosphatidylserine synthase-2; CTβ, CTP-phosphocholine cytidylyltransferase β.
Given the critical role that PEMT appears to play in LD stability, we assessed mRNA levels of these PS-metabolizing enzymes in liver and adipose tissue of 5-day-old Pemt−/− and Pemt+/+ mice (before weaning the mice consume high fat milk) (Fig. 7B). Because WAT is virtually absent in pups of this age, brown adipose tissue (BAT) was examined. The level of PS decarboxylase mRNA was similar in Pemt+/+ and Pemt−/− mice (1.2-fold higher level in liver of Pemt−/− mice compared with Pemt+/+ mice, and 0.8-fold lower level in BAT). However, PS synthase-1 mRNA expression was significantly reduced by PEMT deficiency in both the liver (50%) and BAT (55%). PS synthase-2 mRNA levels in the liver were only slightly (20%) lower in Pemt−/− mice compared with Pemt+/+ mice but were 55% lower in BAT. Together, these results suggest a regulatory loop between PE-consuming and -synthesizing processes during LD formation in adipose tissue.
DISCUSSION
Fat cells constitute the major cell type of adipose tissue and represent under physiological conditions 10–20% of total body mass in human. LD contain both surface proteins and lipids. The best characterized proteins of the LD shell are perilipin, adipose differentiation-related protein, and tail-interacting protein of 47 kilodaltons (TIP47), the so-called PAT family members (7, 49–51).
Comparatively little is known about the nature of the LD monolayer, and only sparse information is available regarding the biological significance of specific monolayer PL subspecies. PE is the second most abundant PL in the LD fraction of adipocytes (12). PE is a conical lipid and is therefore compatible with induction of membrane curvatures (16). In mammals, the conversion of PE to PC is catalyzed by PEMT (18). Previous studies have shown that PEMT-derived PC represent quite distinct molecular PC subspecies (52); they contain preferentially polyunsaturated fatty acids in the sn-2-position. Recently, PEMT expression in differentiated adipocytes has been demonstrated (40).
To define, whether changes in the PL pattern of aminophospholipid occur during the differentiation process of adipocyte cell lines, we quantitated the masses of glycero-PL of fat cells at different stages of differentiation. A highly significant and profound decrease in the PC/PE ratios at the start of differentiation not only of 3T3-L1 cells, but also of OP9 cells, indicated constitutive changes of PC, PE, and PS. The early phase of differentiation of 3T3-L1 adipocytes was also characterized by a relative decrease in PC, whereas the total cellular PE content increased in absolute and relative terms, indicative of very active PL remodeling events during this phase. Although we observed in 3T3-L1 cells a gradual decrease in the PC/PE ratio during the course of differentiation, an intermediate peak at day 3 emerged in OP9 cells, indicating that the differentiation modes of the two cell line are not identical. To our knowledge, such a shift in the overall cellular PL pattern as a function of the metabolic state of mammalian cells has no precedent in the current literature.
Aiming to explore the pathways responsible to meet this dramatic demand for PE at the start of adipogenesis, we used radiolabeled headgroup precursors to trace their incorporation into the corresponding PL. Our results demonstrate that an ethanolamine precursor was readily incorporated into PE, but not further converted into PC. The low ability of fat cells to process significant amounts of labeled ethanolamine indicates metabolic inaccessibility of PE, produced via the Kennedy pathway. Hence, the decarboxylation of PS in mitochondrial membranes (53) might play a role for providing PE during the onset of LD biogenesis. Data from this study support this hypothesis. Using a serine precursor for PL synthesis in 3T3-L1 cells, the label was detected in PS, PE, PC, and sphingomyelin. Labeled PC (produced via PS and PE) increased from day 0 to day 2, most likely at the expense of PE, the amount of which decreased during this period. Label incorporation into PS increased ∼2-fold from day 0 to day 7. At day 7, the amount of labeled PC was only half that of day 2, indicating that conversion of newly produced (serine-derived) PE was attenuated, presumably because of competition with the intracellular pool of unlabeled PE at this stage of differentiation (see Fig. 1). This might explain why intracellular levels of labeled PE (derived from PS) also remained high at day 7. Although the possibility exists that serine incorporation into PL reflects headgroup as well as serine-derived fatty acids, addition of this precursor during onset of LD growth in 3T3 adipocytes demonstrated a broad spectrum of PL interconversions.
The combined view of our data with the [14C]serine precursor indicated the following: (i) PE was produced from PS by the mitochondrial PSD reaction; (ii) the conversion of PE to PC via PEMT occurred subsequently, and (iii) this methylation process is essentially restricted to PE produced by the PSD pathway in mitochondria. The serine-derived PL pattern indicated that production of PC from PS-derived PE was significantly higher in the LD monolayer than in the cell pellet fraction.
Our finding that PEMT activity emerges around day 4/5 in 3T3-L1 adipocytes is in agreement with recent data on expression of PEMT in these cells (40). We have further established that an increase in PEMT mRNA expression during differentiation was not only observed in 3T3-L1 (40) but also in murine OP9 adipocytes. In addition, the PEMT mRNA signal was also visible in tissue samples from mouse and human WAT, which indicates that PEMT is expressed in differentiating/differentiated fat cells. That PEMT expression increases in WAT samples of mice on a high fat diet in comparison with those on a chow diet is striking and points toward a specific requirement of PEMT during LD growth and processing.
PEMT is an integral membrane protein localized in the PL bilayer of ER or MAM (54, 55) and represents a marker protein for MAM (32). This topology makes it unlikely to integrate into monolayered structures. However, mechanisms of how such proteins may become otherwise integrated into either a hairpin-monolayer shell during LD birth (56) or in egg cup-like structures (57) have been proposed recently. Remarkably, previous studies have demonstrated that PEMT protein was abundant in a LD fraction isolated from rat liver cells, indicating strong association of ER/MAM preparations with LD (58). It is important to note that also caveolin, a membrane protein, has previously been identified as a LD-associated protein (3, 59). Caveolin−/− adipocytes form only small LD, and the LD monolayer contains less PS and lyso-PL (60).
Our experiments using PEMT-EGFP fluorescence microscopy and three-dimensional reconstruction clearly visualized abundance of PEMT either directly on, or in close vicinity to LD, and at sites representing ER/MAM regions. Taken together, our data indicated the presence of PEMT in intracellular membranes, presumably at membrane contact sites (43, 61) closely adjoining or wrapping the LD monolayer as proposed previously (3). The combined data therefore allow the conclusion that PEMT is directly involved in the biogenesis of adipocyte LD.
Because of the presumed role of PEMT for LD growth and processing, we investigated potential effects on LD stability, when PEMT activity was attenuated in 3T3-L1 adipocytes or completely absent from fat cells of PEMT-KO mice. In differentiated adipocytes, in which PEMT expression was down-regulated by ∼90%, the TG value decreased significantly by about one-third, and the overall PC content dropped moderately. This is most likely due to (i) defective synthesis of PC from PE via methylation and (ii) consumption of residual PC by the PSS1 reaction to generate PS. Therefore, attenuation of PE conversion had significant impact on LD processing.
Adipocytes express adenosine A1 receptors (62) that effectively suppress adenylate cyclase, thereby inhibiting catecholamine-mediated triglyceride hydrolysis (63). Previous results suggested that adenosine-receptor agonists (such as PIA) effectively reduce the levels of both plasma triglycerides and unesterified fatty acids when infused into rats (63). Our results emphasize that 3T3-L1 adipocytes with a down-regulated PEMT gene exhibit considerably increased basal lipolysis, and fat pads from Pemt−/− mice show a moderate increase in basal lipolysis in comparison with those of WT animals. Intriguingly, relief of the adenosine-mediated lipolysis blockade was significantly attenuated in fat pads of Pemt−/− in comparison with WT mice, when adenosine was removed by ADA (64). One possible interpretation is that PEMT knock-out animals developed desensitization (tolerance) against adenosine signaling, and therefore relief of the adenosine-mediated lipolysis blockade by adenosine-degrading compounds (63) was ineffective. However, characterization of details of this mechanism is ongoing and beyond the scope of this study.
Previous studies have demonstrated that perilipin A restricts the access of intracellular lipases to the LD, thereby suppressing the rate of basal lipolysis under adipogenic conditions (65). Our finding of increased basal lipolysis under PEMT knockdown conditions, which correlates with decreased perilipin A association with LD, is in line with reports on the phenotype of perilipin null mice (66, 67). These animals have greatly diminished WAT stores and show constitutive basal lipolysis in adipocytes. In summary, our observations point toward a pivotal role of PEMT in maintaining a “stable” LD phenotype.
Reasoning that the absence of PEMT might cause a WAT phenotype, PEMT-KO mice and WT littermates were fed either normal chow or a high fat diet for 3 weeks. The results from this experiment demonstrated that PEMT-KO mice are resistant to high fat diet-induced fat cell hypertrophy. Their fat cell diameters remained significantly smaller than those of WT animals under identical high fat conditions, in line with a recent report (46).
Notably, a previous report demonstrated that CTα increased substantially in a late stage of 3T3-L1 adipogenesis (68). Moreover, a recent study (69) showed in Drosophila Schneider 2 (S2) cells that genes encoding enzymes of PL biosynthesis were regulators of LD size and abundance and suggested that LD morphology is affected by its monolayer PL composition.
To address this issue, we used adipose tissue samples from Pemt−/− animals to investigate regulatory effects of PL-synthesizing enzymes. In the virtual absence of WAT in these 5-day-old animals, experiments were performed with BAT. The most striking effect in liver and BAT of PEMT-KO animals was a profound down-regulation of PSS1, the first enzyme of the PS/PE loop, and of PSS2 in BAT. Therefore, it is tempting to speculate that both PE-producing and -consuming processes during early LD formation are interconnected.
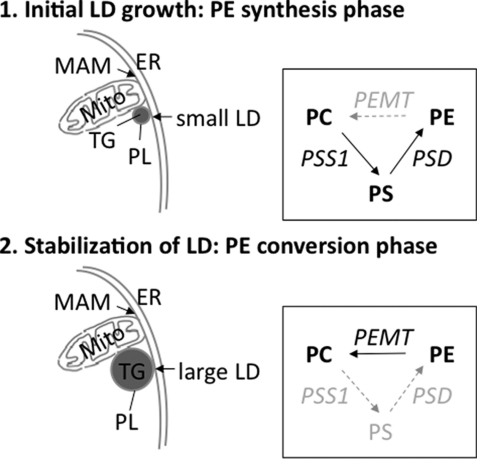
The key findings of the present study are as follows. (i) The increased need for PE at onset of fat cell differentiation is not met via the CDP-ethanolamine pathway that produces PE at ER membranes (first step) (Fig. 8). (ii) Rather, pre-existing ER-residing PC is used as a substrate to be converted to PS, shifted to mitochondria, and decarboxylated to PE (PE synthesis phase). (iii) Only this mitochondria/MAM-localized PE pool is accessible and available for incorporation into the LD monolayer. (iv) Our data further imply that MAM/LD contact sites (43) play a central role during early events of LD formation in that the enzyme PEMT, a MAM marker protein, converts PE to PC again in a second step (PE conversion phase), which in combination with the first phase constitutes a “PL interconversion triad.”
FIGURE 8.
Proposed interconnection between PE synthesis and conversion (PL interconversion triad) during LD biosynthesis. PE represent conically shaped PL molecules and are required for initiation of membrane curvatures in LD sprouts from the outer ER monolayer during early adipogenesis. Compartmentalization of PE produced via the CDP-ethanolamine pathway (i.e. ER resident) results in unavailability of PE for further conversion by the enzyme PEMT during LD biosynthesis. Rather, PE is preferentially produced via PS at onset of differentiation (1, PE synthesis phase; upper panel). This PSS1-mediated reaction requires serine and ER resident PC as substrates. PS is then translocated to mitochondria, where it is decarboxylated by PSD, resulting in PE. This concerted action of PSS1/PSD shifts PE into the MAM/LD compartment, which allows subsequent methylation by PEMT (2, PE conversion phase; lower panel). The resulting serine-derived PC is finally a component of the LD monolayer. In the absence of PEMT from mouse adipose tissue, the PSS1 pathways is attenuated. CT, CTP:phosphocholine cytidylyltransferase; DAG, diacylglycerol; EA, ethanolamine; ET, CTP:phosphoethanolamine cytidylyltransferase; SS1, phosphatidylserine synthase-1.
This leads to our concept that PEMT has a functional role for the decay of local LD-PE abundance at the onset of differentiation and is actively involved in maintaining a stable LD phenotype in adipocytes.
Acknowledgments
We appreciate the technical assistance of Margarete Schadler (Department of Molecular Biology and Biochemistry, University of Graz, Graz, Austria), Silvia Schauer, and Palaniappam Tamilarasan Kuppusamy (Department of Pathology, University of Graz). We thank Andrea Groselj-Strele (ZMF Biostatistics, Medical University of Graz) for help with statistical analyses, Dr. Jean E. Vance (University of Alberta, Edmonton, Alberta, Canada) for fruitful discussions and editorial work, and Drs. Robert Zimmermann and Rudolf Zechner (Institute of Molecular Biosciences, University of Graz) for their helpful advice with lipolysis assays. We also thank Dr. Dagmar Kratky (Department of Molecular Biology and Biochemistry, University of Graz) for providing adipose tissue from Perilipin−/− mice.
This work was supported by Austrian Science Funds P-15.358 (to E. S.), F3007 SFB LIPOTOX (to W. S.), F3002-B05 SFB LIPTOX (to G. H.), and P-19473 (to S. F.), Austrian Heart Foundation Project 01/08 (to E. S.), Franz-Lanyar Foundation (Graz, Austria) Grants P-308 (to G. Hörl) and P-323 (to E. S.), Austrian Ministry for Science and Research GEN-AU Project GOLDIII (to J. G. B.-S.), and the Canadian Institutes of Health Research (to D. E. V.).
- LD
- lipid droplet
- ADA
- adenosine deaminase
- CT
- CTP:phosphocholine cytidylyltransferase
- EGFP
- enhanced GFP
- ER
- endoplasmic reticulum
- MAM
- mitochondria-associated membrane
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PEMT
- phosphatidylethanolamine-N-methyltransferase
- PIA
- ((R)-N6-[l-2-phenylisopropyl]adenosine)
- PL
- phospholipid
- PS
- phosphatidylserine
- TG
- triacylglycerol
- WAT
- white adipose tissue
- BAT
- brown adipose tissue
- fw
- forward
- rev
- reverse
- qPCR
- quantitative PCR
- PSD
- phosphatidylserine decarboxylase.
REFERENCES
- 1. Badman M. K., Flier J. S. (2007) Gastroenterology 132, 2103–2115 [DOI] [PubMed] [Google Scholar]
- 2. Zechner R., Strauss J. G., Haemmerle G., Lass A., Zimmermann R. (2005) Curr. Opin. Lipidol. 16, 333–340 [DOI] [PubMed] [Google Scholar]
- 3. Martin S., Parton R. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 4. Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. (2002) J. Biol. Chem. 277, 44507–44512 [DOI] [PubMed] [Google Scholar]
- 5. Murphy D. J., Vance J. (1999) Trends Biochem. Sci. 24, 109–115 [DOI] [PubMed] [Google Scholar]
- 6. Prattes S., Hörl G., Hammer A., Blaschitz A., Graier W. F., Sattler W., Zechner R., Steyrer E. (2000) J. Cell Sci. 113, 2977–2989 [DOI] [PubMed] [Google Scholar]
- 7. Brasaemle D. L. (2007) J. Lipid Res. 48, 2547–2559 [DOI] [PubMed] [Google Scholar]
- 8. Liu P., Bartz R., Zehmer J. K., Ying Y. S., Zhu M., Serrero G., Anderson R. G. (2007) Biochim. Biophys. Acta 1773, 784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boström P., Andersson L., Rutberg M., Perman J., Lidberg U., Johansson B. R., Fernandez-Rodriguez J., Ericson J., Nilsson T., Borén J., Olofsson S. O. (2007) Nat. Cell Biol. 9, 1286–1293 [DOI] [PubMed] [Google Scholar]
- 10. Kuerschner L., Moessinger C., Thiele C. (2008) Traffic 9, 338–352 [DOI] [PubMed] [Google Scholar]
- 11. Kasturi R., Wakil S. J. (1983) J. Biol. Chem. 258, 3559–3564 [PubMed] [Google Scholar]
- 12. Bartz R., Li W. H., Venables B., Zehmer J. K., Roth M. R., Welti R., Anderson R. G., Liu P., Chapman K. D. (2007) J. Lipid Res. 48, 837–847 [DOI] [PubMed] [Google Scholar]
- 13. Marchesan D., Rutberg M., Andersson L., Asp L., Larsson T., Borén J., Johansson B. R., Olofsson S. O. (2003) J. Biol. Chem. 278, 27293–27300 [DOI] [PubMed] [Google Scholar]
- 14. Andersson L., Boström P., Ericson J., Rutberg M., Magnusson B., Marchesan D., Ruiz M., Asp L., Huang P., Frohman M. A., Borén J., Olofsson S. O. (2006) J. Cell Sci. 119, 2246–2257 [DOI] [PubMed] [Google Scholar]
- 15. Nakamura N., Banno Y., Tamiya-Koizumi K. (2005) Biochem. Biophys. Res. Commun. 335, 117–123 [DOI] [PubMed] [Google Scholar]
- 16. Dowhan W., Bogdanov M., Mileykovskaya E. (2008) in Biochemistry of Lipids, Lipoproteins and Membranes (Vance D. E., Vance J. E. eds) 5th Ed., pp. 1–37, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 17. Sprong H., van der Sluijs P., van Meer G. (2001) Nat. Rev. Mol. Cell Biol. 2, 504–513 [DOI] [PubMed] [Google Scholar]
- 18. Vance D. E., Vance J. E. (2008) in Biochemistry of Lipids, Lipoproteins and Membranes (Vance D. E., Vance J. E. eds) 5th Ed., pp. 213–244, Elsevier Science Publishers B.V., Amsterdam [Google Scholar]
- 19. Vance J. E., Vance D. E. (1986) J. Biol. Chem. 261, 4486–4491 [PubMed] [Google Scholar]
- 20. Fast D. G., Vance D. E. (1995) Biochim. Biophys. Acta 1258, 159–168 [DOI] [PubMed] [Google Scholar]
- 21. Verkade H. J., Fast D. G., Rusiñol A. E., Scraba D. G., Vance D. E. (1993) J. Biol. Chem. 268, 24990–24996 [PubMed] [Google Scholar]
- 22. Nishimaki-Mogami T., Yao Z., Fujimori K. (2002) J. Lipid Res. 43, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 23. Noga A. A., Zhao Y., Vance D. E. (2002) J. Biol. Chem. 277, 42358–42365 [DOI] [PubMed] [Google Scholar]
- 24. Zhu X., Song J., Mar M. H., Edwards L. J., Zeisel S. H. (2003) Biochem. J. 370, 987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Walkey C. J., Donohue L. R., Bronson R., Agellon L. B., Vance D. E. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12880–12885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schrader M. (2001) J. Histochem. Cytochem. 49, 1421–1429 [DOI] [PubMed] [Google Scholar]
- 27. Wolins N. E., Quaynor B. K., Skinner J. R., Tzekov A., Park C., Choi K., Bickel P. E. (2006) J. Lipid Res. 47, 450–460 [DOI] [PubMed] [Google Scholar]
- 28. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 29. Sattler W., Reicher H., Ramos P., Panzenboeck U., Hayn M., Esterbauer H., Malle E., Kostner G. M. (1996) Lipids 31, 1302–1310 [DOI] [PubMed] [Google Scholar]
- 30. Karim M., Jackson P., Jackowski S. (2003) Biochim. Biophys. Acta 1633, 1–12 [DOI] [PubMed] [Google Scholar]
- 31. Brügger B., Erben G., Sandhoff R., Wieland F. T., Lehmann W. D. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cui Z., Vance J. E., Chen M. H., Voelker D. R., Vance D. E. (1993) J. Biol. Chem. 268, 16655–16663 [PubMed] [Google Scholar]
- 33. Ridgway N. D., Vance D. E. (1992) Methods Enzymol. 209, 366–374 [DOI] [PubMed] [Google Scholar]
- 34. Trenker M., Malli R., Fertschai I., Levak-Frank S., Graier W. F. (2007) Nat. Cell Biol. 9, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., Zechner R. (2004) Science 306, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 36. Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 37. Rasband W. S. (1997–2011) ImageJ, Ver. 1.44, National Institutes of Health, Bethesda, MD [Google Scholar]
- 38. Voelker D. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 9921–9925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Voelker D. R. (1989) J. Biol. Chem. 264, 8019–8025 [PubMed] [Google Scholar]
- 40. Cole L. K., Vance D. E. (2010) J. Biol. Chem. 285, 11880–11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vance J. E. (1990) J. Biol. Chem. 265, 7248–7256 [PubMed] [Google Scholar]
- 42. Scow R. O., Blanchette-Mackie E. J. (1991) Brain Res. Bull. 27, 487–491 [DOI] [PubMed] [Google Scholar]
- 43. Kornmann B., Walter P. (2010) J. Cell Sci. 123, 1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Viswanadha S., Londos C. (2006) J. Lipid Res. 47, 1859–1864 [DOI] [PubMed] [Google Scholar]
- 45. Tansey J. T., Huml A. M., Vogt R., Davis K. E., Jones J. M., Fraser K. A., Brasaemle D. L., Kimmel A. R., Londos C. (2003) J. Biol. Chem. 278, 8401–8406 [DOI] [PubMed] [Google Scholar]
- 46. Jacobs R. L., Zhao Y., Koonen D. P., Sletten T., Su B., Lingrell S., Cao G., Peake D. A., Kuo M. S., Proctor S. D., Kennedy B. P., Dyck J. R., Vance D. E. (2010) J. Biol. Chem. 285, 22403–22413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arikketh D., Nelson R., Vance J. E. (2008) J. Biol. Chem. 283, 12888–12897 [DOI] [PubMed] [Google Scholar]
- 48. Steenbergen R., Nanowski T. S., Nelson R., Young S. G., Vance J. E. (2006) Biochim. Biophys. Acta 1761, 313–323 [DOI] [PubMed] [Google Scholar]
- 49. Tansey J. T., Sztalryd C., Hlavin E. M., Kimmel A. R., Londos C. (2004) IUBMB Life 56, 379–385 [DOI] [PubMed] [Google Scholar]
- 50. Wolins N. E., Brasaemle D. L., Bickel P. E. (2006) FEBS Lett. 580, 5484–5491 [DOI] [PubMed] [Google Scholar]
- 51. Chang B. H., Chan L. (2007) Am. J. Physiol. Gastrointest. Liver Physiol. 292, G1465–G1468 [DOI] [PubMed] [Google Scholar]
- 52. DeLong C. J., Shen Y. J., Thomas M. J., Cui Z. (1999) J. Biol. Chem. 274, 29683–29688 [DOI] [PubMed] [Google Scholar]
- 53. Vance J. E. (2008) J. Lipid Res. 49, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 54. Shields D. J., Altarejos J. Y., Wang X., Agellon L. B., Vance D. E. (2003) J. Biol. Chem. 278, 35826–35836 [DOI] [PubMed] [Google Scholar]
- 55. Shields D. J., Lehner R., Agellon L. B., Vance D. E. (2003) J. Biol. Chem. 278, 2956–2962 [DOI] [PubMed] [Google Scholar]
- 56. Fujimoto T., Ohsaki Y., Cheng J., Suzuki M., Shinohara Y. (2008) Histochem. Cell Biol. 130, 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robenek H., Hofnagel O., Buers I., Robenek M. J., Troyer D., Severs N. J. (2006) J. Cell Sci. 119, 4215–4224 [DOI] [PubMed] [Google Scholar]
- 58. Lehner R., Cui Z., Vance D. E. (1999) Biochem. J. 338, 761–768 [PMC free article] [PubMed] [Google Scholar]
- 59. Ost A., Ortegren U., Gustavsson J., Nystrom F. H., Strålfors P. (2005) J. Biol. Chem. 280, 5–8 [DOI] [PubMed] [Google Scholar]
- 60. Blouin C. M., Le Lay S., Eberl A., Köfeler H. C., Guerrera I. C., Klein C., Le Liepvre X., Lasnier F., Bourron O., Gautier J. F., Ferré P., Hajduch E., Dugail I. (2010) J. Lipid Res. 51, 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Levine T. (2004) Trends Cell Biol. 14, 483–490 [DOI] [PubMed] [Google Scholar]
- 62. Børglum J. D., Vassaux G., Richelsen B., Gaillard D., Darimont C., Ailhaud G., Négrel R. (1996) Mol. Cell. Endocrinol. 117, 17–25 [DOI] [PubMed] [Google Scholar]
- 63. Hoffman B. B., Chang H., Dall'Aglio E., Reaven G. M. (1986) J. Clin. Invest. 78, 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ciruela F., Albergaria C., Soriano A., Cuffí L., Carbonell L., Sánchez S., Gandía J., Fernández-Dueñas V. (2010) Biochim. Biophys. Acta 1798, 9–20 [DOI] [PubMed] [Google Scholar]
- 65. Brasaemle D. L., Rubin B., Harten I. A., Gruia-Gray J., Kimmel A. R., Londos C. (2000) J. Biol. Chem. 275, 38486–38493 [DOI] [PubMed] [Google Scholar]
- 66. Martinez-Botas J., Anderson J. B., Tessier D., Lapillonne A., Chang B. H., Quast M. J., Gorenstein D., Chen K. H., Chan L. (2000) Nat. Genet. 26, 474–479 [DOI] [PubMed] [Google Scholar]
- 67. Tansey J. T., Sztalryd C., Gruia-Gray J., Roush D. L., Zee J. V., Gavrilova O., Reitman M. L., Deng C. X., Li C., Kimmel A. R., Londos C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kast H. R., Nguyen C. M., Anisfeld A. M., Ericsson J., Edwards P. A. (2001) J. Lipid Res. 42, 1266–1272 [PubMed] [Google Scholar]
- 69. Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. (2008) Nature 453, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]